Abstract
Squamous cell carcinomas of head and neck (HNSCC) are associated with immune, inflammatory and angiogenic responses involving interleukin-6 (IL-6). This paper reports an ultrasensitive electrochemical immunosensor for human IL-6 and proof-of-concept studies of IL-6 detection in HNSCC cells. Single wall carbon nanotube (SWNT) forests with attached capture antibodies (Ab1) for IL-6 were used in an electrochemical sandwich immunoassay protocol using enzyme label horseradish peroxidase (HRP) to measure very low (≤30 pg mL−1) and elevated levels of IL-6. Two levels of multi-enzyme labeling were used to measure a broad concentration range of IL-6 in a representative panel of HNSCC cells. Secondary antibodies (Ab2) attached to carboxylated multiwall carbon nanotubes with 106 HRP labels per 100 nm gave the highest sensitivity of 19.3 nA-mL (pg IL-6)−1 cm−2 and the best detection limit (DL) of 0.5 pg mL−1 (25 fM) for IL-6 in 10 µL calf serum. For more concentrated samples, biotinylated Ab2 bound to streptavidin-HRP to provide 14–16 labels per antigen was used. These immunosensors accurately measured secreted IL-6 in a wide range of HNSCC cells demonstrated by excellent correlations with standard enzyme-linked immunosorbent assays (ELISA), suggesting that SWNT immunosensors combined with multilabel detection have excellent promise for detecting IL-6 in research and clinical applications.
INTRODUCTION
Development of devices for sensitive and reliable point-of-care measurement of biomarker proteins for early cancer detection and treatment monitoring is a significant challenge. However, the potential payoff is large since point-of-care analyses would reduce costs, minimize sample decomposition, facilitate on-the-spot diagnosis, and alleviate patient stress. Ideally, these measurements should be done cheaply, at high accuracy and sensitivity, and require minimal technical expertise and system maintenance.
Interleukin-6 (IL-6), a multifunctional cytokine characterized as a regulator of immune and inflammatory responses,1 is a suitable biomarker overexpressed by several types of cancer, including head and neck squamous cell carcinoma (HNSCC). HNSCC affects nearly 44,000 patients and results in ~11,000 deaths per year in the U.S.2 Despite general advances in cancer treatment, outcome remains poor for HNSCC patients primarily due to lack of measurable biomarkers for early detection, and patients are often diagnosed at advanced stages.3 HNSCC is associated with high IL-6 levels.4,5 Mean serum IL-6 in patients with HNSCC is ≥20 pg mL−1 compared to ≤6 pg mL−1 in healthy individuals. Compared to other secreted cancer biomarkers such as prostate specific antigen (PSA) with normal patient serum levels in the ng mL−1 range,6 normal IL-6 levels are nearly 1000-fold lower, presenting a significant analytical challenge. Both normal and elevated levels of IL-6 need to be measured accurately for reliable early detection and monitoring of HNSCC.
Another complication is that single biomarkers often have inadequate predictive value, e.g. ~75% for PSA.6 Predictive success approaching 100% can be achieved by measuring 5 to 10 biomarkers for a given cancer.7–11 Thus, low-cost, accurate, multiprotein arrays for serum analysis will be required for point-of-care cancer detection. Sensor development for IL-6 is addressed in the present study to achieve the necessary ultrahigh sensitivity along the way to development of electrochemical immunosensor arrays for simultaneous measurement of many biomarkers.
Alternative methods for detection of protein biomarkers have yet to meet all requirements for point-of-care use. Enzyme-linked immunosorbent assay (ELISA) is an important commercial method with detection limits (DL)12–14 approaching 1 pg mL−1, but is difficult to adapt to multiplexing and point-of-care. Bead-based immunoassays using electrochemiluminescence, chemiluminescence or fluorescence provide DL approaching several pg mL−1 but require costly, high maintenance instruments for automated analyses.15–17 Modern LC-MS proteomics can achieve multiple biomarker measurements approaching the necessary sensitivity and DL,18,19 but current technology is too expensive, labor intensive, and complex for routine diagnostics. Emerging methods for sensitive protein measurements,16 including arrays based on optical,20 electrochemical21,22 and nanotransistor23 detection, have been reported, but most are in developmental stages and have yet to address IL-6 in real samples.
