1. Abstract
Animal models have enriched understanding of the physiological basis of metabolic disorders and advanced identification of genetic risk factors underlying the metabolic syndrome (MetS). Murine models are especially appropriate for this type of research, and are an excellent resource not only for identifying candidate genomic regions, but also for illuminating the possible molecular mechanisms or pathways affected in individual components of MetS. In this review, we briefly discuss findings from mouse models of metabolic disorders, particularly in light of issues raised by the recent flood of human genome-wide association studies (GWAS) results. We describe how mouse models are revealing that genotype interacts with environment in important ways, indicating that the underlying genetics of MetS is highly context dependant. Further we show that epistasis, imprinting and maternal effects each contribute to the genetic architecture underlying variation in metabolic traits, and mouse models provide an opportunity to dissect these aspects of the genetic architecture that are difficult if not impossible to ascertain in humans. Finally we discuss how knowledge gained from mouse models can be used in conjunction with comparative genomic methods and bioinformatic resources to inform human MetS research.
Keywords: Metabolic Syndrome, hypertension, obesity, type-2 diabetes, cardiovascular disease, murine models, bioinformatics, comparative genomics, genome-wide association studies
2. Overview
The metabolic syndrome (MetS) is a suite of conditions, including impaired glucose tolerance, dyslipidemia, high blood pressure and central obesity that tend to occur in conjunction. This suite of conditions puts an individual at risk for developing type-2 diabetes (T2D) and cardiovascular disease (CVD) [1]. As such, the MetS classification is of great epidemiological interest: T2D has increased in prevalence and currently afflicts ~7% of the US adult population [2]. Approximately 90% of individuals with T2D are considered overweight, and ~ 70% will develop cardiovascular disease [2–4]. In the US alone, the prevalence of MetS exceeds 20%, and it is becoming more and more common in developing countries [5, 6]. Many hypotheses have been proposed to explain this epidemic in terms of environmental factors, for example the thrifty gene and the sedentary lifestyle hypotheses, which posit that over-consumption of high caloric foods and inactive lifestyles are major causes of the metabolic disorders that comprise MetS [7]. Indeed, dietary and lifestyle modification have proven therapeutic [8]. However, it is becoming increasingly evident that some populations are more prone to metabolic disorders than others and, in general, that women are less prone than men of the same body mass [1, 9]. Clearly there is a genetic component to MetS. Yet therapeutic dietary and lifestyle guidelines generally do not take into account underlying genetic variation, which can effect an individual’s response to treatment [10]. Understanding the pathogenesis of the metabolic disorders comprising MetS is an enormous biomedical challenge because phenotypic variation is caused by complex interactions of many genes of small effects, by environmental factors, and by the interplay between the two. Thus animal models are particularly important because both genetic and environmental influences can be controlled for and monitored in a population of known genetic structure.
2.1 Murine models for human disease
Animal models have enriched understanding of the physiological basis of complex disease and advanced identification of genetic risk factors. In using animal models, a researcher is able to plan crosses between animal strains with measurable phenotypic differences and to generate large numbers of offspring from a single set of founders of known genomic background, overcoming the confounding factor of genetic heterogeneity present in human studies. In genetic mapping studies, this increases the power to detect quantitative trait loci (QTL) having small effects, those that are most likely to underlie complex disease and that are difficult to identify in human studies. Additionally, because phenotypes are ascertained in a controlled environment, animal models allow for detailed analysis of the architecture of gene-by-environment interactions, which for both practical and ethical reasons is not possible in human studies. Once a QTL has been identified in an animal model, fine-scale mapping of the genomic region can break a locus down into the quantitative trait genes (QTG) or the quantitative trait nucleotides (QTN) responsible for the QTL effect [11]. When an animal’s DNA sequence has been identified, the human homolog can be isolated by DNA hybridization or by homology alignment to the human genome, and research can be conducted to determine if variation in the candidate sequence is associated with variation in the same phenotype in humans. Finally, returning to the animal model to obtain experimental proof through expression or intervention analysis can determine if and how the variants identified contribute to the disease.
Mouse models are especially appropriate for this type of research, and mouse studies have made major contributions to our knowledge of complex disease etiology [12]. This is, in part, because mice have relatively short gestation times and are relatively cheaper to breed and maintain than other mammalian models. In addition, mouse development and physiology is very well characterized and dense panels of polymorphic genetic markers are readily available. The initial sequence of the mouse genome was released in 2002 and, as of July 2007, is considered “essentially complete” with NCBI build 37 [13]. The only other species’ genome that is considered “complete” is human [14]. Mouse–human chromosomal homology is well characterized and syntenic maps are available allowing for easy translation of linkage maps between the species. The Jackson Laboratories house the world’s largest repository of inbred laboratory mouse strains and genetically engineered mouse stocks, including a large subset used in research of MetS components: T2D, obesity, hypertension, and cardiovascular disease [15]. Online resources are freely available for mouse queries, ranging from biology to genomic sequence to expression, providing valuable information for study design and potential generation of new mouse models that recapitulate the pathology of human diseases (see Table 1). In many instances mouse models represent monogenic changes with major effects for a disease, despite the fact that a common disease is generally the result of contributions from multiple genes. To circumvent this problem, different teams have worked with mice to combine recombinant congenic strains with multiple contributing genes for a disease. Consequently, existing mouse models have evolved to better approximate human disease models, and have become ideal tools for the genetic dissection of complex traits.
Table 1.
Useful online resources for mouse model study design.
| Resource | Information |
|---|---|
| The Jackson Labs (http://www.jax.org/) |
Repository of inbred laboratory mouse strains and genetically engineered mouse stocks |
| UCSC Genome Browser (http://genome.ucsc.edu/) |
Sequence information, multi-species whole-genome alignments, tools for visualizing genome-wide data sets |
| Ensemble (http://www.ensembl.org/index.html) |
Sequence information, data mining and exporting via BioMart |
| National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) |
Gateway to NCBI mouse genome resources including dbSNP, Clone Finder, Full-Length cDNAs, e-PCR and more |
| Mouse Genotype Informatics (http://www.informatics.jax.org/) |
Integrated resource for molecular, genomic and biological data and their applications to human disease research |
| Mouse Phenome Database (http://phenome.jax.org/pub-cgi/phenome/mpdcgi) |
Phenotype data organized by subject, by mouse strain and by project with the aim of enabling investigators to find research-appropriate mouse strains |
| Gene Paint (http://www.genepaint.org/) |
Atlas of gene expression patterns in the mouse embryo |
| Galaxy (http://main.g2.bx.psu.edu/) |
Online tool for retrieving sequences and alignments and performing simple statistics and evolutionary analyses |
2.2 The search for a murine model of metabolic syndrome
The physiological features of MetS components vary across species and MetS is not well defined in mouse models. For example, absolute criteria for defining extreme blood serum levels in mice do not exist, and mice transport most of their cholesterol in high density lipoprotein cholesterol (HDLC) rather than in low-density lipoprotein cholesterol (LDLC) as humans do [1]. Mouse models for diabetes are generally resistant to many of the complications common to humans, e.g. diabetic neuropathy, nephropathy and retinopathy. When such complications do occur in mice, they do not reflect all the characteristics seen in human pathology [16–18]. Further, mice do not express certain genes affecting development of diabetic complications in humans, such as cholesterol ester transport protein [15]. Moreover, despite humans and mice sharing most of the same transcription factors, there are significant differences in the targets with which they bind [19]. These dissimilarities reflect genus-specific metabolic differences resulting from 65–85 million years of divergent evolution to fill different ecological niches [13, 20]. Nevertheless, mouse models have expanded our understanding of MetS pathophysiology and have highlighted genomic regions that are fruitful candidates for further study.
Most candidate genomic regions are identified in mice by crossing two phenotypically distinct inbred strains, within each of which animals are genetically identical, and then intercrossing the resulting F1 hybrid offspring to create an F2 generation where all possible combinations of genotypes (homozygotes for each parental allele and the heterozygote) have the potential to be represented at each locus. This method facilitates QTL mapping and when matings are carried out to the F10 generation or beyond to create an advanced intercross line (AIL), resolution improves due to the accumulation of recombination over generations (e.g. an F10 generation has approximately 5 times the recombination of an F2 generation). For example, Ehrich et al. (2005) identified several QTL affecting serum glucose and insulin levels in an F16 AIL of LG/J x SM/J (Wustl:LG,SM-G16) [21], and recently Fawcett et al. (2009) fine-mapped QTL contributing to obesity and organ weights using a combined F9/F10 population (Wustl:LG,SM-G19 and Wustl:LG,SM-G10) [22]. A key advantage of using inbred crosses and AILs is that the architecture of genetic by environment interactions can be fully explored, and this is discussed further in Section 4.
Another common method for identifying candidate genomic regions is the use of recombinant inbred lines (RIL), which are produced by brother-sister mating F2 intercross offspring for at least 20 generations. At this point all individuals are genetically identical and homozygous across the genome with a unique mosaic of fixed random alleles from the original parental genomes. Cheverud et al. (2004) demonstrated genetic independence of some diabetes and obesity related traits in a study of genetic correlations among LGXSM RILs [23]. Recently, Koutnikova et al. (2009) identified a QTL for hypertension in a BXD RILs, and subsequent association studies confirmed the human syntenic region’s involvement in both systolic and diastolic blood pressure [24]. An advantage of using RILs is that results are replicable because the panel needs only to be genotyped once, yet the strains can be phenotyped across time by a diverse group of researchers.
The Jackson Laboratories provide catalogs of commonly used laboratory mouse models for T2D, obesity and cardiovascular research. As of this writing, 53 strains are used frequently to model T2D and obesity, and 48 to model diabetes without obesity. Approximately 250 mouse strains are used in CVD research, including 27 to model hypertension, 57 to model hypercholesterolemia, and 17 to model hypertriglyceremia (www.jaxmice.org/research/index/html). C57BL/6J features prominently in each of these research areas due to its physiological sensitivity to experimental diets, and much recent research has utilized this strain to explore MetS components [25–28]. Additionally, there are 10 strains that display a suite of characteristics resembling MetS. For example, studies indicate that low-density lipoprotein receptor-deficient mice, B6.129S7-Ldlrtm1Her/J, may be a good model of dietary induced MetS [5, 29–33]. Leiter (2009) discusses how one could go about selecting a mouse model when designing a T2D and/or MetS study [15].
3. Murine models of metabolic disorders
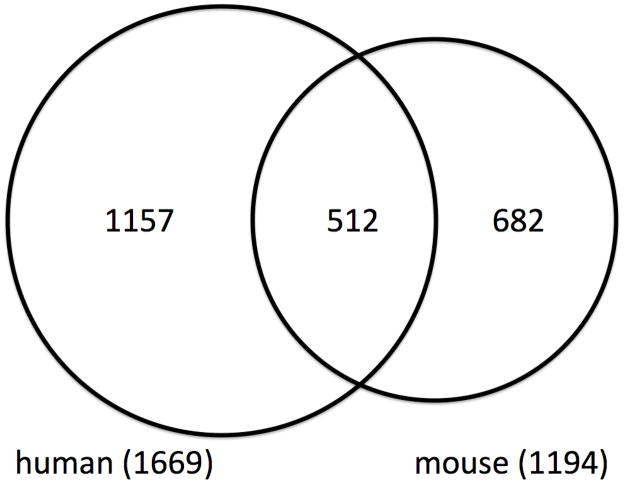
Figure 1 shows the number of unique genes found associated with one or more components of MetS for humans, for mice, and for their intersection. These numbers were ascertained for mouse by utilizing the “Phenotype/Human Disease” section of the advanced search for “Genes/Markers” and conditioning on “Gene” through the Mouse Genome Informatics database. For human, these numbers were obtained through the “Entrez Gene” database at the National Center for Biotechnology Information website. Both databases were queried on October 10, 2009 with keywords: “obesity”, “type-2 diabetes”, “insulin resistance”, “impaired glucose tolerance”, “hypertension”, “dislipidemia”, “cholesterol”, “triglycerides”, and “free-fatty acids”. Supplemental Table 1 provides the gene names for the three full sets of these genes. Table 2 presents a list of the 28 genes out of the 512 that intersect in which mutations in both human and mouse orthologs are associated with phenotypes diagnostic of the risk factors of MetS in humans.
