Abstract
Background.
Genome-wide association studies (GWAS) may yield insights into longevity.
Methods.
We performed a meta-analysis of GWAS in Caucasians from four prospective cohort studies: the Age, Gene/Environment Susceptibility-Reykjavik Study, the Cardiovascular Health Study, the Framingham Heart Study, and the Rotterdam Study participating in the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium. Longevity was defined as survival to age 90 years or older (n = 1,836); the comparison group comprised cohort members who died between the ages of 55 and 80 years (n = 1,955). In a second discovery stage, additional genotyping was conducted in the Leiden Longevity Study cohort and the Danish 1905 cohort.
Results.
There were 273 single-nucleotide polymorphism (SNP) associations with p < .0001, but none reached the prespecified significance level of 5 × 10−8. Of the most significant SNPs, 24 were independent signals, and 16 of these SNPs were successfully genotyped in the second discovery stage, with one association for rs9664222, reaching 6.77 × 10−7 for the combined meta-analysis of CHARGE and the stage 2 cohorts. The SNP lies in a region near MINPP1 (chromosome 10), a well-conserved gene involved in regulation of cellular proliferation. The minor allele was associated with lower odds of survival past age 90 (odds ratio = 0.82). Associations of interest in a homologue of the longevity assurance gene (LASS3) and PAPPA2 were not strengthened in the second stage.
Conclusion.
Survival studies of larger size or more extreme or specific phenotypes may support or refine these initial findings.
Keywords: Longevity, Genome-wide association study, Meta-analysis
INCREASES in longevity of the general population worldwide are an unprecedented phenomenon with significant health and social impact. Although environmental factors have led to an increase in life span, there is ample evidence that genetic factors are involved in extreme longevity both in humans (1–7) and in other organisms (8). The protective genetic factors that lead to longevity are likely to involve fundamental processes of aging that may be different from those associated with early mortality or premature onset of age-related diseases in younger individuals. The mechanisms of aging in humans are far from understood, but available evidence suggests that several pathways—inflammation, oxidative stress and stress responses, cellular senescence, DNA damage and repair, and the growth hormone or insulin-like growth factor and insulin (GH, IGF, INS) axis—may play key roles (9–12). Model organisms suggest that inhibiting the GH, IGF, or INS axis, which is involved in regulating cell proliferation, cell death, wound repair, and metabolism, may promote longevity by reducing oxidative stress and slowing the rate of cell replication and the accumulation of somatic-cell DNA mutations (13). There is also evidence for other important pathways such as the heat-shock proteins and heat-shock factors that are highly conserved across species and play a role in prolongevity transcription pathways. Clinical and epidemiological investigations, including candidate gene studies, have suggested that inflammation pathways may affect life span and risk of age-related conditions such as cardiovascular disease (CVD) and its risk factors (14–19). A combination of multiple genetic variants may be required for an individual to achieve exceptional longevity, which may account in part for its rarity.
Two previous studies have used whole-genome screening to identify genetic variants associated with longevity (20,21). In a linkage analysis, the earliest report (20) identified a locus on chromosome 4 that has not been replicated. A recent report from the Framingham Heart Study (FHS) (22) identified modest associations between longevity (or age at death) and single-nucleotide polymorphisms (SNPs) in or near important candidate genes, including FOXO1A, GAPDH, KL, LEPR, PON1, PSEN1, SOD2, and WRN, but none of the associations achieved conventional levels of statistical significance; the sample size was modest, and the genotyping platform did not cover the genome well by current standards. The advent of genome-wide association studies (GWAS) has successfully led to the discovery of novel genetic variants that have strong evidence for replication and that are outside of traditional candidate gene regions for several common diseases (23–29). The detection of novel genetic variants associated with longevity holds the promise to provide important insights to biologic pathways in the aging process and thus the potential to develop innovative strategies to promote a long and healthy life.
We conducted a meta-analysis of GWAS findings for longevity within an international consortium of four longitudinal community-based cohort studies that followed adults over many years. Longevity was defined as survival to age 90 years or older, and a comparison group was drawn from each cohort. Furthermore, we identified two independent cohorts of long-lived individuals, the Leiden Longevity Study cohort and the Danish 1905 cohort, to evaluate initial findings for the strongest allelic associations for longevity in a second discovery stage.