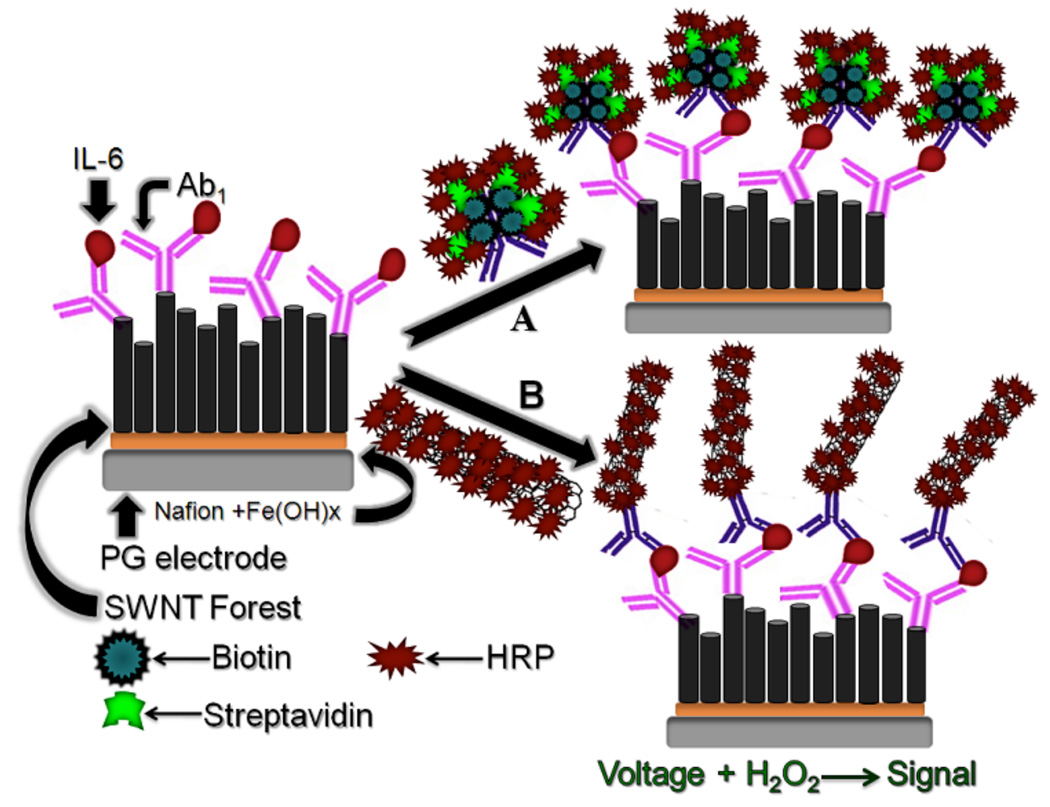
The present work utilizes electrochemical immunosensor protocols for detecting very low and elevated cancer-related levels of IL-6 in experimental HNSCC cells. High sensitivity is achieved by coupling multilabel amplification with nanostructured single wall nanotube (SWNT) forest platforms. We reported on these strategies as previously developed for PSA,24 and also employed for 4-protein arrays.25 The immunosensors are constructed on an electrically conductive, high surface area, conductive platform featuring densely-packed, upright SWNT forests with capture antibodies (Ab1) attached to their ends. Antigen from the sample binds to these Ab1 molecules, and then an enzyme-labeled secondary antibody (Ab2) bioconjugate is added to bind to the antigen. Electrochemical detection of the label gives signals proportional to the amount of antigen.
We combined several multilabel strategies to achieve moderate and ultrahigh sensitivity as necessary for IL-6. The first approach utilizes bioconjugates denoted Ab2-biotin-streptavidin-HRP. Here, after binding to IL-6 on the sensor surface, biotinylated Ab2 specifically binds streptavidin-HRP to provide 14–16 labels per antigen. We previously reported a detection limit (DL) for this method of 30 pg mL−1 (1.5 fmol mL−1) for human IL-6 in calf serum.26 For ultrahigh sensitivity detection, we employed a multilabeled HRP-multiwall carbon nanotube (MWNT)-HPR-Ab2 bioconjugate to obtain a 60-fold lower DL for IL-6. We show below that these two approaches accurately measure IL-6 secreted from a panel of HNSCC cells over a broad concentration range, and gave excellent correlations with standard ELISA assays. Most importantly, our work demonstrates strategies to detect a biomarker both well above and well below normal serum levels in the range ≤6 pg mL−1, suggesting the feasibility for incorporating nanotube-based immunosensors into future multiprotein immunoarrays.
EXPERIMENTAL SECTION
Chemicals and Materials
Monoclonal anti-human Interleukin-6 (IL-6) antibody (clone no. 6708), biotinylated anti-human IL-6 antibody, recombinant human IL-6 (carrier-free) in calf serum and streptavidin-horseradish peroxidase (HRP) were from R&D systems, Inc. (Minneapolis, MN, USA). HRP (MW 44000 Da), lyophilized 99% bovine serum albumin (BSA) and Tween-20 were from Sigma Aldrich. Single-walled carbon nanotubes (HiPco) were from Carbon Nanotechnologies, Inc. Carboxylated multiwalled carbon nanotubes were obtained from Nanocs, Inc. 2,2’-Azino-Bis(3-ethylbenzthiazoline-6-sulfonic acid) was from Sigma. Immunoreagents were dissolved in pH 7.2 phosphate saline (PBS) buffer (0.01 M phosphate, 0.14 M NaCl, 2.7 mM KCl). 1-(3-(dimethylamino)-propyl)-3-ethylcarbodiimidehydrochloride (EDC) and N-hydroxysulfosuccinimide (NHSS) were dissolved in water immediately before use.
Cell Lines and Conditions
The representative HNSCC cell lines (Cal27, HEp2, HN4, HN12, HN13, HN30) and immortalized human skin keratinocyte, HaCaT cells27 are described elsewhere.28 OSCC3 and UMSCC17B cells were kindly provided by Drs. N. J. D'Silva and T. E. Carey, respectively, University of Michigan. Normal oral keratinocytes (NOKsi) established from resected oral gingival tissue and maintained in supplemented defined K-SFM media (Invitrogen) were used as the controls. All cell lines were cultured as previously described in DMEM supplemented with 10% fetal bovine serum (FBS), at 37°C in 95% air/5% CO2. For the collection of conditioned media, cells were initially grown to 60–70% confluence, followed by serum starvation (12–16 h) and then maintained in media without serum or supplements for 48 h before collection, after which a brief centrifugation was used to remove cellular debris and the samples was stored at −70°C. Similarly, conditioned media from serum starved HaCaT cells with and without stimulation with TNF-alpha (20 ng/mL; Cell Sciences, MA, USA) were also collected.