Figure 1.
Venn diagram illustrating the number of unique genes associated with a MetS component for humans, for mice, and their intersection
Table 2.
Genes where mutations in both human and mouse orthologs are associated with phenotypesdiagnostic of the human metabolic disorder.
| Metabolic Disorder (Human Disease Query) |
OMIM ID | Human Gene RefSeq accession |
Human Genome Coordinates (hg19; build 37.1) |
Mouse Genome Coordinates (mm9; build 37) |
Mouse Genetic Background (Strain) |
Reference (First author) |
|---|---|---|---|---|---|---|
| Obesity | 601665 | MC4R NM_005912 |
chr18: 58038564-58040001 | chr18: 67017362-67020126 | 129S4/SvJae*C57BL/6 | Huszar D. Cell.1997;88:131–141 |
| SIM1 NM_005068 |
chr6: 100836751-100911551 | chr10: 50615457-50708958 | 129/Sv*C57BL/6 | Goshu E. Mol Cell Biol. 2002;22:4147–4157 | ||
| 609734 | POMC NM_000939 |
chr2: 25383722-25391559 | chr12: 3954951-3960618 | 129S1/SvEv*129X1/SvJ | Yaswen L. Nat Med. 1999;5:1066–1077 | |
| Type-2 Diabetes/ | 125800 | AQP2 NM_000486 |
chr12: 50344524-50352662 | chr15: 99409487-99414976 | C57BL/6*Aqp2cph | McDill B. Proc Natl Acad Sci USA. 2006; 103:6952–6957 |
| Insulin Resistance/ | 125700 | AVP NM_000490 |
chr20: 3063203-3065370 | chr2: 130406413-130408277 | 129S1/Sv*129X1/SvJ | Russell TA. J Clin Invest. 2003;112:1697–1706 |
| Impaired Glucose | 222100 | HLA-DQB1 NM_002123 |
chr6: 32627657-32634466 | chr17: 34401704-34404980 | 129S2/SvPas*C57BL/6*CBA*SJL | Wen L. J Clin Invest. 2001;107:871–880 |
| Tolerance | 125853 | AKT2 NM_001626 |
chr19: 40736225-40791265 | chr7: 28390879-28424472 | 129P2/O1aHsd*C57BL/6 | Cho H. Science. 2001; 292:1728–1731 |
| GCK NM_000162 |
chr7: 44183870-44229022 | chr11: 5800825-5849602 | 129X1/SvJ*C57BL/6*CBA*ICR | Toye AA. Diabetes. 2004; 111:1147–1160 | ||
| HNF1A NM_000545 | chr12: 121416549-121440312 | chr5: 115398370-115421047 | 129X1/SvJ*C57BL/6 | Lee Y. Mol Cell Biol. 1998;18:3059–3068 | ||
| IRS1 NM_005544 |
chr2: 227304277-227371750 | chr1: 82229680-82288014 | 129X1/SvJ*C57BL/6*CBA*ICR | Lin X. J Clin Invest. 2004; 114:908–916 | ||
| IRS2 NM_003749 |
chr13: 109204185-109236915 | chr8: 10986964-11008430 | 129S1/Sv*129X1/SvJ*C57BL/6 | Withers DJ. Nature. 1994; 372:182–186 | ||
| PDX1 NM_000209 |
chr13: 27392168-27398451 | chr5: 148081707-148086725 | 129S6/SvEvTac*C57BL/6*129T/Sv | Kim SK. Nat Genet; 2002; 30:430–435 | ||
| SLC2A4 NM_001042 |
chr17: 7185054-7191366 | chr11: 69755788-69761692 | 129/Sv*C57BL/6J*CD-1*SJL | Stenbit AE. Nat Med. 1997;3:1096–1101 | ||
| 304800 | AVPR2 NM_000054 |
chrX: 153170529-153172619 | chrX: 71137441-71139766 | 129S1/Sv*129X1/SvJ*CF-1 | Yun J. J Clin Invest. 2000;106:1361–1371 | |
| 610199 | GLIS3 NM_001042413 |
chr9: 3824128-4300035 | chr19: 28336938-28754567 | C57BL/6*CBA | Watanabe N. FEBS Lett. 2009; 583:2108–2113 | |
| 226980 | EIF2AK3 NM_004836 |
chr2: 88856261-88926994 | chr6: 70794521-70855234 | 129S6/SvEvTac*Swiss Webster | Harding HP. Mol Cell. 2001; 7:1153–1163 | |
| 304790 | FOXP3 NM_014009 |
chrX: 49106898-49121288 | chrX: 7156819-7172360 | 101/RI*C3Hf/RI*STOCK MR | Wildin RS. Nat Genet. 2001; 27:18–20 | |
| 125850 | HNF4A NM_178849 |
chr20: 43029924-43060029 | chr2: 163372924-163398643 | 129X1/SvJ*C57BL/6*DBA | Miura A. J Biol Chem. 2006;281:5246–5247 | |
| 249270 | SLC19A2 NM_006996 |
chr1: 169433151-169455208 | chr1: 166179186-166195499 | 129X1/SvJ | Oishi K. Hum Mol Genet. 2002;11:2951–2960 | |
| Hypertension | 145500 | NOS3 NM_000603 |
chr7: 150688144-150711686 | chr5: 23870637-23890292 | 129P2/o1aHsd*C57BL/6 | Duplain H. Circulation. 2001; 104:342–345 |
| 178600 | SMAD9 NM_001127217 |
chr13: 37422207-37494409 | chr13: 54559504-54605179 | 129S4/SvJaeSor | Huang Z. HumMol Genet. 2009; 18:2791–2801 | |
| Dyslipidemia/ | 136120 | LCAT NM_000229 |
chr16: 67973787-67978015 | chr8: 108463452-108467282 | 129X1/SvJ*C57BL/6J | Sakai N. J Biol Chem. 1997; 272:7506–7510 |
| 143890 | LDLR NM_000527 |
chr19: 11200057-11244503 | chr9: 21528038-21554362 | 129S7/SvEvBrd*C57BL/6 B6.Lg-LepobLdlrTm1Her | Hasty AH. J Biol Chem. 2001; 276:37402–37408 | |
| 603813 | LDLRAP1 NM_015627 |
chr1: 25870076-25895375 | chr4: 134301327-134323919 | 129S5/SvEvBrd*C57BL/6 | Harada-Shiba M. Circ Res. 2004; 95: 945–952 | |
| Cholesterol/ | 200100 | MTTP NM_000253 |
chr4: 100485240-100545153 | chr3: 137752819-137794341 | 129S1/Sv*129X1/SvJ*C57BL/6 | Chang BH. J Biol Chem. 1999; 274: 6051–6055 |
| Triglycerides/ | 201470 | ACADS NM_000017 |
chr12: 121163571-1221177810 | chr5: 115560308-115569322 | BALB/cByJ | Schiffer SP. Biochem Genet. 1989; 27:47–58 |
| Free-Fatty Acids | 301500 | GLA NM_0000169 |
chrX: 100652779-100663001 | chrX: 131122708-131135544 | 129S4/SvJae*C57BL/6 | Ohshima T. Proc Natl Acad Sci USA. 1997; 94: 2540–2544 |
| 144250 | LPL NM_000237 |
chr8: 19796582-19824769 | chr8: 71404454-71430831 | 129P2/O1aHsd | Xian X. Biochem Biophys Res Commun. 2009; 385:563–569 |
This illustrates that mouse models generally do not point to the same genes affecting the same phenotype in the same way in humans. Rather, their power lies in that they can aid in identification of genes acting in the same pathway and/or physiological system. As such, mouse models are an excellent resource not only for identifying candidate genomic regions, but also for illuminating the possible molecular mechanisms or pathways affected in individual components of metabolic disease. For example, the leptin pathway was first characterized in studies of the ob [34] and db [35] genes in mice. Subsequent studies in leptin (LEP)[36] and the leptin receptor (LEPR)[37] in humans revealed mutations in these genes underlie obesity [38]. Additionally, characterization of the melanocortin-AGRP pathway in studies of the mouse agouti gene[39] led to the discovery of mutations in MC4R, shown to similarly affect body fat content in mice and humans [40]. Recently, the T2D susceptibility gene sortilin-related domain containing receptor1 (SORCS1) was identified in humans based on candidate status from mouse studies [41], joining the ranks of calpain-10 (CAPN10) [42], peroxisome proliferator-activated receptor γ (PPARG), and transcription factor 7-like 2 (TCF7L2) [43] as genes that confer diabetes risk. Kraja et al. (2008) constructed gene networks for both humans and mouse models and identified 859 mouse genes with a corresponding human counterpart in one or more MetS components [44]. Further analysis of the interconnection of these genes in pathways, especially those affecting multiple disease components, could prove effective in identifying new genetic associations with MetS in humans.
In the following sections we briefly review QTL findings from mouse models of disease components of MetS, particularly in light of issues raised by the recent flood of human genome-wide association studies (GWAS) results. We also discuss how such mouse models have illuminated gene-by-environment interactions contributing to metabolic disease. Finally, we reflect on how these findings can be used in conjunction with genomic and bioinformatic resources to inform future study design.
3.1 Obesity
The overall prevalence of the MetS has increased in parallel with increases in obesity. The prevalence rate of obesity has increased steadily among US adults (>20 years of age) since 1960 from 13.3 to 32.1 percent [3]. Twin studies reveal estimates of broad sense heritability of BMI to be up to 70% in both children and adults, and admixture mapping shows that obesity is highly correlated with an individual’s relative percentages of ethnic ancestries [7, 45]. Mouse models have aided in identification of major variants in single genes underlying the obese phenotype in humans, for example proopiomelanocortin (POMC) [46], and the LEP [36], LEPR [37], and MC4R [47] genes discussed above. Genome-wide linkage analysis and positional cloning have identified several other genes contributing to obesity in human populations, for example CAPN10, also associated with T2D [42], and PPARG [43], ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1) [48], also associated with T2D, as well as solute carrier family 6 (SLC6A14) [49], and glutamate decarboxylase 2 (GAD2) [50].
GWAS have successfully identified novel loci associated with obesity, for example the fat mass and obesity associated gene (FTO) [51], catenin beta-like 1 (CTNNBL1) [52], and fibrillin 2 (FBN2) [53]. More recent GWAS have found novel chromosomal loci in addition to replicating previous obesity associations with genes such as FTO, MC4R [54] and brain-derived neurotrophic factor (BDNF) [55]. The office of population genomics at the National Human Genome Research Institute has made available a hand-curated catalog of published human genome-wide association studies (www.genome.gov/gwastudies) [56]. As of this writing, there are 19 unique GWAS reported loci associated with obesity.
While GWAS have successfully identified some obesogenic genes, these genes account for a very small percentage of the overall heritable variation of obesity in humans. For example FTO variants reported explain only ~1% of the heritability of BMI, despite having been identified as an obesogenic gene in multiple ethnic populations [51, 57], and despite obesity’s high heritability [45]. This is a common result in GWAS of complex traits and the paucity of results is due to a lack of power to detect genes of small effect. Indeed, the The Wellcome Trust Case Control Consortium recently demonstrated that enormous sample sizes are required to have adequate power to detect genomic regions having even large disease effects [58]. This is partly because GWAS were designed with the common disease–common variant hypothesis in mind [59–61]. While the method is successful at identifying associations between common phenotypes and common allelic variants in homogeneous populations, GWAS lack of power is reflective of humans’ heterogeneity in both genotypic and environmental influences. McCarthy et al. (2008) provide a good review of issues related to GWAS [62]. Although obesity is a common disease, the alleles underlying the phenotype are many and relatively rare, and most will not pass the stringent multiple testing criteria necessary to claim association, hence the “missing” heritability. The same holds true for other metabolic disorders: T2D, CVD and hypertension. Thus a candidate gene approach, where candidates are identified independently in mouse models, can be used to protect genomic regions from strict thresholds and increase the power of these studies.