METHODS
Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium
The Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium was convened to promote the discovery of new genomic loci involved in multiple complex traits in population-based follow-up studies using genome-wide association analysis (30). This meta-analysis used data from the CHARGE Consortium, which includes the Age, Gene/Environment Susceptibility-Reykjavik Study (AGES-Reykjavik) (31), the Cardiovascular Health Study (CHS) (32), FHS (33–36), and the Rotterdam Study (RS) (37).
The AGES-Reykjavik was funded by the National Institute on Aging (NIA) and was designed to examine genetic susceptibility and environmental interactions as risk factors for disease and disability in old age. Detailed phenotyping of the cardiovascular, neurocognitive, musculoskeletal, and body composition and metabolism was conducted in 5,764 men and women enrolled in 2002–2006 who were sampled from the 11,549 survivors of the AGES-Reykjavik of 30,000 men and women sampled from the 1907–1935 birth cohort (31). The CHS is a National Heart Lung and Blood Institute (NHLBI) contract-funded cohort study designed to evaluate risk factors for coronary heart disease (CHD) and stroke in older adults (32). Participants (n = 5,201) were recruited in 1989–1990, with an additional 687 minorities recruited in 1992–1993. The FHS is an NHLBI contract-funded cohort study initiated in 1948 to study determinants of CVD and other major illnesses. The original cohort comprised 5,209 men and women aged 28–62 years at enrollment who have undergone routine biennial examinations (33,34). In 1971, 5,124 offspring of the original cohort participants and offspring spouses, aged 5–70 years, were enrolled into the Framingham Offspring Study. Offspring participants have been examined approximately every 4–8 years (35,36). In the 1990s, DNA was obtained for genetic studies from surviving original cohort and offspring participants. The RS was planned and designed in the early 1990s as a longitudinal study investigating the incidence and progression of diseases in the elderly participants. From 1991 to 1995, all inhabitants of Ommoord, a district of Rotterdam in the Netherlands, who were aged 55 years or older, were invited to participate in this study (38). Of 10,275 eligible individuals, 7,983 agreed to participate (78%). The participants in the CHARGE studies are Caucasian by self-report. In each CHARGE study, population structure was assessed using principal components analysis, and outliers were removed. Any remaining within-study structure was adjusted for using appropriate methods (39). The details of each participating cohort study’s genotyping platform, imputation algorithm, and quality control procedures used by each study are summarized in Supplementary Table 1. Each study was approved by the respective Institutional Review Board, and all participants provided consent.
Longevity and Comparison Group Definitions
In the present study, achievement of longevity was defined as reaching age 90 years or older, regardless of whether the participants were still living or had since died. Genotyped participants from these studies who died between the ages of 55 and 80 years were used as the comparison group. The comparison group was limited to deceased participants to ensure that no one in the comparison group could subsequently achieve longevity. The minimum age at death was set to match the minimum age at enrollment in the RS to promote age comparability of the comparison group across the four cohorts. The maximal age at death in the comparison group was set arbitrarily at age 80 years to include the majority of deaths, to maximize the overlap between birth cohorts, and to exclude those persons who survived far beyond average life expectancy for their respective birth cohort, that is persons who nearly reached longevity. Because of the timing of recruitment, DNA collection, and death, there was only partial overlap of the birth cohorts included in the comparison groups and the group of persons achieving longevity. Only Caucasian participants were included. Across the four studies, there were 1,836 persons who achieved longevity (144 from AGES-Reykjavik, 557 from CHS, 362 from FHS, and 773 from the RS), and the comparison group had 1,955 participants (122, 544, 355, and 934 participants from the AGES-Reykjavik, CHS, FHS, and RS, respectively). To facilitate comparison of results across studies, we imputed to 2.5 million SNPs using the HapMap Centre d’Etude du Polymorphisme Humain European Ancestry–genotyped samples as a reference. The effective sample size for all but one of the top SNPs was more than 80% of the full sample size of 3,791, indicating that the SNPs that were not directly genotyped were imputed well in most studies.
Second Discovery Stage Genotyping
Among the top 24 independent regions with the strongest associations for longevity in the four-study meta-analysis (p < 10−4), we selected the 22 SNPs that had been tested in all four CHARGE cohorts in two additional Caucasian cohorts: the Leiden Longevity Study cohort and the Danish 1905 cohort. We excluded the two SNPs that could not be genotyped or imputed in all four CHARGE cohorts. Of the 22 SNPs selected for genotyping, 2 could not be genotyped and 4 did not pass quality control procedures; thus, 16 SNPs were analyzed in the second stage.