Transfections and Real-time Polymerase Chain Reaction Analysis
Pre-designed siRNA oligonucleotides were utilized to target human (Hs) IL-6 for knock-down experiments. Two IL-6 sequences (si-IL-6-1: 5'-CTCAAATAAATGGCTAACTTA-3'; si-IL-6-2: 5'- CAGAACTTATGTTGTTCTCTA-3') together with AllStars Control siRNA (all from Qiagen, Valencia, CA) were used to transfect (30 nM) into exponentially growing HN13 cells using HiPerFect transfection reagent and following the protocol recommended by the manufactures (Qiagen). Transfected cells were incubated for 24 h followed by change of media to serum free and left for an additional 48 h, when the conditioned media was collected and processed as previously and, the attached cells were immediately lysed with Trizol (Invitrogen, Carlsbad, CA) and processed for RNA extraction following the provided protocol. Next, 300 ng of total RNA was used as input for generating first-strand cDNA using the Superscript III first-Strand Synthesis SuperMix kit and following the provided protocol (Invitrogen). The resulting DNA product was used as template (50 ng) for quantitative reverse transcription real-time polymerase chain reaction (RT-PCR) of IL-6 using iQ SYBER Green Supermix, specific primers and the iCycler apparatus (Bio-Rad, Hercules, CA). Primers were designed using Primer-BLAST (NCBI) and included Hs. IL-6 (Forward: 5'-AGTTCCTGCAGAAAAAGGCA-3'; Reverse: 5'-AAAGCTGCGCAGAATGAGAT-3') and Hs. GAPDH (Forward: 5'-GAGTCAACGGATTTGGTCGT-3'; Reverse: 5'-TTGATTTTGGAGGGATCTCG-3').
Fabrication of Immunosensors
SWNT forests were assembled on abraded, ordinary basal plane pyrolytic graphite disks (A= 0.14 cm2) from shortened SWNT dispersions in DMF on a thin iron oxide-Nafion layer as reported previously,24,29 except that the iron oxide underlayer was deposited from pH 1.8, 31 mM FeCl3 solution. Quality of SWNT forests was confirmed by atomic force microscopy and Raman spectroscopy as described previously.29 Capture antibody was attached by spotting 30 µL of freshly prepared 400 mM EDC and 100 mM NHSS in water onto the SWNT forest sensors, washing after 10 min, then incubating 3 h at room temperature with 20 uL 0.67 nmol L−1 (100 µg mL−1) primary anti-IL-6 antibody in pH 7.2 PBS buffer. The sensor was washed with 0.05 % Tween-20 in PBS, and then PBS for 3 min each. The immunosensor was then incubated for 1 h with 20 µL 1% BSA in PBS, followed by washing with 0.05 % Tween-20 in PBS and PBS for 3 min each. All washing steps were optimized to minimize non specific binding (NSB) to achieve the necessary sensitivity.
For standardization, the immunosensor prepared as above was incubated for 1 h with 10 µL calf serum containing human IL-6, followed by washing with 0.05 % Tween-20 in PBS and PBS for 3 min each. Then, the sensor was incubated with 10 µL of 1.1 pmol L−1 biotinylated secondary antibody (Ab2) in 0.1% BSA in pH 7.2 PBS buffer for 1 h, followed by washing with 0.05 % Tween-20 in PBS and PBS for 3 min each. For moderate sensitivity, the sensor was incubated with 10 µL streptavidin-HRP for 30 min, followed by washing with 0.05 % Tween-20 in PBS and PBS for 3 min each. For high sensitivity, incubation with multilabel Ab2-MWNT-HRP (described below) was substituted. The immunosensor was then placed in an electrochemical cell containing 10 mL pH 7.2 PBS with 1 mM hydroquinone as mediator. Rotating disk amperometry at 3000 rpm at −0.3 V vs SCE was done, and 0.4 mM H2O2 was injected to develop the signal.
The same approach was used to analyze 10 µL samples of the conditioned media from the cell cultures described above. These samples were also analyzed by a standard human IL-6 duo set ELISA assay (DL 8 pg mL−1).30
Synthesis of Ab2-MWNT-HRP Bioconjugates
Multiple HRPs and Ab2 were attached to carboxylated MWNTs using an EDC/NHSS amidization protocol with a reaction mixture 400:1 HRP:Ab2 molar ratio as described previously.24 Briefly, 1.5 mg oxidized MWNTs in 2 mL pH 7.0 0.01 M 2-(N-morpholino) ethanesulfonic acid buffer (MES buffer) were sonicated 10 min to obtain a homogenous dispersion. This dispersion was mixed with 1 mL 400 mM EDC and 100 mM NHSS in pH 7.0 PBS and vortexed 5 min, then centrifuged at 15000 rpm for 5 min and the supernatant discarded. The buffer wash was repeated several times to remove excessive EDC and NHSS. 150 µL Ab2 at 2.5 µg mL−1 and 150 µL HRP at 1 mg mL−1 was added to the mixture and stirred for 6 h, then centrifuged at 12000 rpm at 4°C for 10 min and the supernatant discarded. Washing was crucial to remove free Ab2 and HRP, and was repeated 4 times. 1 mL of 0.05 % Tween-20 in pH 7.2 PBS buffer was added to the bioconjugate precipitate collected and vortexed to form a homogenous dispersion and stored in refrigerator at 4°C and diluted 10-fold with 0.1 % BSA in PBS immediately before use.