There have been a great many studies of the genetics of obesity and body weight in mice, spanning over a century [63], and mice have proven to be excellent models for complementing our understanding of the biology of obesity in humans. Several papers provide good reviews of single mutant mouse models of obesity [64–66]. However, polygenic obesity is the most common pattern of inheritance, and the most relevant in terms of modeling a component of MetS. A total of 536 records are produced when one searches the term “obesity” in the “Mouse Phenotypes and Models of Human Disease” section of the “Genes and Markers” query form in the Mouse Genome Informatics Database (Accessed October 9, 2009) [67]. Of these records, 172 are for genes identified in studies of major mutants and transgenics, and 326 are for QTL that have been mapped in many different inbred strain crosses, each segregating for its own unique set of obesity QTL. Most of these QTL were identified in F2 populations and Brockmann and Bevova (2002) provide a good review [68] of the most common strains used in these crosses. Wuschke et al. (2007) report results of a meta-analysis of QTL associated with obesity in mice, wherein they compiled a list of candidate regions comprised of 162 non-redundant QTL for body weight and 117 QTL for adiposity (measured as fatpad weight) [69].
The largest number of obesity and body size QTL mapped in any single cross is in the LG/J x SM/J advanced intercross line [21–23, 70–76]. LG/J and SM/J differ genetically for a number of MetS related phenotypic traits, described in Ehrich et al. (2003), and genome-wide scans in the F2 generation identified eight QTL for adiposity (Adip1–Adip8)[71, 72]. Fine-mapping efforts, which combined the F2/F3 generations, confirmed seven of these eight original QTL (Adip1–Adip6 and Adip8), and found eleven more loci affecting adiposity (Adip9–Adip20) [70]. Further mapping of the F9/F10 generations of the LG/J x SM/J AIL identified 31 adiposity QTL, 13 of which replicated from previous generations [22]. Additional mapping efforts in the F16 LG/J x SM/J AIL are ongoing, and preliminary results not only replicate previously identified adiposity QTL, but also identify novel locations at high resolution because the genetic map is 8 times that of the original F2 map due to accumulated recombination (Cheverud et al. in preparation).
3.2 Type-2 diabetes
Like obesity, T2D is highly heritable, ranging from ~50–90% in twin studies, and disease incidence varies with an individuals’ percent ethnic ancestry [77, 78]. Linkage analysis and positional cloning have led to identification of monogenic forms of T2D, generally through studies of relevant phenotypes such as pancreatic β-cell function, for example the ATP-binding cassette (ABCC8) [79], insulin resistance, for example insulin-degrading enzyme (IDE) [80], and obesity, for example PPARG [81]. However, most cases of T2D do not exhibit Mendelian inheritance and GWAS attempt to identify common variants through hypothesis-free testing [82]. As of this writing, there are 30 unique GWAS reported loci associated with T2D [56].
A total of 212 QTL are associated with “type 2 diabetes” in the Mouse Genome Informatics Database (Accessed October 9, 2009) [67]. Of these, 56 QTL are also retrieved when “obesity” is queried, demonstrating the strong genetic association between these two components of MetS. Clee and Attie (2007) provide an excellent review of the genetic and physiological background of various mouse strains used to model T2D and its co-morbidities such as obesity [78].
While C57BL/6 is the most commonly used and, hence best characterized, mouse strain used in T2D research, results generated, whether “T2D-susceptible” or “T2D-resistant”, are contingent on the strain it is being compared with, because it shows intermediate glucose and insulin levels compared with other strains [15]. Mice do not get diabetes per se therefore it is important to understand both the physiology and the genetic background of the inbred strains when evaluating a mouse model of T2D. The LG/J and SM/J have been well characterized with respect to T2D related traits and compared with SM/J, LG/J animals have lower basal glucose levels and respond better to a glucose challenge [72]. Strains from a set of LGXSM RILs show variation in development of hyperglycemia and hyperinsulinemia [83]. Additionally, ninety-one QTL mapping to 39 unique genomic locations were identified for obesity, glucose response, and serum glucose and insulin levels in an LGXSM RIL [23]. Cheverud et al. (2004) found discordance among the loci affecting obesity, insulin and glucose, suggesting there are genotypes that are protective for T2D in the presence of obesity [84]. While obesity is a major risk factor for T2D, approximately 20% of individuals with T2D are not obese [82], and Cheverud et al.’s results suggest it is possible to dissect the relationship between these two components of MetS. Ongoing research using an LG/J x SM/J F16 AIL is mapping QTL for serum glucose and insulin levels as well as response to a glucose challenge. Preliminary results identify 70 QTL mapping to 64 unique genomic locations (Lawson et al., in preparation).
3.3 Cardiovascular disease
CVD has a multifactorial etiology involving metabolic, neuro-endocrine and genetic interactions [85]. It shares many common risk factors with obesity and T2D, including dyslipidemia and elevated blood pressure, and it is the leading cause of death in the United States [86]. Heritability estimates for risk factors of CVD are variable among studies across populations dependent on whether they are twin studies or family based studies. For example estimates range from 20–66% for diastolic blood pressure, from 8–72% for fasting total cholesterol, from 21–79% for fasting HDLC, from 31–68% for fasting LDLC, and from 19–72% for fasting triglycerides [87]. After the leptin and the leptin receptor pathways were characterized in mouse models, studies in human families have found >600 mutations in the LDLR gene [88], and mutations in genes in the LDL and LDLR pathways, such as apolipoprotein B (APOB) [89] and the ATP-binding cassette (ABCG5) [90] account for a large majority of monogenetic dyslipidemia [91]. However, as with obesity and T2D, single genes with large effects account for a small minority of cases of CVD, and most of the genes underlying CVD remain unknown. As of this writing, there are 114 unique GWAS reported loci associated with the CVD domains of total cholesterol, HDLC, LDLC, triglycerides, blood pressure and systolic blood pressure [56].
A total of 340 QTL are associated with “cholesterol”, “triglycerides” and “blood pressure” in the Mouse Genome Informatics Database (Accessed October 9, 2009) [67]. Of these, 99 are also associated with obesity, 34 with diabetes and 18 with all three components of MetS. Mouse models have significantly contributed to our knowledge of CVD and, despite being characteristically resistant to CVD per se, many genetically altered strains respond to high-cholesterol feeding with CVD pathologies [92]. The body of literature on this topic is rich: a search of PubMed for “mouse models” and “cardiovascular disease” limited to the last five years yields 4,672 results (www.ncbi.nlm.nih.gov/sites/entrez; Accessed October 17, 2009), entire books are dedicated to the role of the laboratory mouse in CVD research [93], and the Jackson Laboratories currently offer 250 mouse strains appropriate for CVD studies. Again the most commonly referenced strain is C57BL/6 [1, 27, 94], and the Mouse Phenome Database provides complete blood serum lipid profiles as well as atherosclerotic phenotypes characterized for this strain [95]. The LG/J and SM/J strains have also been well characterized with respect to CVD related traits. Compared with SM/J, LG/J animals have higher cholesterol and free fatty acid levels [72]. Ongoing research using an LG/J x SM/J F16 AIL is mapping QTL for variation in cholesterol, free fatty acid and triglyceride levels, and their variation in response to dietary fat. Preliminary results identify 25 trait-specific QTL mapping to 24 unique genomic locations (Lawson et al., in preparation).
3.4 Hypertension
Hypertension is the most common CVD risk factor, with an approximate 27% world-wide prevalence [96]. Estimates of heritable variation vary, as discussed above, and while some genes associated with hypertension have shown Mendelian patterns of inheritance, for example mineralocorticoid (NR3C2) [97] and PPARG (also associated with obesity and T2D)[98], relatively fewer causal genes than other risk factors have been identified. As of this writing, there are 23 unique GWAS reported loci associated with hypertension [56]. Cowly (2006) provides a good review of the genetic analysis of the etiology of hypertension [99].
Historically, the rat has been the preferred rodent model for the genetics of hypertension, however the mouse is beginning to take a prominent role with the development of equipment capable of measuring blood pressure in small animals [100]. Recently, a QTL associated with blood pressure was identified in a mouse BXD RIL panel, and this candidate genomic region was used to identify the gene ureidopropionase (UPB1). Subsequent analysis in humans revealed the syntenic region to be a determinant of both systolic and diastolic blood pressure [24]. A total of 33 QTL are associated with “hypertension” in the Mouse Genome Informatics Database (Accessed October 9, 2009) [67]. Of these, 9 are also associated with obesity, 3 with diabetes, and 7 with CVD. Three QTL, Bpq21–Bpq23, are associated with all four components of MetS. These three loci were identified in the males of an F2 generation NZO/HILtJ x C3H/HeJ intercross [101]. However, a previous analysis of the F2 progeny of this same intercross found no relation between blood pressure and other MetS components [102]. Supplementary Table 2 lists the mouse QTL identified for each of the metabolic disorders discussed above.
4. Metabolic syndrome components: gene-by-gene and gene-by-environment interactions
The genetic effects of most of these individual QTL discussed above are predominantly additive, where the combined phenotypic effects of two alleles at a locus is equal to the sum of their individual effects, and dominance deviations, where the heterozygote phenotype at a locus deviates from the midpoint of the two homozygotes, i.e. due to interaction between alleles at the same locus, also occur quite frequently. However, mouse-based QTL studies of MetS components are uncovering a more complex genetic architecture, wherein QTL effects are modified by diet and by sex [23, 103]. Further, other characteristics such as epistasis [22, 70], imprinting [104], and maternal effects [105], each of which are difficult to ascertain in human populations, are being illuminated through mouse models.
4.1 Gene-by-diet and gene-by-sex effects
Characterizing variation in response to diet is essential for testing hypotheses of the environmental contribution to metabolic disorders and to understanding the etiology of MetS. This is a challenging endeavor in studies of human populations because it is difficult to control and/or record diet over time. Some human population studies have successfully examined gene-by-environmental interactions [106–108], but typically, gene-by-environmental interactions are regarded as nuisance factors, despite the fact that they may underlie the increasing worldwide prevalence of MetS. Hence mouse models can be used to elucidate gene-by-environmental effects that can be further extrapolated to humans. Svenson et al. (2007) present results of a study assessing response to a high-fat diet in 43 inbred strains for 10 traits, including blood serum levels and body composition including weight and adiposity [109]. The Mouse Phenome Database [95] has compiled an inventory of high-fat diet intervention studies, including sex effects where applicable.
A number of QTL have been found in crosses between inbred strains fed a high-fat diet [110–115], some of which showed sex-specificity [68, 111, 112, 115]. While these studies are valuable in characterizing response to a high-fat environment, most do not examine this response relative to a low-fat diet. As such, the context dependence of genetic by environmental interactions is missed. Cheverud et al. have taken advantage of the genotypic and phenotypic differences between LG/J and SM/J to identify genetic variation in dietary response for multiple MetS component traits (obesity (adiposity), glucose tolerance, and serum insulin, glucose, cholesterol, free fatty acid, and triglyceride levels) both in the LGXSM RIL panel [84] and in the F16 LG/J x SM/J AIL, dividing litters into high- and low-fat diet treatments [21]. Of the 91 QTL mapped in the RIL, 34% were only observed in individuals fed a high-fat diet and, for 58% of the traits examined, having the LG/J allele led to higher values. Eight sex-specific QTL were found, and in general females showed a greater genetic response to high-fat diet for obesity related traits, whereas males showed a greater response in fasting glucose levels [23]. Ongoing QTL mapping efforts in the F16 LG/J x SM/J AIL for these same MetS component traits confirm that genotype interacts with environment in important ways, indicating that the underlying genetics of MetS is highly context dependant (Cheverud et al., in preparation; Lawson et al., in preparation).