In the “Leiden Longevity Study” (7,40), a total of 950 long-lived proband siblings (mean age 94 years, range 89–104 years), 1,750 offspring (mean age 61 years, range 39–81 years), and 758 partners of offspring (mean age 60 years, range 36–79 years) were included. The additional genotyping of selected SNPs was undertaken in all 950 long-lived probands, and these were compared with the 744 partners of their offspring and an additional 680 blood bank donors (60% men, mean age 31 years, range 18–40 years). All long-lived individuals and the comparison groups were from the Leiden area in the Netherlands and of European ancestry.
Participants in the “Danish 1905 Cohort Survey” are from the Danish 1905 birth cohort ascertained in 1998 when they were aged 92–93 years (41). Of the 3,600 participants alive from that cohort, 2,262 participants enrolled in the study. Participants underwent a home-based interview on health and lifestyle parameters, physical and cognitive tests, and collection of biologic material. The current genetic study comprises a total of 1,644 participants from this survey, mean age 93 years (range 92–93 years), 28% men. A comparison group included 2,007 Caucasians who were twins (one twin per pair) collected from all over Denmark, with a mean age of 57 years (range 46–68 years), 45% men.
Second Discovery Stage Genotyping Methods
Genotyping of the selected SNPs was performed using an iPLEX genotyping assay developed for use with the MassARRAY platform (Sequenom, Inc., San Diego, CA) (42). The iPLEX genotyping assay is based on mass spectrometry and enables genotyping of 25–36 custom SNPs on a sample in a single reaction. For the purposes of quality control, the system first automatically calls the genotypes and then generates cluster plots for all SNPs that are inspected individually by experienced technicians who check whether the plots show clear separation of the genotype clusters. There were two SNPs that did not pass quality control and two SNPs where no heterozygotes could not be detected; thus, lack of Hardy–Weinberg equilibrium was the quality control. Negative controls were included in the genotyping procedure (8 per 384-well plate), and importantly, 4% of samples were genotyped twice to confirm reproducibility (reproducibility was ≥99.7%).
Statistical Analysis
Using logistic regression, each imputed and observed HapMap SNP was tested for association with the longevity outcome using an additive genetic model adjusting for sex. The mean dosage of one of the alleles (a value between 0 and 2) was the predictor for imputed SNPs. The CHS additionally adjusted for field study site in the regression model, and the FHS used generalized estimating equations to account for familial correlations. We used the ratio of observed to expected variance in the imputed SNP genotype counts as a quality control metric for imputed SNPs (43). This ratio, multiplied by the sample size, is an estimate of the effective sample size. In the imputation software MaCH, this ratio is called r2 as it is an estimate of the allelic correlation between the imputed genotypes and the true genotypes for the SNP. A total of 2,287,520 SNPs that had average minor allele frequency greater than 0.01 and were genotyped or imputed in all studies with variance ratio greater than 0.1 were meta-analyzed. The study-specific inflation factors (λGC) were computed using the set of chi-square statistics used for the meta-analysis for each study. The inflation factor is computed as the median of all chi-square statistics divided by the expected median of the statistics (approximately 0.456) for a chi-square distribution with 1 df. We calculated a meta-analysis odds ratio (OR) for each SNP using a fixed-effects model that combined logistic regression parameters and standard errors across the studies using inverse variance weights. The meta-analysis OR represents the increase in log-odds of surviving to age 90 years or older versus dying between ages 55 and 80 years for each additional copy of the minor allele of the SNP. SNP associations were considered to be significant on a genome-wide level at p < 5 × 10−8. The 16 SNPs in the second discovery phase effort were analyzed in the two study samples using an additive model. The results were added to the previous meta-analysis using a fixed-effects model as described earlier. Finally, using the top 24 results, we conducted a pathway analysis with the Database for Annotation, Visualization and Integrated Discovery (http://david.abcc.ncifcrf.gov/).