Average height of atomic force microscopy (AFM) images of oxidized MWNTs on mica before bioconjugation was 25±2 nm, and length was 700±50 nm (see Supporting Information, Figure 1). AFM size analysis showed an increase of ~14 nm in height for the fully bioconjugated Ab2-MWNT-HRP giving a diameter of 39±5 nm, consistent with the average thickness of a monolayer of the major coating component HRP (4.0 × 6.7 × 11.7 nm, Brookhaven Protein Database)31 on the 25 nm nanotubes.
Figure 1.
Two strategies for multilabel detection in amperometric immunosensor (A) immunosensor after treating with Ab2-biotin-streptavidin-HRP, providing 14 to 16 HRP’s on one Ab2; (B) immunosensor after treating with HRP-MWNT-Ab2 bioconjugate having 106 active HRPs (enzyme labels) per 100 nm of carboxylated carbon nanotubes. The final detection step involves immersing the immunosensor in an electrochemical cell containing PBS buffer and mediator, applying voltage and injecting a small amount of hydrogen peroxide.
Bioconjugate dispersions were stable for ~10 days at 4 °C as shown by reproducible signals for IL-6 standards in immunoassays. The amount of HRP per unit length MWNTs was determined by reacting the Ab2-MWNT-HRP dispersion with substrate 2,2’-azino-bis-(3-ethylbenz-thiazoline-6-sulfonic acid) (ABTS) and H2O2.32 The rate of formation of the soluble product with an absorbance at 405 nm was monitored. This provided a linear increase in absorbance of the product at 405nm. From these data, the concentration of active HRP in the stock biotin-Ab2-MWNT-HRP dispersions was determined as 5.48 µg mL−1. Additionally, the amount of active HRP was found to be 137 pmol mL−1 of stock dispersion. Considering a MWNT with an average diameter of 25 nm and an average length of 700 nm (from AFM) with the enzyme activity data,33 106 active HRP labels per 100 nm length of the CNTs were estimated.
RESULTS
Immunosensor Calibration
Our strategy (Figure 1) employed the moderate sensitivity Ab2-biotin-streptavidin-HRP label system for most of the immunoassays. We previously presented calibration results for SWNT immunosensors using this 14–16 label system,26 reporting a detection limit (DL) of 30 pg mL−1 and sensitivity 3.6 nA-mL cm−2 (pg IL-6)−1. However, if levels of IL-6 in samples fell near or below the detection limit of this assay, we reanalyzed using an ultrahigh sensitivity Ab2-MWNT-HRP label system. Standardization of the assays employed pure IL-6 dissolved in calf serum, which gives a good approximation to human serum in electrochemical immunoarrays.24
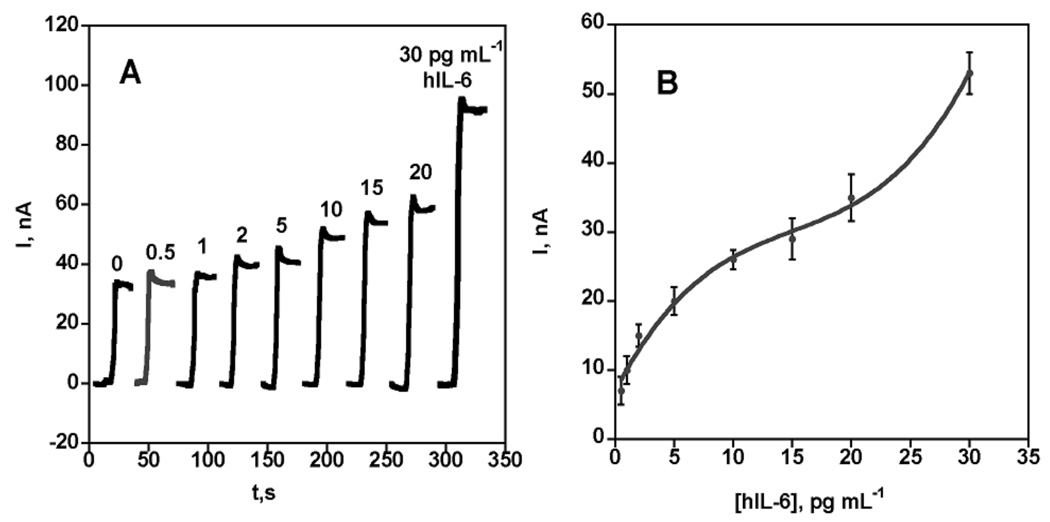
Calibration data for SWNT immunosensors featuring Ab2-MWNT-HRP bioconjugates with 106 HRPs per 100 nm of CNTs is shown in Figure 2. As in most immunoassays, a key to achieving high sensitivity was minimizing non-specific binding (NSB), in this case utilizing competitive binding of bovine serum albumin (BSA) and detergent along with an optimized concentration of Ab2-MWNT-HRP (see experimental section). Figure 2A shows that the steady state sensor current increased with IL-6 concentration in serum over a range of 0.5–30 pg mL−1. Although this calibration is non-linear, it remains appropriate for samples up to and slightly above 30 pg mL−1. Sensitivity as the slope of the calibration plot in the nearly linear 0.5–5 pg mL−1 region, (Figure 2B) was 19.3 nA-mL cm−2 (pg IL-6)−1, an increase of ~5 fold compared with the Ab2-biotin-streptavidin-HRP system. Good device-to-device reproducibility is illustrated by the small error bars. This approach provided a detection limit of 0.5 pg mL−1 (25 fM) as 3 times average noise plus the zero IL-6 control.