4.2 Epistasis
Epistasis occurs when the interaction among the alleles at two or more loci cause a phenotypic effect. In humans, epistasis between genetic variants of PPARG2 and proprotein convertase subtilisin/kexin (PCSK1) effects individual susceptibility to insulin resistance [116]. Additionally, the contribution of the apolipoprotein epsilon locus (ApoE) to serum cholesterol levels is mediated by an individual’s genotype at the LDLR locus [117]. However, in general, studies of the contribution of epistasis to components of MetS in humans are under-represented in the literature. Mouse models of MetS components are revealing that epistatic interactions among QTL are commonplace, and that even QTL having small individual effects contribute considerably to epistatic interactions [101, 118–121]. Recently, Fawcett et al. (2009) found that epistasis contributed significantly to genetic variation in fatpad (8.1%) and body weight (12.3%) in an analysis of a combined F9/F10 LG/J x SM/J AIL [22]. Sometimes, an individual QTL will interact with many others. For example, Brockman et al. (2000)found a QTL, Lepq1, on chromosome 14, interacting with seven other QTL associated with adiposity in an F2 DU6i x DBA/2 intercross [118]. Cheverud et al. (2001) found that Adip8, on chromosome 18, interacted with all seven other adiposity QTL identified in a LG/J x SM/J intercross [71]. These results point to QTL that may contain major regulators of the genetic pathways underlying MetS.
4.3 Imprinting
Our knowledge of the influence of epigenetic factors, cell-specific heritable changes in gene expression occurring in the absence of DNA mutation, on metabolism is limited. However, the various disease components comprising MetS show non-Mendelian features, such as some discordance in twins, male and female differences in prevalence, and individual variation in both healthy and disease states, each of which are consistent with epigenetic mechanisms [122]. Genomic imprinting can be generally defined as the unequal expression of maternally and paternally derived copies of a gene, and it is becoming apparent that imprinting is an important aspect of the genetic architecture of many complex traits, including growth and metabolism [123]. More than 80 imprinted genes have been identified in humans and mice [124], and genome-wide bioinformatic analyses have trained algorithms with imprinting signatures, such as methylation and histone modification, to predict that several hundred genes are likely to be imprinted across the genome [125, 126]. QTL mapping can also identify imprinted loci [104, 127, 128], and loci with imprinting effects on obesity have been mapped in human populations [129]. Using an F16 population of LG/J x SM/J AIL fed both high- and low-fat diet treatments, we have mapped genome-wide imprinting values for various MetS component traits (adiposity, serum lipid levels of cholesterol, free fatty acids, and triglycerides, serum insulin and glucose, and glucose tolerance). The imprinting genotypic value is defined as half the difference between the reciprocal heterozygotes, LS and SL, where the first allele is derived from the father and the second from the mother (Cheverud et al., in preparation; Lawson et al., in preparation).
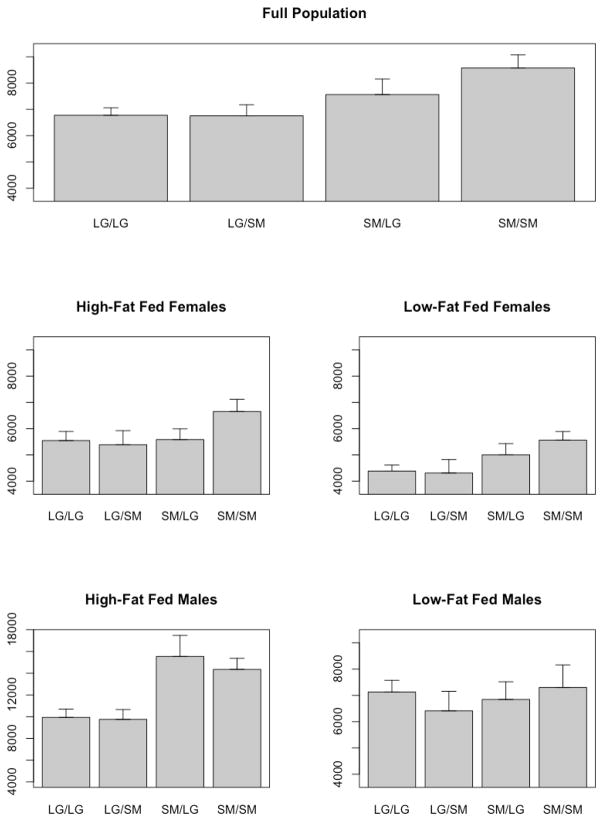
In this study, one thousand two animals were partitioned by sex and fed low- (247 males; 254 females) and high- (253 males; 248 females) fat diets. Animals were scored at 1,402 autosomal SNPs using the Illumina Golden Gate Assay. Details of the pedigree and of the phenotyping and genotyping process are described in Ehrich et al., (2005) [21]. Additive, dominance, and imprinting scores were estimated at each marker and additional markers were imputed at 1 cM intervals. Analyses were performed using the SAS PROC mixed model with additive, dominance, and imprinting genotypic scores, their interactions with sex, diet, and with sex by diet as the fixed effects and with family, family by sex, diet and by sex and diet interactions as the random effects. Figure 2 shows the genotypic means for a significant QTL found on chromosome 8 for glucose tolerance, measured as the area under the curve (AUC) calculated from results of an intra-peritoneal glucose tolerance test performed at 10 weeks of age. This QTL has significant additive effects in the full population, and significant additive and imprinting effects when the QTL is deconstructed into sex-by-diet cohort. In males fed a high-fat diet, there is paternal imprinting whereby individuals inheriting the SM/J allele from their father have a relatively poor response to glucose stress. The implications of this result are two-fold: genomic imprinting is not only a dynamic contributor to variation, but also an effect that is highly context dependent. These results support previous findings that imprinting patterns are labile [104, 130] and not consistent across all genotypes and environments. Further, if sex-by-diet cohorts are not considered individually, the imprinting effect is washed out in the full population. Such context goes unmeasured in human GWAS and by not being considered, results are missing an important aspect of the genetic architecture underlying variation in metabolic traits.
Figure 2.
Genotypic means for a significant QTL for glucose tolerance in an F16 LG/J and SM/J AIL (Wustl:LG,SM-G16). The QTL has significant additive effects in the full population, and significant additive and imprinting effects in males fed a high-fat diet. Note the scale is different in the high-fat fed males to accommodate that cohort’s higher mean values.
4.4 Maternal effects
Maternal effects can be generally defined as the influence of a mother’s phenotype on the phenotypes of her offspring [131], and human studies indicate that maternal BMI is significantly associated with MetS in offspring [132, 133]. While maternal phenotype is considered environmental with respect to offspring genotype, there is a genetic basis to variation in the maternal generation including the mitochondria (that generate energy and are referred to as the “powerhouse” of the cells), transmitted by mother’s ovum. Because it is difficult to quantify the genetic and environmental components of maternal phenotype on MetS in offspring in humans, the mouse has proved invaluable for illuminating this aspect of the genetic architecture. For example, Wolf et al. (2002) identified maternal effect QTL that accounted for 31.5% of among litter variance in offspring early growth in a cross-fostered F3 LG/J x SM/J intercross [134]. Jarvis et al. (2005) measured genetic maternal effects on offspring lipid, obesity and diabetes related traits in reciprocal crosses between C57BL/6J and 10 LGXSM RILs [105]. The authors found that genetic maternal effects accounted for as much as 10% of the phenotypic variance in offspring at 17 or more weeks after weaning. These results indicate that variation in metabolic traits not only is dependent on an individual’s own genotype, but also is independently influenced by the individual’s mother’s genotype.
5. Promise of comparative genomics and bioinformatics
The wealth of information provided by recently available whole-genome sequences promises to expand our knowledge of MetS. Direct examination of the human syntenic regions for mouse QTL has already led to identification of QTG for plasma lipids [135–137] and hypertension [24, 138, 139]. Bioinformatic tools are developed that can aid this research [140], and several databases catalog positional information for human disease mutations, including both coding and non-coding variations [141]. For example, bioinformatic analysis of multiple-species protein coding sequence alignments finds that heritable human disease-associated mutations overwhelmingly occur at phylogentically conserved amino acid sites [142–144]. However, comparative genomic analyses of mammalian molecular evolution have also found evidence that genes related to some known Mendelian disorders have evolved differently in humans than in lineages leading to other closely related contemporary mammals [145, 146]. Comparison among primates has identified several instances of a human disease-associated coding variant corresponding to the wild-type amino acid in the chimpanzee, the macaque, or the reconstructed ancestral primate genome [147], implying that the protective variant seen in humans is derived. Thus a non-trivial proportion of disease genetic risk is associated with ancestral alleles [148], and knowledge of a QTL associated with a disease phenotype in mouse can allow one to use whole genome multiple-species alignments to reconstruct the evolution of a genomic region and to make functional predictions, which, in a biomedical context, have obvious therapeutic implications.
Comparing multiple species’ genomes has led to the discovery of polymorphisms and molecules that have been shown to contribute to disease risk. For example, copy number variants (CNVs), DNA segments that vary in number among individuals, are a recently identified source of genetic variation in mammals and have been shown to affect gene expression, presumably through altered gene dosage [149–151]. Recently, CNVs were found to be highly associated with variation in gene expression levels in mouse adipose tissue, and QTL for metabolic traits were identified at or near several CNV genes in an F2 C57BL/6J x C3H/HeJ intercross [152]. Non-coding microRNAs have been shown to affect many biological processes, including metabolism [153, 154], and mouse models have recently been used to characterize microRNA-dependent regulation of glucose homeostasis and lipid metabolism in mouse models of obesity and T2D [155, 156].
A particularly promising area of research where mouse models are proving invaluable is the characterization of coding and non-coding DNA sequence in gene networks and regulatory pathways. Genes do not function singly, or singly affect a trait. A recent experiment that focused on susceptibility loci for metabolic disease traits to identify gene networks validated lipoprotein lipase (Lpl), lactamase beta (Lactb), and protein phosphatae 1-like (Ppm1l) as genes underlying obesity in mice [157]. Some recent bioinformatic approaches have taken advantage of genes and QTL found through mouse models to identify sequences affecting multiple components of MetS. Both these methods, experimental and bioinformatic, point to focal candidates for further analysis in human populations [44, 69]. Further, under a comparative genomics framework, one could examine co-evolution of genes that interact within and between disease components to identify patterns that could guide future research into the etiology of MetS. Such an approach should incorporate insights gained from mouse models about the complexity and the context-dependency of gene-by-environment interactions if it is to lead to rational treatments and prevention strategies in humans.
Supplementary Material
Acknowledgments
This work was supported by NIDDK DK055736 to James M. Cheverud and by NHLBI T32-HL091823, program director Dr D.C. Rao, to Heather A. Lawson.
References
- 1.Cornier MA, Dabelea D, Hernandez TL, Lindstrom RC, Steig AJ, Stob NR, Van Pelt RE, Wang H, Eckel RH. The metabolic syndrome. Endocr Rev. 2008;29:777–822. doi: 10.1210/er.2008-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.National_Diabetes_Information_Clearinghouse. National Diabetes Statistics. Bethesda: 2005. [Google Scholar]
- 3.National_Center_for_Health_Statistics. Public Health Service. Hyattsville: 2006. [Google Scholar]
- 4.The_Jackson_Laboratory. Type_2_Diabetes_and_Obesity_Research_and_the_Laboratory_Mouse. Bar Harbor: 2007. [Google Scholar]
- 5.Polotsky VY. Mouse model of the metabolic syndrome: the quest continues. J Appl Physiol. 2007;102:2088–9. doi: 10.1152/japplphysiol.00219.2007. [DOI] [PubMed] [Google Scholar]
- 6.Abegunde DO, Mathers CD, Adam T, Ortegon M, Strong K. The burden and costs of chronic diseases in low-income and middle-income countries. Lancet. 2007;370:1929–38. doi: 10.1016/S0140-6736(07)61696-1. [DOI] [PubMed] [Google Scholar]
- 7.Walley AJ, Asher JE, Froguel P. The genetic contribution to non-syndromic human obesity. Nat Rev Genet. 2009;10:431–42. doi: 10.1038/nrg2594. [DOI] [PubMed] [Google Scholar]
- 8.Perusse L, Rice T, Province MA, Gagnon J, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C. Familial aggregation of amount and distribution of subcutaneous fat and their responses to exercise training in the HERITAGE family study. Obes Res. 2000;8:140–50. doi: 10.1038/oby.2000.15. [DOI] [PubMed] [Google Scholar]
- 9.Williams CM. Lipid metabolism in women. Proc Nutr Soc. 2004;63:153–60. doi: 10.1079/PNS2003314. [DOI] [PubMed] [Google Scholar]
- 10.Ordovas JM, Shen J. Gene-environment interactions and susceptibility to metabolic syndrome and other chronic diseases. J Periodontol. 2008;79:1508–13. doi: 10.1902/jop.2008.080232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mackay TF, Stone EA, Ayroles JF. The genetics of quantitative traits: challenges and prospects. Nat Rev Genet. 2009;10:565–77. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- 12.Boguski MS. Comparative genomics: the mouse that roared. Nature. 2002;420:515–6. doi: 10.1038/420515a. [DOI] [PubMed] [Google Scholar]
- 13.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O’Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 14.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 15.Leiter EH. In: Type 2 Diabetes, Methods in Molecular Biology. Stocker C, editor. Vol. 560. Springer; New York: 2009. pp. 1–17. [DOI] [PubMed] [Google Scholar]
- 16.Brosius FC, 3rd, Alpers CE, Bottinger EP, Breyer MD, Coffman TM, Gurley SB, Harris RC, Kakoki M, Kretzler M, Leiter EH, Levi M, McIndoe RA, Sharma K, Smithies O, Susztak K, Takahashi N, Takahashi T. Mouse Models of Diabetic Nephropathy. J Am Soc Nephrol. 2009 doi: 10.1681/ASN.2009070721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breyer MD, Bottinger E, Brosius FC, Coffman TM, Fogo A, Harris RC, Heilig CW, Sharma K. Diabetic nephropathy: of mice and men. Adv Chronic Kidney Dis. 2005;12:128–45. doi: 10.1053/j.ackd.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Breyer MD, Bottinger E, Brosius FC, 3rd, Coffman TM, Harris RC, Heilig CW, Sharma K. Mouse models of diabetic nephropathy. J Am Soc Nephrol. 2005;16:27–45. doi: 10.1681/ASN.2004080648. [DOI] [PubMed] [Google Scholar]
- 19.Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet. 2007;39:730–2. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foote M, Hunter JP, Janis CM, Sepkoski JJ., Jr Evolutionary and preservational constraints on origins of biologic groups: divergence times of eutherian mammals. Science. 1999;283:1310–4. doi: 10.1126/science.283.5406.1310. [DOI] [PubMed] [Google Scholar]
- 21.Ehrich TH, Kenney-Hunt JP, Pletscher LS, Cheverud JM. Genetic variation and correlation of dietary response in an advanced intercross mouse line produced from two divergent growth lines. Genet Res. 2005;85:211–22. doi: 10.1017/S0016672305007603. [DOI] [PubMed] [Google Scholar]
- 22.Fawcett GL, Jarvis JP, Roseman CC, Wang B, Wolf JB, Cheverud JM. Fine-Mapping of Obesity-Related Quantitative Trait Loci in and F9/10 Advanced Intercross Line. Obesity (Silver Spring) 2009 doi: 10.1038/oby.2009.411. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheverud JM, Ehrich TH, Hrbek T, Kenney JP, Pletscher LS, Semenkovich CF. Quantitative trait loci for obesity-and diabetes-related traits and their dietary responses to high-fat feeding in LGXSM recombinant inbred mouse strains. Diabetes. 2004;53:3328–36. doi: 10.2337/diabetes.53.12.3328. [DOI] [PubMed] [Google Scholar]
- 24.Koutnikova H, Laakso M, Lu L, Combe R, Paananen J, Kuulasmaa T, Kuusisto J, Haring HU, Hansen T, Pedersen O, Smith U, Hanefeld M, Williams RW, Auwerx J. Identification of the UBP1 locus as a critical blood pressure determinant using a combination of mouse and human genetics. PLoS Genet. 2009;5:e1000591. doi: 10.1371/journal.pgen.1000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondo H, Minegishi Y, Komine Y, Mori T, Matsumoto I, Abe K, Tokimitsu I, Hase T, Murase T. Differential regulation of intestinal lipid metabolism-related genes in obesity-resistant A/J vs. obesity-prone C57BL/6J mice. Am J Physiol Endocrinol Metab. 2006;291:E1092–9. doi: 10.1152/ajpendo.00583.2005. [DOI] [PubMed] [Google Scholar]
- 26.Clee SM, Yandell BS, Schueler KM, Rabaglia ME, Richards OC, Raines SM, Kabara EA, Klass DM, Mui ET, Stapleton DS, Gray-Keller MP, Young MB, Stoehr JP, Lan H, Boronenkov I, Raess PW, Flowers MT, Attie AD. Positional cloning of Sorcs1, a type 2 diabetes quantitative trait locus. Nat Genet. 2006;38:688–93. doi: 10.1038/ng1796. [DOI] [PubMed] [Google Scholar]
- 27.Mundy AL, Haas E, Bhattacharya I, Widmer CC, Kretz M, Hofmann-Lehmann R, Minotti R, Barton M. Fat intake modifies vascular responsiveness and receptor expression of vasoconstrictors: implications for diet-induced obesity. Cardiovasc Res. 2007;73:368–75. doi: 10.1016/j.cardiores.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 28.Adeghate E, Schattner P, Dunn E. An update on the etiology and epidemiology of diabetes mellitus. Ann N Y Acad Sci. 2006;1084:1–29. doi: 10.1196/annals.1372.029. [DOI] [PubMed] [Google Scholar]
- 29.Merat S, Casanada F, Sutphin M, Palinski W, Reaven PD. Western-type diets induce insulin resistance and hyperinsulinemia in LDL receptor-deficient mice but do not increase aortic atherosclerosis compared with normoinsulinemic mice in which similar plasma cholesterol levels are achieved by a fructose-rich diet. Arterioscler Thromb Vasc Biol. 1999;19:1223–30. doi: 10.1161/01.atv.19.5.1223. [DOI] [PubMed] [Google Scholar]
- 30.Towler DA, Bidder M, Latifi T, Coleman T, Semenkovich CF. Diet-induced diabetes activates an osteogenic gene regulatory program in the aortas of low density lipoprotein receptor-deficient mice. J Biol Chem. 1998;273:30427–34. doi: 10.1074/jbc.273.46.30427. [DOI] [PubMed] [Google Scholar]
- 31.Schreyer SA, Vick C, Lystig TC, Mystkowski P, LeBoeuf RC. LDL receptor but not apolipoprotein E deficiency increases diet-induced obesity and diabetes in mice. Am J Physiol Endocrinol Metab. 2002;282:E207–14. doi: 10.1152/ajpendo.2002.282.1.E207. [DOI] [PubMed] [Google Scholar]
- 32.Seidelmann SB, De Luca C, Leibel RL, Breslow JL, Tall AR, Welch CL. Quantitative trait locus mapping of genetic modifiers of metabolic syndrome and atherosclerosis in low-density lipoprotein receptor-deficient mice: identification of a locus for metabolic syndrome and increased atherosclerosis on chromosome 4. Arterioscler Thromb Vasc Biol. 2005;25:204–10. doi: 10.1161/01.ATV.0000149146.32385.1b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lloyd DJ, Helmering J, Cordover D, Bowsman M, Chen M, Hale C, Fordstrom P, Zhou M, Wang M, Kaufman SA, Veniant MM. Antidiabetic effects of 11beta-HSD1 inhibition in a mouse model of combined diabetes, dyslipidaemia and atherosclerosis. Diabetes Obes Metab. 2009;11:688–99. doi: 10.1111/j.1463-1326.2009.01034.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–32. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- 35.Clement K, Vaisse C, Lahlou N, Cabrol S, Pelloux V, Cassuto D, Gourmelen M, Dina C, Chambaz J, Lacorte JM, Basdevant A, Bougneres P, Lebouc Y, Froguel P, Guy-Grand B. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature. 1998;392:398–401. doi: 10.1038/32911. [DOI] [PubMed] [Google Scholar]
- 36.Green ED, Maffei M, Braden VV, Proenca R, DeSilva U, Zhang Y, Chua SC, Jr, Leibel RL, Weissenbach J, Friedman JM. The human obese (OB) gene: RNA expression pattern and mapping on the physical, cytogenetic, and genetic maps of chromosome 7. Genome Res. 1995;5:5–12. doi: 10.1101/gr.5.1.5. [DOI] [PubMed] [Google Scholar]
- 37.Chung WK, Power-Kehoe L, Chua M, Leibel RL. Mapping of the OB receptor to 1p in a region of nonconserved gene order from mouse and rat to human. Genome Res. 1996;6:431–8. doi: 10.1101/gr.6.5.431. [DOI] [PubMed] [Google Scholar]
- 38.Montague CT, Farooqi IS, Whitehead JP, Soos MA, Rau H, Wareham NJ, Sewter CP, Digby JE, Mohammed SN, Hurst JA, Cheetham CH, Earley AR, Barnett AH, Prins JB, O’Rahilly S. Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature. 1997;387:903–8. doi: 10.1038/43185. [DOI] [PubMed] [Google Scholar]
- 39.Bultman SJ, Michaud EJ, Woychik RP. Molecular characterization of the mouse agouti locus. Cell. 1992;71:1195–204. doi: 10.1016/s0092-8674(05)80067-4. [DOI] [PubMed] [Google Scholar]
- 40.Meehan TP, Tabeta K, Du X, Woodward LS, Firozi K, Beutler B, Justice MJ. Point mutations in the melanocortin-4 receptor cause variable obesity in mice. Mamm Genome. 2006;17:1162–71. doi: 10.1007/s00335-006-0073-z. [DOI] [PubMed] [Google Scholar]
- 41.Goodarzi MO, Lehman DM, Taylor KD, Guo X, Cui J, Quinones MJ, Clee SM, Yandell BS, Blangero J, Hsueh WA, Attie AD, Stern MP, Rotter JI. SORCS1: a novel human type 2 diabetes susceptibility gene suggested by the mouse. Diabetes. 2007;56:1922–9. doi: 10.2337/db06-1677. [DOI] [PubMed] [Google Scholar]