Figure 2.
Amperometric response for SWNT immunosensors incubated with human IL-6 in 10 µL calf serum, then biotin-Ab2-MWNT-HRP (A) current at −0.3 V and rotating the electrode at 3000 rpm in PBS buffer containing 1 mM hydroquinone mediator and then injecting H2O2 to 0.04 mM to develop the signal; (B) influence of IL-6 concentration on steady-state current (corrected for background) for immunosensor using the bioconjugate. Error bars represent device-to-device standard deviation (n = 3).
IL-6 Secreted by Human Squamous cells
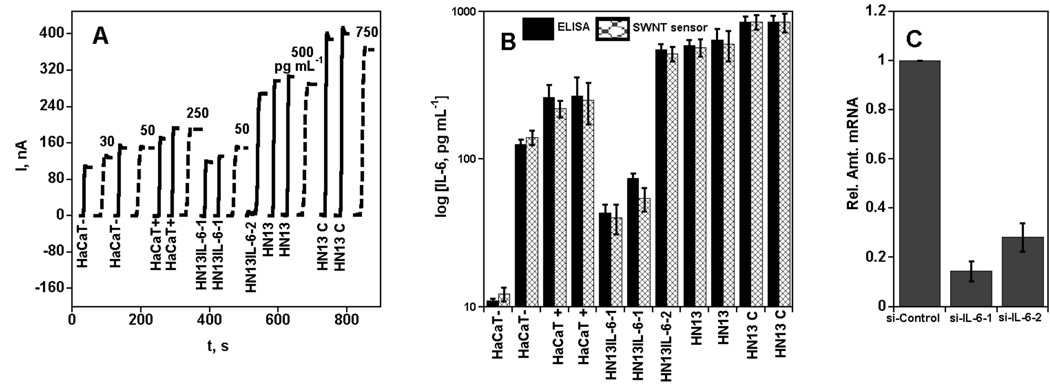
We next used the immunosenors to determine secreted levels of IL-6 from in vitro cell preparations. For the first study, HaCaT cells were stimulated with TNF-alpha, a cytokine known to stimulate IL-6 secretion.34 As controls, we employed HN13 cells with high endogenous levels of IL-6.27 that were subsequently transfected with predesigned siRNAs to knock down human IL-6.27 Specifically, we used siRNA oligonucleotides designated si-IL-6-1 and si-IL-6-2 to knock down IL-6 in HN13 as well as HN13 treated with a control siRNA that does not affect IL-6. HN13 cell lines knocked down by si-IL-6-1 are designated as HN13IL-6-1 and those knocked down by si-IL-6-2 as HN13IL-6-2. SWNT immunosensors were used to detect IL-6 in the resulting conditioned media as shown in Figure 3(A). HaCaT treated with TNF-alpha (HaCaT+) showed 15-fold increased IL-6 levels when compared to control cells (HaCaT−) that gave minimal levels. The siRNA IL-6 knock down in HN13 cells resulted in 18-fold decreased IL-6 levels for HN13 treated with si-IL-6-1 (HN13IL-6-1) and 1.5-fold decreased levels for HN13 treated with si-IL-6-2 (HN13IL-6-2), compared to control cells without IL-6 knockdown, consisting of HN13 cells alone, and HN13 cells treated with siRNA (HN13 C). Conditioned media were also analyzed by ELISA for IL-6 and levels were comparable to those determined by the immunosensors (Figure 3B). Results of a correlation plot of the SWNT sensor data vs. ELISA (Supporting Information, Figure S2A) was analyzed by linear regression and gave a slope of 0.99 ± 0.02 and an intercept of −10.6 ± 9.2, demonstrating very good correlation.
Figure 3.
Amperometric response for SWNT immunosensors incubated with IL-6 in 10 µL calf serum (pg mL−1 labeled on curves, dashed lines) or conditioned media from cells treated with TNF-alpha (HaCaT −/+) and those transfected with indicated siRNA oligonucleotides (HN13IL-6-1, HN13IL-6-2, HN13 [non transfected] and HN13 C [control siRNA]; solid lines) for 1 h. Conditioned media samples (HaCaT−, HaCaT+, HN13IL-6-1, HN13IL-6-2, HN13, HN13 C) were analyzed using 10 µL 1.1 pmol L−1 biotinylated secondary antibody (Ab2) in 0.1 % BSA in pH 7.2 PBS buffer and 10 µL streptavidin-HRP except the one below 30 pg mL−1 (HaCaT−), which was analyzed using 10 µL Ab2-MWNT-HRP bioconjugates (A) current at −0.3 V and 3000 rpm using 1 mM hydroquinone as mediator in PBS buffer, then injecting H2O2 to 0.4 mM; (B) SWNT sensor results for conditioned media shown with results from ELISA for the same samples; (C) qRT-PCR analysis of RNA extracted from control and IL-6 siRNA transfected cells and expression levels indicated (fold) are after normalization to levels of GAPDH.
As a further measure of effectiveness of IL-6 knock down, total RNA extracted from HN13 cells treated with control siRNA and IL-6 siRNAs was subjected to reverse transcription real-time quantitative polymerase chain reaction (RT-qPCR) analysis. As shown in Figure 3C, mRNA levels decreased in those cells that were treated with siRNA to IL-6 in comparison to control cells. Levels were normalized to mRNA expression levels of the house keeping gene GAPDH, to reflect the amount of protein secreted into the conditioned media. These results are consistent with the lower levels of IL-6 measured by the immunosensors in HN13 cells subjected to IL-6 knock down.