- 42.Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, Lindner TH, Mashima H, Schwarz PE, del Bosque-Plata L, Oda Y, Yoshiuchi I, Colilla S, Polonsky KS, Wei S, Concannon P, Iwasaki N, Schulze J, Baier LJ, Bogardus C, Groop L, Boerwinkle E, Hanis CL, Bell GI. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet. 2000;26:163–75. doi: 10.1038/79876. [DOI] [PubMed] [Google Scholar]
- 43.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Gudnason V, Sigurdsson G, Thorsteinsdottir U, Gulcher JR, Kong A, Stefansson K. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–3. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 44.Kraja AT, Province MA, Huang P, Jarvis JP, Rice T, Cheverud JM, Rao DC. Trends in metabolic syndrome and gene networks in human and rodent models. Endocr Metab Immune Disord Drug Targets. 2008;8:198–207. doi: 10.2174/187153008785700145. [DOI] [PubMed] [Google Scholar]
- 45.Bogardus C. Missing heritability and GWAS utility. Obesity (Silver Spring) 2009;17:209–10. doi: 10.1038/oby.2008.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krude H, Biebermann H, Luck W, Horn R, Brabant G, Gruters A. Severe early-onset obesity, adrenal insufficiency and red hair pigmentation caused by POMC mutations in humans. Nat Genet. 1998;19:155–7. doi: 10.1038/509. [DOI] [PubMed] [Google Scholar]
- 47.Vaisse C, Clement K, Guy-Grand B, Froguel P. A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat Genet. 1998;20:113–4. doi: 10.1038/2407. [DOI] [PubMed] [Google Scholar]
- 48.Abate N, Chandalia M, Satija P, Adams-Huet B, Grundy SM, Sandeep S, Radha V, Deepa R, Mohan V. ENPP1/PC-1 K121Q polymorphism and genetic susceptibility to type 2 diabetes. Diabetes. 2005;54:1207–13. doi: 10.2337/diabetes.54.4.1207. [DOI] [PubMed] [Google Scholar]
- 49.Suviolahti E, Oksanen LJ, Ohman M, Cantor RM, Ridderstrale M, Tuomi T, Kaprio J, Rissanen A, Mustajoki P, Jousilahti P, Vartiainen E, Silander K, Kilpikari R, Salomaa V, Groop L, Kontula K, Peltonen L, Pajukanta P. The SLC6A14 gene shows evidence of association with obesity. J Clin Invest. 2003;112:1762–72. doi: 10.1172/JCI17491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boutin P, Dina C, Vasseur F, Dubois S, Corset L, Seron K, Bekris L, Cabellon J, Neve B, Vasseur-Delannoy V, Chikri M, Charles MA, Clement K, Lernmark A, Froguel P. GAD2 on chromosome 10p12 is a candidate gene for human obesity. PLoS Biol. 2003;1:E68. doi: 10.1371/journal.pbio.0000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loos RJ, Bouchard C. FTO: the first gene contributing to common forms ofhuman obesity. Obes Rev. 2008;9:246–50. doi: 10.1111/j.1467-789X.2008.00481.x. [DOI] [PubMed] [Google Scholar]
- 52.Liu YJ, Liu XG, Wang L, Dina C, Yan H, Liu JF, Levy S, Papasian CJ, Drees BM, Hamilton JJ, Meyre D, Delplanque J, Pei YF, Zhang L, Recker RR, Froguel P, Deng HW. Genome-wide association scans identified CTNNBL1 as a novel gene for obesity. Hum Mol Genet. 2008;17:1803–13. doi: 10.1093/hmg/ddn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cotsapas C, Speliotes EK, Hatoum IJ, Greenawalt DM, Dobrin R, Lum PY, Suver C, Chudin E, Kemp D, Reitman M, Voight BF, Neale BM, Schadt EE, Hirschhorn JN, Kaplan LM, Daly MJ. Common body mass index-associated variants confer risk of extreme obesity. Hum Mol Genet. 2009;18:3502–7. doi: 10.1093/hmg/ddp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyre D, Delplanque J, Chevre JC, Lecoeur C, Lobbens S, Gallina S, Durand E, Vatin V, Degraeve F, Proenca C, Gaget S, Korner A, Kovacs P, Kiess W, Tichet J, Marre M, Hartikainen AL, Horber F, Potoczna N, Hercberg S, Levy-Marchal C, Pattou F, Heude B, Tauber M, McCarthy MI, Blakemore AI, Montpetit A, Polychronakos C, Weill J, Coin LJ, Asher J, Elliott P, Jarvelin MR, Visvikis-Siest S, Balkau B, Sladek R, Balding D, Walley A, Dina C, Froguel P. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–9. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 55.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, Jonsdottir T, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Jonsson F, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Lauritzen T, Aben KK, Verbeek AL, Roeleveld N, Kampman E, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Becker DM, Gulcher J, Kiemeney LA, Pedersen O, Kong A, Thorsteinsdottir U, Stefansson K. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 56.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association locifor human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, McCarthy MI. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–94. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.The_Wellcome_Trust_Case_Control_Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Collins A, Lonjou C, Morton NE. Genetic epidemiology of single-nucleotide polymorphisms. Proc Natl Acad Sci U S A. 1999;96:15173–7. doi: 10.1073/pnas.96.26.15173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–7. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 61.Lander ES. The new genomics: global views of biology. Science. 1996;274:536–9. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 62.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 63.Cuenot G. Pure strains and their combination in the mouse. Arch Zool Exp Genet. 1905;4:122–123. [Google Scholar]
- 64.Justice MJ. Capitalizing on large-scale mouse mutagenesis screens. Nat Rev Genet. 2000;1:109–15. doi: 10.1038/35038549. [DOI] [PubMed] [Google Scholar]
- 65.Tschop M, Heiman ML. Rodent obesity models: an overview. Exp Clin Endocrinol Diabetes. 2001;109:307–19. doi: 10.1055/s-2001-17297. [DOI] [PubMed] [Google Scholar]
- 66.Speakman J, Hambly C, Mitchell S, Krol E. Animal models of obesity. Obes Rev. 2007;8(Suppl 1):55–61. doi: 10.1111/j.1467-789X.2007.00319.x. [DOI] [PubMed] [Google Scholar]
- 67.Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–8. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brockmann GA, Bevova MR. Using mouse models to dissect the genetics of obesity. Trends Genet. 2002;18:367–76. doi: 10.1016/s0168-9525(02)02703-8. [DOI] [PubMed] [Google Scholar]
- 69.Wuschke S, Dahm S, Schmidt C, Joost HG, Al-Hasani H. A meta-analysis of quantitative trait loci associated with body weight and adiposity in mice. Int J Obes (Lond) 2007;31:829–41. doi: 10.1038/sj.ijo.0803473. [DOI] [PubMed] [Google Scholar]
- 70.Fawcett GL, Roseman CC, Jarvis JP, Wang B, Wolf JB, Cheverud JM. Genetic architecture of adiposity and organ weight using combined generation QTL analysis. Obesity (Silver Spring) 2008;16:1861–8. doi: 10.1038/oby.2008.300. [DOI] [PubMed] [Google Scholar]
- 71.Cheverud JM, Vaughn TT, Pletscher LS, Peripato AC, Adams ES, Erikson CF, King-Ellison KJ. Genetic architecture of adiposity in the cross of LG/J and SM/J inbred mice. Mamm Genome. 2001;12:3–12. doi: 10.1007/s003350010218. [DOI] [PubMed] [Google Scholar]
- 72.Ehrich TH, Kenney JP, Vaughn TT, Pletscher LS, Cheverud JM. Diet, obesity, and hyperglycemia in LG/J and SM/J mice. Obes Res. 2003;11:1400–10. doi: 10.1038/oby.2003.189. [DOI] [PubMed] [Google Scholar]
- 73.Kramer MG, Vaughn TT, Pletscher LS, King-Ellison K, Adams E, Erikson C, Cheverud JM. Genetic variation in body weight gain and composition in the intercross of Large (LG/J) and Small (SM/J) inbred strains of mice. Genetics and Molecular Biology. 1998;21:211–218. [Google Scholar]
- 74.Kenney-Hunt JP, Vaughn TT, Pletscher LS, Peripato A, Routman E, Cothran K, Durand D, Norgard E, Perel C, Cheverud JM. Quantitative trait loci for body size components in mice. Mamm Genome. 2006;17:526–37. doi: 10.1007/s00335-005-0160-6. [DOI] [PubMed] [Google Scholar]
- 75.Cheverud JM, Fawcett GL, Jarvis JP, Norgard EA, Pavlicev M, Pletscher LS, Polonsky KS, Ye H, Bell GI, Semenkovich CF. Calpain-10 is a component of the obesity-related quantitative trait locus, Adip1. J Lipid Res. 2009 doi: 10.1194/jlr.M900128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Perusse L, Bouchard C. The human obesitygene map: the 2005 update. Obesity (Silver Spring) 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- 77.Permutt MA, Wasson J, Cox N. Genetic epidemiology of diabetes. J Clin Invest. 2005;115:1431–9. doi: 10.1172/JCI24758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clee SM, Attie AD. The genetic landscape of type 2 diabetes in mice. Endocr Rev. 2007;28:48–83. doi: 10.1210/er.2006-0035. [DOI] [PubMed] [Google Scholar]
- 79.Huopio H, Otonkoski T, Vauhkonen I, Reimann F, Ashcroft FM, Laakso M. A new subtype of autosomal dominant diabetes attributable to a mutation in the gene for sulfonylurea receptor 1. Lancet. 2003;361:301–7. doi: 10.1016/S0140-6736(03)12325-2. [DOI] [PubMed] [Google Scholar]
- 80.Farris W, Mansourian S, Chang Y, Lindsley L, Eckman EA, Frosch MP, Eckman CB, Tanzi RE, Selkoe DJ, Guenette S. Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc Natl Acad Sci U S A. 2003;100:4162–7. doi: 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, Tuomi T, Gaudet D, Hudson TJ, Daly M, Groop L, Lander ES. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 82.Wolfs MG, Hofker MH, Wijmenga C, van Haeften TW. Type 2 Diabetes Mellitus: New Genetic Insights will Lead to New Therapeutics. Curr Genomics. 2009;10:110–8. doi: 10.2174/138920209787847023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hrbek T, de Brito RA, Wang B, Pletscher LS, Cheverud JM. Genetic characterization of a new set of recombinant inbred lines (LGXSM) formed from the inter-cross of SM/J and LG/J inbred mouse strains. Mamm Genome. 2006;17:417–29. doi: 10.1007/s00335-005-0038-7. [DOI] [PubMed] [Google Scholar]
- 84.Cheverud JM, Ehrich TH, Kenney JP, Pletscher LS, Semenkovich CF. Genetic evidence for discordance between obesity-and diabetes-related traits in the LGXSM recombinant inbred mouse strains. Diabetes. 2004;53:2700–8. doi: 10.2337/diabetes.53.10.2700. [DOI] [PubMed] [Google Scholar]