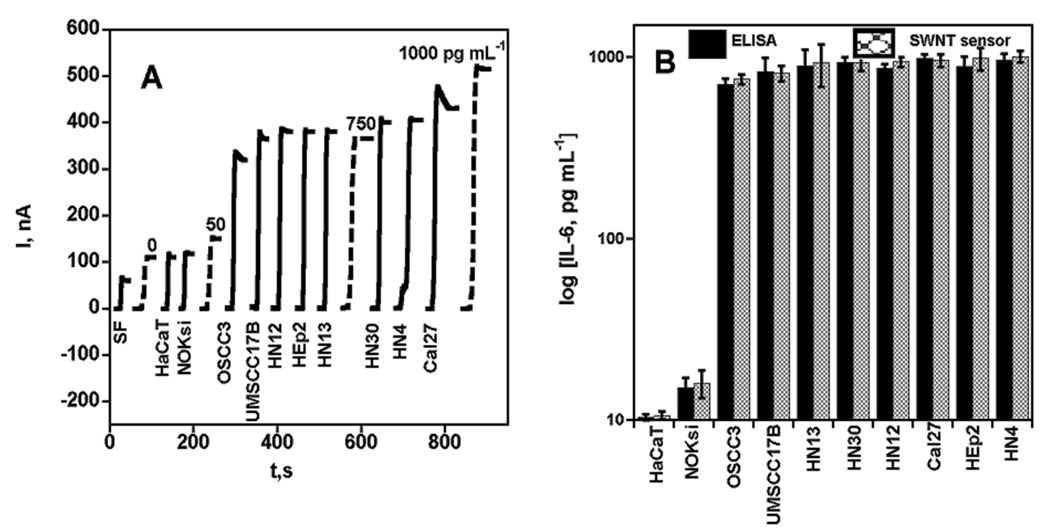
Conditioned media from heterogeneous populations of ten different cell lines were then analyzed to explore the generality of the sensing approach towards IL-6 detection in oral carcinomas. Figure 4A illustrates the results along human IL-6 standards in serum at comparable levels. Most of the cell lines (Cal27, HEp2, HN4, HN12, HN13, HN30) express high levels of IL-6, ranging between 750–1005 pg mL−1, but HaCaT and NOKsi demonstrated low levels of the cytokine, ranging between 10–16 pg mL−1. Samples were also assayed by ELISA and excellent correlation was found (Figure 4B). A linear correlation plot (SI file, Figure S2B) for SWNT immunosensors vs. ELISA gave slope 1.03 ± 0.03 and intercept of 4.8 ± 27.4, indicating excellent correlation. Previous studies on these cell lines showed similar expression levels of IL-6 in a subset of these cells.27 These results suggest the validity of using the immunosensors for determination of IL-6 levels in samples for a range cancer cell types.
Figure 4.
Amperometric response for SWNT immunosensors incubated with IL-6 in 10 µL calf serum (pg mL−1 labeled on curves, dashed lines) and conditioned media (HaCaT, Cal27, HEp2, HN4, HN12, HN13, HN30, NOKsi, OSCC3, UMSCC17B) and serum free media, SF (solid lines) for 1 h. Conditioned media samples (Cal27, HEp2, HN4, HN12, HN13, HN30, OSCC3, UMSCC17B) were analyzed using 10 µL 1.1 pmol L−1 biotinylated secondary antibody (Ab2) in 0.1 % BSA in pH 7.2 PBS buffer and 10 µL streptavidin-HRP except those below 30 pg mL−1 (HaCaT, NOKsi), which were analyzed using 10 µL Ab2-MWNT-HRP bioconjugates (A) current at −0.3 V and 3000 rpm using 1 mM hydroquinone as mediator in PBS buffer, then injecting H2O2 to 0.4 mM; (B) SWNT sensor results for conditioned media shown with results from ELISA (relative standard deviation [ RSD] ± 10%) for the same samples.
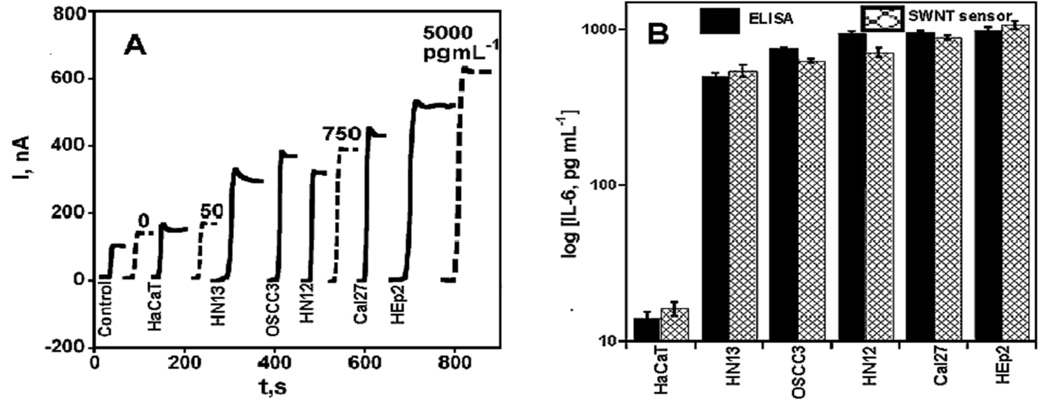
To confirm the IL-6 expression levels of the cells and the repeatability of the SWNT sensor, we then independently grew a sub-set of the 10 cell lines, and analyzed the new conditioned media. The results are shown along with human IL-6 standards (Figure 5A). Results for these cells followed the same trends as in the previous analyses. Here, HaCaT cells were found to express low levels of IL-6, ranging between 17.8 – 20.0 pg mL−1. Very high levels of IL-6 (>1000 pg mL−1) were expressed by HEp2, in accordance with ELISA results. Also, the expression of IL-6 in other cell lines (Cal27, HN12, HN13, OSCC3) measured by SWNT sensors were in excellent agreement with ELISA (Figure 5B). The slope of the correlation graph (SI file, Figure S2C) of the SWNT sensor results vs. ELISA was 0.89 ± 0.14, with an intercept of 18.8 ± 111.9, again showing good correlation. T-tests indicated no significant differences between the concentrations of all conditioned media samples measured by SWNT sensors and ELISA (p> 0.05).
Figure 5.
Amperometric response for SWNT immunosensors incubated with IL-6 in 10 µL calf serum (pg mL−1 labeled on curves, dashed lines) and a second set of conditioned media (Cal27, HaCaT, HEp2, HN12, HN13, OSCC3) and control (solid lines) for 1 h. Conditioned media samples (Cal27, HEp2, HN12, HN13, OSCC3) were analyzed using 10 µL 1.1 pmol L−1 biotinylated secondary antibody (Ab2) in 0.1 % BSA in pH 7.2 PBS buffer and 10 µL streptavidin-HRP except the one below 30 pg mL−1 (HaCaT), which was analyzed using 10 µL Ab2-MWNT-HRP bioconjugates (A) current at −0.3 V and 3000 rpm using 1 mM hydroquinone as mediator in PBS buffer, then injecting H2O2 to 0.4 mM; (B) SWNT sensor results for conditioned media shown with results from ELISA (RSD ± 10%) for the same samples.
DISCUSSION
Results above demonstrate that nanostructured SWNT forest immunosensors combined with multilabel detection strategies can accurately and reproducibly detect cancer biomarkers at levels between 1 to thousands of pg mL−1 in complex biological samples (Figure 3 to Figure 5). This range is also representative of IL-6 in the serum of disease-free and cancer patients.4 Two labeling levels were used to accommodate this wide range of concentrations. Using Ab2-MWNT-HRP with 106 labels per 100 nm of CNT gave extremely high sensitivity of 19.3 nA-mL cm−2 (pg IL-6)−1 and an ultralow detection limit of 0.5 pg mL−1 (Figure 2). This immunosensor protocol is ~5-fold more sensitive compared to the alternative Ab2-biotin-streptavidin-HRP14–16 labeling system, and the detection limit was 60-fold better. The detection limit using Ab2-MWNT-HRP is also 16-fold better than that of the ELISA method at 8 pg mL−1.30
SWNT immunosensors showed very good reproducibility demonstrated by small device to device standard deviations (Figure 2 to Figure 5) with both Ab2-biotin-streptavidin-HRP14–16 and biotin-Ab2-MWNT-HRP detection systems. In addition, good accuracy for IL-6 was demonstrated by good correlation of SWNT immunosensor results for IL-6 with ELISA assays for a broad range of conditioned media samples. Selectivity was confirmed by accurate detection of IL-6 in these media that also contain hundreds of other proteins. We previously showed that SWNT sensor arrays similar to the one described here, but with the low sensitivity protocol only, gave accurate determinations of IL-6 and 3 other protein biomarkers in the serum of a limited set of prostate cancer patients.25
The present approach for IL-6 improved the sensitivity and DL of the method, and should be readily adaptable to multiprotein arrays. Ideally, such an array could be integrated into low cost microfluidic systems for automated point-of-care assays. Such devices are currently being designed in our laboratories, and have the potential to increase predictive power by simultaneously detecting panels of biomarkers for a given cancer.
In summary, we have demonstrated ultrasensitive, selective and accurate electrochemical detection of IL-6 representative of normal patient to high cancer patient levels from a variety of head and neck cancer cells. The detection limit of 0.5 pg mL−1 for IL-6 is 16-fold lower than that of conventional ELISA and 60-fold lower than for our previously reported IL-6 immunosensors.25,26 SWNT immunosensor protocols described are suitable for measuring other biomarkers, and could be easily adapted to array formats25 for the detection of multiple protein biomarkers.
Supplementary Material
ACKNOWLEDGEMENT
This research was supported by US PHS grant ES013557 from National Institutes of Environmental Health Sciences of the National Institutes of Health and by intramural programs of the National Institute of Dental and Craniofacial Research, NIH. JR thanks Science Foundation Ireland for a Walton Research Fellowship in 2009.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Two additional figures illustrating phase contrast AFM images of carboxylated MWNTs before and after bioconjugation, linear correlation plots and a statistical table correlating SWNT sensor results with ELISA are provided.