- 85.de Oliveira CM, Pereira AC, de Andrade M, Soler JM, Krieger JE. Heritability of cardiovascular risk factors in a Brazilian population: Baependi Heart Study. BMC Med Genet. 2008;9:32. doi: 10.1186/1471-2350-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lloyd-Jones D, Adams R, Carnethon M, De Simone G, Ferguson TB, Flegal K, Ford E, Furie K, Go A, Greenlund K, Haase N, Hailpern S, Ho M, Howard V, Kissela B, Kittner S, Lackland D, Lisabeth L, Marelli A, McDermott M, Meigs J, Mozaffarian D, Nichol G, O’Donnell C, Roger V, Rosamond W, Sacco R, Sorlie P, Stafford R, Steinberger J, Thom T, Wasserthiel-Smoller S, Wong N, Wylie-Rosett J, Hong Y. Heart disease and stroke statistics--2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2009;119:480–6. doi: 10.1161/CIRCULATIONAHA.108.191259. [DOI] [PubMed] [Google Scholar]
- 87.Elder SJ, Lichtenstein AH, Pittas AG, Roberts SB, Fuss PJ, Greenberg AS, McCrory MA, Bouchard TJ, Jr, Saltzman E, Neale MC. Genetic and environmental influences on factors associated with cardiovascular disease and the metabolic syndrome. J Lipid Res. 2009 doi: 10.1194/jlr.P900033-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goldstein JL, Hobbs HH, Brown MS. In: The Metabolic and Molecular Basis of Inherited Disease. 8. Scriver CR, Beaudet AL, Sly WS, Vall D, editors. Vol. 2. McGraw-Hill; New York: 2001. pp. 2863–2913. [Google Scholar]
- 89.Goldstein JL, Brown MS. Molecular medicine. The cholesterol quartet. Science. 2001;292:1310–2. doi: 10.1126/science.1061815. [DOI] [PubMed] [Google Scholar]
- 90.Berge KE, Tian H, Graf GA, Yu L, Grishin NV, Schultz J, Kwiterovich P, Shan B, Barnes R, Hobbs HH. Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science. 2000;290:1771–5. doi: 10.1126/science.290.5497.1771. [DOI] [PubMed] [Google Scholar]
- 91.Nabel EG. Cardiovascular disease. N Engl J Med. 2003;349:60–72. doi: 10.1056/NEJMra035098. [DOI] [PubMed] [Google Scholar]
- 92.Russell JC, Proctor SD. Small animal models of cardiovascular disease: tools for the study of theroles of metabolic syndrome, dyslipidemia, and atherosclerosis. Cardiovasc Pathol. 2006;15:318–30. doi: 10.1016/j.carpath.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 93.Xu Q. A Handbook of Mouse Models of Cardiovascular Disease. Wiley; Hoboken: 2006. p. 402. [Google Scholar]
- 94.Adeghate E, Christopher Howarth F, Rashed H, Saeed T, Gbewonyo A. The effect of a fat-enriched diet on the pattern of distribution of pancreatic islet cells in the C57BL/6J mice. Ann N Y Acad Sci. 2006;1084:361–70. doi: 10.1196/annals.1372.002. [DOI] [PubMed] [Google Scholar]
- 95.Grubb SC, Maddatu TP, Bult CJ, Bogue MA. Mouse phenome database. Nucleic Acids Res. 2009;37:D720–30. doi: 10.1093/nar/gkn778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hajjar I, Kotchen TA. Trends in prevalence, awareness, treatment, and control of hypertension in the United States, 1988–2000. JAMA. 2003;290:199–206. doi: 10.1001/jama.290.2.199. [DOI] [PubMed] [Google Scholar]
- 97.Geller DS, Farhi A, Pinkerton N, Fradley M, Moritz M, Spitzer A, Meinke G, Tsai FT, Sigler PB, Lifton RP. Activating mineralocorticoid receptor mutation in hypertension exacerbated by pregnancy. Science. 2000;289:119–23. doi: 10.1126/science.289.5476.119. [DOI] [PubMed] [Google Scholar]
- 98.Barroso I, Gurnell M, Crowley VE, Agostini M, Schwabe JW, Soos MA, Maslen GL, Williams TD, Lewis H, Schafer AJ, Chatterjee VK, O’Rahilly S. Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature. 1999;402:880–3. doi: 10.1038/47254. [DOI] [PubMed] [Google Scholar]
- 99.Cowley AW., Jr The genetic dissection of essential hypertension. Nat Rev Genet. 2006;7:829–40. doi: 10.1038/nrg1967. [DOI] [PubMed] [Google Scholar]
- 100.Feng M, Deerhake ME, Keating R, Thaisz J, Xu L, Tsaih SW, Smith R, Ishige T, Sugiyama F, Churchill GA, DiPetrillo K. Genetic analysis of blood pressure in 8 mouse intercross populations. Hypertension. 2009;54:802–9. doi: 10.1161/HYPERTENSIONAHA.109.134569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nishihara E, Tsaih SW, Tsukahara C, Langley S, Sheehan S, DiPetrillo K, Kunita S, Yagami K, Churchill GA, Paigen B, Sugiyama F. Quantitative trait loci associated with blood pressure of metabolic syndrome in the progeny of NZO/HILtJxC3H/HeJ intercrosses. Mamm Genome. 2007;18:573–83. doi: 10.1007/s00335-007-9033-5. [DOI] [PubMed] [Google Scholar]
- 102.Tsukahara C, Sugiyama F, Paigen B, Kunita S, Yagami K. Blood pressure in 15 inbred mouse strains and its lack of relation with obesity and insulin resistance in the progeny of an NZO/HILtJ × C3H/HeJ intercross. Mamm Genome. 2004;15:943–50. doi: 10.1007/s00335-004-2411-3. [DOI] [PubMed] [Google Scholar]
- 103.Cheverud JM, Routman EJ, Duarte FA, van Swinderen B, Cothran K, Perel C. Quantitative trait loci for murine growth. Genetics. 1996;142:1305–19. doi: 10.1093/genetics/142.4.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wolf JB, Cheverud JM, Roseman C, Hager R. Genome-wide analysis reveals a complex pattern of genomic imprinting in mice. PLoS Genet. 2008;4:e1000091. doi: 10.1371/journal.pgen.1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jarvis JP, Kenney-Hunt J, Ehrich TH, Pletscher LS, Semenkovich CF, Cheverud JM. Maternal genotype affects adult offspring lipid, obesity, and diabetes phenotypes in LGXSM recombinant inbred strains. J Lipid Res. 2005;46:1692–702. doi: 10.1194/jlr.M500073-JLR200. [DOI] [PubMed] [Google Scholar]
- 106.Junyent M, Tucker KL, Smith CE, Garcia-Rios A, Mattei J, Lai CQ, Parnell LD, Ordovas JM. The effects of ABCG5/G8 polymorphisms on plasma HDL cholesterol concentrations depend on smoking habit in the Boston Puerto Rican Health Study. J Lipid Res. 2009;50:565–73. doi: 10.1194/jlr.P800041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Junyent M, Tucker KL, Smith CE, Lane JM, Mattei J, Lai CQ, Parnell LD, Ordovas JM. The effects of ABCG5/G8 polymorphisms on HDL-cholesterol concentrations depend on ABCA1 genetic variants in the Boston Puerto Rican Health Study. Nutr Metab Cardiovasc Dis. 2009 doi: 10.1016/j.numecd.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kabagambe EK, Glasser SP, Ordovas JM, Warodomwichit D, Tsai MY, Hopkins PN, Borecki IB, Wojczynski MK, Arnett DK. TCF7L2 polymorphisms and inflammatory markers before and after treatment with fenofibrate. Diabetol Metab Syndr. 2009;1:16. doi: 10.1186/1758-5996-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Svenson KL, Von Smith R, Magnani PA, Suetin HR, Paigen B, Naggert JK, Li R, Churchill GA, Peters LL. Multiple trait measurements in 43 inbred mouse strains capture the phenotypic diversity characteristic of human populations. J Appl Physiol. 2007;102:2369–78. doi: 10.1152/japplphysiol.01077.2006. [DOI] [PubMed] [Google Scholar]
- 110.York B, Lei K, West DB. Sensitivity to dietary obesity linked to a locus on chromosome 15 in a CAST/Ei × C57BL/6J F2 intercross. Mamm Genome. 1996;7:677–81. doi: 10.1007/s003359900204. [DOI] [PubMed] [Google Scholar]
- 111.Taylor BA, Tarantino LM, Phillips SJ. Gender-influenced obesity QTLs identified in a cross involving the KK type II diabetes-prone mouse strain. Mamm Genome. 1999;10:963–8. doi: 10.1007/s003359901141. [DOI] [PubMed] [Google Scholar]
- 112.Taylor BA, Wnek C, Schroeder D, Phillips SJ. Multiple obesity QTLs identified in an intercross between the NZO (New Zealand obese) and the SM (small) mouse strains. Mamm Genome. 2001;12:95–103. doi: 10.1007/s003350010254. [DOI] [PubMed] [Google Scholar]
- 113.West DB, Waguespack J, McCollister S. Dietary obesity in the mouse: interaction of strain with diet composition. Am J Physiol. 1995;268:R658–65. doi: 10.1152/ajpregu.1995.268.3.R658. [DOI] [PubMed] [Google Scholar]
- 114.Rance KA, Heath SC, Keightley PD. Mapping quantitative trait loci for body weight on the X chromosome in mice. II. Analysis of congenic backcrosses. Genet Res. 1997;70:125–33. doi: 10.1017/s0016672397002929. [DOI] [PubMed] [Google Scholar]
- 115.Rance KA, Hill WG, Keightley PD. Mapping quantitative trait loci for body weight on the X chromosome in mice. I. Analysis of a reciprocal F2 population. Genet Res. 1997;70:117–24. doi: 10.1017/s0016672397002917. [DOI] [PubMed] [Google Scholar]
- 116.Baratta R, Di Paola R, Spampinato D, Fini G, Marucci A, Coco A, Vigneri R, Frittitta L, Trischitta V. Evidence for genetic epistasis in human insulin resistance: the combined effect of PC-1 (K121Q) and PPARgamma2 (P12A) polymorphisms. J Mol Med. 2003;81:718–23. doi: 10.1007/s00109-003-0466-3. [DOI] [PubMed] [Google Scholar]
- 117.Pedersen JC, Berg K. Interaction between low density lipoprotein receptor (LDLR) and apolipoprotein E (apoE) alleles contributes to normal variation in lipid level. Clin Genet. 1989;35:331–7. doi: 10.1111/j.1399-0004.1989.tb02953.x. [DOI] [PubMed] [Google Scholar]
- 118.Brockmann GA, Kratzsch J, Haley CS, Renne U, Schwerin M, Karle S. Single QTL effects, epistasis, and pleiotropy account for two-thirds of the phenotypic F(2) variance of growth and obesity in DU6i × DBA/2 mice. Genome Res. 2000;10:1941–57. doi: 10.1101/gr.gr1499r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cheverud JM. In: The Character Concept in Evolutionary Biology. Wagner G, editor. Academic Press; New York: 2002. pp. 411–434. [Google Scholar]
- 120.Hanlon P, Lorenz WA, Shao Z, Harper JM, Galecki AT, Miller RA, Burke DT. Three-locus and four-locus QTL interactions influence mouse insulin-like growth factor-I. Physiol Genomics. 2006;26:46–54. doi: 10.1152/physiolgenomics.00247.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stylianou IM, Korstanje R, Li R, Sheehan S, Paigen B, Churchill GA. Quantitative trait locus analysis for obesity reveals multiple networks of interacting loci. Mamm Genome. 2006;17:22–36. doi: 10.1007/s00335-005-0091-2. [DOI] [PubMed] [Google Scholar]
- 122.Junien C, Nathanielsz P. Report on the IASO Stock Conference 2006: early and lifelong environmental epigenomic programming of metabolic syndrome, obesity and type II diabetes. Obes Rev. 2007;8:487–502. doi: 10.1111/j.1467-789X.2007.00371.x. [DOI] [PubMed] [Google Scholar]
- 123.Smith FM, Garfield AS, Ward A. Regulation of growth and metabolism by imprinted genes. Cytogenet Genome Res. 2006;113:279–91. doi: 10.1159/000090843. [DOI] [PubMed] [Google Scholar]
- 124.Williamson CR, Blake A, Thomas S, Beechey CV, Hancock J, Cattanach BM, Peters J. [accessed October 17, 2009];World Wide Web Site -Mouse Imprinting Data and References. www.har.mrc.ac.uk/research/genomic_imprinting/citation.html.
- 125.Luedi PP, Dietrich FS, Weidman JR, Bosko JM, Jirtle RL, Hartemink AJ. Computational and experimental identification of novel human imprinted genes. Genome Res. 2007;17:1723–30. doi: 10.1101/gr.6584707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Luedi PP, Hartemink AJ, Jirtle RL. Genome-wide prediction of imprinted murine genes. Genome Res. 2005;15:875–84. doi: 10.1101/gr.3303505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mantey C, Brockmann GA, Kalm E, Reinsch N. Mapping and exclusion mapping of genomic imprinting effects in mouse F2 families. J Hered. 2005;96:329–38. doi: 10.1093/jhered/esi044. [DOI] [PubMed] [Google Scholar]
- 128.Wolf JB, Hager R, Cheverud JM. Genomic imprinting effects on complex traits: a phenotype-based perspective. Epigenetics. 2008;3:295–9. doi: 10.4161/epi.3.6.7257. [DOI] [PubMed] [Google Scholar]
- 129.Lindgren CM, Heid IM, Randall JC, Lamina C, Steinthorsdottir V, Qi L, Speliotes EK, Thorleifsson G, Willer CJ, Herrera BM, Jackson AU, Lim N, Scheet P, Soranzo N, Amin N, Aulchenko YS, Chambers JC, Drong A, Luan J, Lyon HN, Rivadeneira F, Sanna S, Timpson NJ, Zillikens MC, Zhao JH, Almgren P, Bandinelli S, Bennett AJ, Bergman RN, Bonnycastle LL, Bumpstead SJ, Chanock SJ, Cherkas L, Chines P, Coin L, Cooper C, Crawford G, Doering A, Dominiczak A, Doney AS, Ebrahim S, Elliott P, Erdos MR, Estrada K, Ferrucci L, Fischer G, Forouhi NG, Gieger C, Grallert H, Groves CJ, Grundy S, Guiducci C, Hadley D, Hamsten A, Havulinna AS, Hofman A, Holle R, Holloway JW, Illig T, Isomaa B, Jacobs LC, Jameson K, Jousilahti P, Karpe F, Kuusisto J, Laitinen J, Lathrop GM, Lawlor DA, Mangino M, McArdle WL, Meitinger T, Morken MA, Morris AP, Munroe P, Narisu N, Nordstrom A, Nordstrom P, Oostra BA, Palmer CN, Payne F, Peden JF, Prokopenko I, Renstrom F, Ruokonen A, Salomaa V, Sandhu MS, Scott LJ, Scuteri A, Silander K, Song K, Yuan X, Stringham HM, Swift AJ, Tuomi T, Uda M, Vollenweider P, Waeber G, Wallace C, Walters GB, Weedon MN, Witteman JC, Zhang C, Zhang W, Caulfield MJ, Collins FS, Davey Smith G, Day IN, Franks PW, Hattersley AT, Hu FB, Jarvelin MR, Kong A, Kooner JS, Laakso M, Lakatta E, Mooser V, Morris AD, Peltonen L, Samani NJ, Spector TD, Strachan DP, Tanaka T, Tuomilehto J, Uitterlinden AG, van Duijn CM, Wareham NJ, Hugh W, Waterworth DM, Boehnke M, Deloukas P, Groop L, Hunter DJ, Thorsteinsdottir U, Schlessinger D, Wichmann HE, Frayling TM, Abecasis GR, Hirschhorn JN, Loos RJ, Stefansson K, Mohlke KL, Barroso I, McCarthy MI. Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet. 2009;5:e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hager R, Cheverud JM, Wolf JB. Relative contribution of additive, dominance, and imprinting effects to phenotypic variation in body size and growth between divergent selectionlines of mice. Evolution. 2009;63:1118–28. doi: 10.1111/j.1558-5646.2009.00638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cheverud JM, Wolf JB. In: Maternal Effects in Mammals. Maestripieri D, Mateo J, editors. University of Chicago Press; Chicago: 2009. pp. 11–37. [Google Scholar]
- 132.Ekelund U, Anderssen S, Andersen LB, Riddoch CJ, Sardinha LB, Luan J, Froberg K, Brage S. Prevalence and correlates of the metabolic syndrome in a population-based sample of European youth. Am J Clin Nutr. 2009;89:90–6. doi: 10.3945/ajcn.2008.26649. [DOI] [PubMed] [Google Scholar]
- 133.Hirschler V, Roque MI, Calcagno ML, Gonzalez C, Aranda C. Maternal waist circumference and the prediction of children’s metabolic syndrome. Arch Pediatr Adolesc Med. 2007;161:1205–10. doi: 10.1001/archpedi.161.12.1205. [DOI] [PubMed] [Google Scholar]
- 134.Wolf JB, Vaughn TT, Pletscher LS, Cheverud JM. Contribution of maternal effectQTL to genetic architecture of early growth in mice. Heredity. 2002;89:300–10. doi: 10.1038/sj.hdy.6800140. [DOI] [PubMed] [Google Scholar]
- 135.Wang X, Ishimori N, Korstanje R, Rollins J, Paigen B. Identifying novel genes for atherosclerosis through mouse-human comparative genetics. Am J Hum Genet. 2005;77:1–15. doi: 10.1086/431656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang X, Paigen B. Genome-wide search for new genes controlling plasma lipid concentrations in mice and humans. Curr Opin Lipidol. 2005;16:127–37. doi: 10.1097/01.mol.0000162317.09054.9d. [DOI] [PubMed] [Google Scholar]
- 137.Wang X, Ria M, Kelmenson PM, Eriksson P, Higgins DC, Samnegard A, Petros C, Rollins J, Bennet AM, Wiman B, de Faire U, Wennberg C, Olsson PG, Ishii N, Sugamura K, Hamsten A, Forsman-Semb K, Lagercrantz J, Paigen B. Positional identification of TNFSF4, encoding OX40 ligand, as a gene that influences atherosclerosis susceptibility. Nat Genet. 2005;37:365–72. doi: 10.1038/ng1524. [DOI] [PubMed] [Google Scholar]
- 138.Stoll M, Kwitek-Black AE, Cowley AW, Jr, Harris EL, Harrap SB, Krieger JE, Printz MP, Provoost AP, Sassard J, Jacob HJ. New target regions for human hypertension via comparative genomics. Genome Res. 2000;10:473–82. doi: 10.1101/gr.10.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sugiyama F, Churchill GA, Higgins DC, Johns C, Makaritsis KP, Gavras H, Paigen B. Concordance of murine quantitative trait loci for salt-induced hypertension with rat and human loci. Genomics. 2001;71:70–7. doi: 10.1006/geno.2000.6401. [DOI] [PubMed] [Google Scholar]
- 140.Taylor J, Schenck I, Blankenberg D, Nekrutenko A. Using galaxy to perform large-scale interactive data analyses. Curr Protoc Bioinformatics. 2007;19:10.5.1–10.5.25. doi: 10.1002/0471250953.bi1005s19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, Cooper DN. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shastry BS. SNP alleles in human disease and evolution. J Hum Genet. 2002;47:561–6. doi: 10.1007/s100380200086. [DOI] [PubMed] [Google Scholar]
- 143.Shastry BS. SNPs in disease gene mapping, medicinal drug development and evolution. J Hum Genet. 2007;52:871–80. doi: 10.1007/s10038-007-0200-z. [DOI] [PubMed] [Google Scholar]
- 144.Subramanian S, Kumar S. Evolutionary anatomies of positions and types of disease-associated and neutral amino acid mutations in the human genome. BMC Genomics. 2006;7:306. doi: 10.1186/1471-2164-7-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Clark AG, Glanowski S, Nielsen R, Thomas PD, Kejariwal A, Todd MA, Tanenbaum DM, Civello D, Lu F, Murphy B, Ferriera S, Wang G, Zheng X, White TJ, Sninsky JJ, Adams MD, Cargill M. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–3. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- 146.Nielsen R, Bustamante C, Clark AG, Glanowski S, Sackton TB, Hubisz MJ, Fledel-Alon A, Tanenbaum DM, Civello D, White TJ, JJS, Adams MD, Cargill M. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, Batzer MA, Bustamante CD, Eichler EE, Hahn MW, Hardison RC, Makova KD, Miller W, Milosavljevic A, Palermo RE, Siepel A, Sikela JM, Attaway T, Bell S, Bernard KE, Buhay CJ, Chandrabose MN, Dao M, Davis C, Delehaunty KD, Ding Y, Dinh HH, Dugan-Rocha S, Fulton LA, Gabisi RA, Garner TT, Godfrey J, Hawes AC, Hernandez J, Hines S, Holder M, Hume J, Jhangiani SN, Joshi V, Khan ZM, Kirkness EF, Cree A, Fowler RG, Lee S, Lewis LR, Li Z, Liu YS, Moore SM, Muzny D, Nazareth LV, Ngo DN, Okwuonu GO, Pai G, Parker D, Paul HA, Pfannkoch C, Pohl CS, Rogers YH, Ruiz SJ, Sabo A, Santibanez J, Schneider BW, Smith SM, Sodergren E, Svatek AF, Utterback TR, Vattathil S, Warren W, White CS, Chinwalla AT, Feng Y, Halpern AL, Hillier LW, Huang X, Minx P, Nelson JO, Pepin KH, Qin X, Sutton GG, Venter E, Walenz BP, Wallis JW, Worley KC, Yang SP, Jones SM, Marra MA, Rocchi M, Schein JE, Baertsch R, Clarke L, Csuros M, Glasscock J, Harris RA, Havlak P, Jackson AR, Jiang H, Liu Y, Messina DN, Shen Y, Song HX, Wylie T, Zhang L, Birney E, Han K, Konkel MK, Lee J, Smit AF, Ullmer B, Wang H, Xing J, Burhans R, Cheng Z, Karro JE, Ma J, Raney B, She X, Cox MJ, Demuth JP, Dumas LJ, Han SG, Hopkins J, Karimpour-Fard A, Kim YH, Pollack JR, Vinar T, Addo-Quaye C, Degenhardt J, Denby A, Hubisz MJ, Indap A, Kosiol C, Lahn BT, Lawson HA, Marklein A, Nielsen R, Vallender EJ, Clark AG, Ferguson B, Hernandez RD, Hirani K, Kehrer-Sawatzki H, Kolb J, Patil S, Pu LL, Ren Y, Smith DG, Wheeler DA, Schenck I, Ball EV, Chen R, Cooper DN, Giardine B, Hsu F, Kent WJ, Lesk A, Nelson DL, O’Brien WE, Prufer K, Stenson PD, Wallace JC, Ke H, Liu XM, Wang P, Xiang AP, Yang F, Barber GP, Haussler D, Karolchik D, Kern AD, Kuhn RM, Smith KE, Zwieg AS. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–34. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 148.Di Rienzo A, Hudson RR. An evolutionary framework for common diseases: the ancestral-susceptibility model. Trends Genet. 2005;21:596–601. doi: 10.1016/j.tig.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 149.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lee AS, Gutierrez-Arcelus M, Perry GH, Vallender EJ, Johnson WE, Miller GM, Korbel JO, Lee C. Analysis of copy number variation in the rhesus macaque genome identifies candidate loci for evolutionary and human disease studies. Hum Mol Genet. 2008;17:1127–36. doi: 10.1093/hmg/ddn002. [DOI] [PubMed] [Google Scholar]
- 151.Graubert TA, Cahan P, Edwin D, Selzer RR, Richmond TA, Eis PS, Shannon WD, Li X, McLeod HL, Cheverud JM, Ley TJ. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 2007;3:e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Orozco LD, Cokus SJ, Ghazalpour A, Ingram-Drake L, Wang S, van Nas A, Che N, Araujo JA, Pellegrini M, Lusis AJ. Copy number variation influences gene expression and metabolic traits in mice. Hum Mol Genet. 2009;18:4118–29. doi: 10.1093/hmg/ddp360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Krutzfeldt J, Stoffel M. MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab. 2006;4:9–12. doi: 10.1016/j.cmet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 154.Tang X, Tang G, Ozcan S. Role of microRNAs in diabetes. Biochim Biophys Acta. 2008;1779:697–701. doi: 10.1016/j.bbagrm.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tang X, Muniappan L, Tang G, Ozcan S. Identification of glucose-regulated miRNAs from pancreatic {beta} cells reveals a role for miR-30d in insulin transcription. RNA. 2009;15:287–93. doi: 10.1261/rna.1211209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhao E, Keller MP, Rabaglia ME, Oler AT, Stapleton DS, Schueler KL, Neto EC, Moon JY, Wang P, Wang IM, Lum PY, Ivanovska I, Cleary M, Greenawalt D, Tsang J, Choi YJ, Kleinhanz R, Shang J, Zhou YP, Howard AD, Zhang BB, Kendziorski C, Thornberry NA, Yandell BS, Schadt EE, Attie AD. Obesity and genetics regulate microRNAs in islets, liver, and adipose of diabetic mice. Mamm Genome. 2009;20:476–85. doi: 10.1007/s00335-009-9217-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, Leonardson A, Castellini LW, Wang S, Champy MF, Zhang B, Emilsson V, Doss S, Ghazalpour A, Horvath S, Drake TA, Lusis AJ, Schadt EE. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–35. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.