REFERENCES
- 1.Kishimoto T. Annu. Rev. Immunol. 2005;23:1–21. doi: 10.1146/annurev.immunol.23.021704.115806. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Murray T, Ward E. CA Cancer J. Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 3.Thomas GR, Nadiminti H, Regalado J. Int. J. Exp. Pathol. 2005;86:347–363. doi: 10.1111/j.0959-9673.2005.00447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riedel F, Zaiss I, Herzog D, Götte K, Naim R, Hörmann K. Anticancer Res. 2005;25:2761–2765. [PubMed] [Google Scholar]
- 5.Hong DS, Angelo LS, Kurzrock R. Cancer. 2007;110:1911–1928. doi: 10.1002/cncr.22999. [DOI] [PubMed] [Google Scholar]
- 6.Lilja H, Ulmert D, Vickers AJ. Nat. Rev. Cancer. 2008;8:268–278. doi: 10.1038/nrc2351. [DOI] [PubMed] [Google Scholar]
- 7.Xiao Z, Prieto D, Conrads TP, Veenstra TD, Issaq HJ. Mol. Cell Endocrinol. 2005;230:95–106. doi: 10.1016/j.mce.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 8.Weston AD, Hood L. J. Proteome Res. 2004;3:179–196. doi: 10.1021/pr0499693. [DOI] [PubMed] [Google Scholar]
- 9.Wagner PD, Verma M, Srivastava S. Ann. NY Acad. Sci. 2004;1022:9–16. doi: 10.1196/annals.1318.003. [DOI] [PubMed] [Google Scholar]
- 10.Hanash SM, Pitteri SJ, Faca VM. Nature. 2008;452:571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 11.Stevens EV, Liotta LA, Kohn EC. Int. J. Gynecol. Cancer. 2003;13:133–139. doi: 10.1111/j.1525-1438.2003.13358.x. [DOI] [PubMed] [Google Scholar]
- 12.Williams TI, Toups KL, Saggese DA, Kalli KR, Cliby WA, Muddiman DC. J. Proteome Res. 2007;6:2936–2962. doi: 10.1021/pr070041v. [DOI] [PubMed] [Google Scholar]
- 13.Kingsmore SF. Nat. Rev. Drug Discov. 2006;5:310–320. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ward MA, Catto JWF, Hamdy FC. Ann. Clin. Biochem. 2001;38:633–651. doi: 10.1258/0004563011901055. [DOI] [PubMed] [Google Scholar]
- 15.Hendricks HA, Kortlandt W, Verweij WM. Clin. Chem. 2000;46:105–111. [PubMed] [Google Scholar]
- 16.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 17.Yurkovetsky ZR, Kirkwood JM, Edington HD, Marrangoni AM, Velikokhatnaya L, Winans MT, Gorelik E, Lokshin AE. Clin. Cancer Res. 2007;13:2422–2428. doi: 10.1158/1078-0432.CCR-06-1805. [DOI] [PubMed] [Google Scholar]
- 18.Ferrari M. Nat. Rev. Cancer. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 19.Debad JB, Glenzer EN, Wholstadter J, Sigal GB, Leland JK. In: “Electrogenerated Chemiluminescence”, Chapter 8. Bard AJ, editor. NY: Marcel Dekker; 2004. [Google Scholar]
- 20.Lee HJ, Wark A, Corn RM. Analyst. 2008;133:975–983. doi: 10.1039/b717527b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J. Electroanal. 2007;19:769–776. [Google Scholar]
- 22.Kim SN, Rusling JF, Papadimitrakopoulos F. Adv. Mater. 2007;19:3214–3228. doi: 10.1002/adma.200700665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.(a) Patolsky F, Zheng G, Lieber CM. Anal. Chem. 2006;78:4260–4269. doi: 10.1021/ac069419j. [DOI] [PubMed] [Google Scholar]; (b) Zhang H, Zhao Q, Li XF, Le XC. Analyst. 2007;132:724–737. doi: 10.1039/b704256f. [DOI] [PubMed] [Google Scholar]
- 24.Yu X, Munge B, Patel V, Jensen G, Bhirde A, Gong JD, Kim SN, Gillespie J, Gutkind JS, Papadimitrakopoulos F, Rusling JF. J. Am. Chem. Soc. 2006;128:11199–11205. doi: 10.1021/ja062117e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chikkaveeraiah BV, Bhirde A, Malhotra R, Patel V, Gutkind JS, Rusling JF. Anal. Chem. 2009;81:9129–9134. doi: 10.1021/ac9018022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munge BS, Krause CE, Malhotra R, Patel V, Gutkind JS, Rusling JF. Electrochem. Comm. 2009;11:1009–1012. doi: 10.1016/j.elecom.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sriuranpong V, Park JI, Amornphimoltham P, Patel V, Nelkin BD, Gutkind JS. Cancer Res. 2003;63:2948–2956. [PubMed] [Google Scholar]
- 28.Jeon GA, Lee JS, Patel V, Gutkind JS, Thorgeirsson SS, Kim EC, Chu IS, Amornphimoltham P, Park MH. Int. J. Cancer. 2004;112:249–258. doi: 10.1002/ijc.20399. [DOI] [PubMed] [Google Scholar]
- 29.Yu X, Kim SN, Papadimitrakopoulos F, Rusling JF. Mol. BioSyst. 2005;1:70–78. doi: 10.1039/b502124c. [DOI] [PubMed] [Google Scholar]
- 30. http://www.rndsystems.com/pdf/dy206.pdf.
- 31. http://www.rcsb.org/pdb/explore/explore.do?structureId=1ATJ.
- 32.Gallati H. J. Clin. Chem. Clin. Biochem. 1979;17:1–7. [PubMed] [Google Scholar]
- 33.Jensen GC, Yu X, Gong JD, Munge B, Bhirde A, Kim SN, Papadimitrakopoulos F, Rusling JF. J. Nanosci. Nanotechnol. 2009;9:249–255. doi: 10.1166/jnn.2008.J016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu B, Elias PM, Feingold KR. “Skin Barrier”, Chapter 19. Taylor and Francis Group; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.