RESULTS
Table 1 provides the characteristics of the persons achieving longevity and the comparison group in each of the four CHARGE discovery cohorts at the time of DNA collection. In line with the design of the study, persons achieving longevity were 10–20 years older than participants in the comparison group at baseline and were more likely to be women. Between 45% and 83% of those achieving longevity were still alive at the time that longevity status was ascertained. Among those who had died, the distributions of causes of death differed between those achieving longevity and the comparison group. Whereas 6%–12% of those achieving longevity died of cancer, more than 30% of the comparison group had death attributed to cancer. The prevalence of diabetes and a history of ever smoking were higher in the comparison group than in persons achieving longevity. The baseline prevalence of other cardiovascular risk factor levels showed substantial overlap between the two groups.
Table 1.
Characteristics of Longevity Cases and Comparison Group at DNA Collection
| CHS |
Framingham Heart Study |
Rotterdam Study |
AGES-Reykjavik |
|||||
| Characteristic, M (SD) or % | Survival Age to >90 y, n = 557 | Comparison Group, n = 544 | Survival Age to >90 y, n = 362 | Comparison Group, n = 355 | Survival Age to >90 y, n = 773 | Comparison Group, n = 934 | Survival Age to >90 y, n = 144 | Comparison Group, n = 122 |
| Age at DNA draw, y | 79.6 (4.5) | 69.5 (3.0) | 87.3 (3.8) | 66.5 (6.9) | 83.7 (5.53) | 66.5 (5.37) | 88.0 (2.4) | 73.8 (3.2) |
| Women, % | 61 | 54 | 70 | 34 | 79 | 41 | 56 | 43 |
| Alive, % | 45 | 0 | 36 | 0 | 33 | 0 | 83 | 0 |
| Cause of death* | ||||||||
| CVD, % | 39 | 33 | 22 | 23 | 34 | 32 | 48 | 39 |
| Cancer, % | 10 | 40 | 9 | 45 | 6 | 39 | 12 | 38 |
| Other, % | 50 | 27 | 57 | 25 | 52 | 27 | 40 | 23 |
| Unknown, % | 0.3 | 0.2 | 12 | 6 | 7 | 2 | 0 | 0 |
| Body mass index, kg/m2 | 25.5 (3.9) | 26.6 (5.2) | 26.0 (4.1) | 28.0 (5.5) | 26.8 (3.81) | 26.3 (3.75) | 25.9 (4.0) | 27.4 (4.7) |
| Ever smoker, % | 40 | 70 | 54 | 81.0 | 29 | 43 | 49.3 | 80 |
| Hypertension, % | 57 | 53 | 68 | 75 | 40 | 40 | 83 | 80 |
| Diabetes, % | 8 | 20 | 8 | 22 | 6 | 8 | 8 | 11 |
| Total cholesterol, mg/dL | 210.5 (40.2) | 212.2 (38.7) | 198.8 (38.1) | (204.7 (47.1) | 248 (49.4) | 254 (46.8) | 207.6 (44.3) | 224.44 (42.7) |
Notes: In the CHS, ever smoking was defined as having smoked more than 100 cigarettes or five packs during the participant's lifetime; hypertension was defined as a systolic blood pressure 140 mmHg or more or a diastolic blood pressure 90 mmHg or more or a history of hypertension and taking antihypertensive medication; diabetes was defined as fasting glucose more than 125 mg/dL or use of insulin or oral hypoglycemic medications. In Framingham Heart Study, ever smoking was defined as self-reported cigarette smoking of at least 1 cigarette/d for a year at any attended examination; total serum cholesterol was measured using an automated enzymatic procedure (44); hypertension was defined as blood pressure 140/90 mmHg or more or on antihypertensive medication; diabetes was defined as fasting blood glucose more than 125 mg/dL, a random blood glucose of more than 200 mg/dL, or use of insulin or oral hypoglycemic agents. In the Rotterdam Study, ever smoking was defined as self-reported ever smoking (cigarette, cigar, or pipe); hypertension was defined as systolic blood pressure 160 mmHg or more and/or diastolic blood pressure 100 mmHg or more and/or blood pressure–lowering medication with an indication for hypertension; total serum cholesterol was measured using an automated enzymatic procedure (40); diabetes was defined as self-reported diabetes at baseline. In the AGES-Reykjavik, ever smoking was defined as having smoked more than 100 cigarettes in one’s lifetime; total serum cholesterol was measured using an automated enzymatic procedure (40); hypertension was defined as systolic blood pressure 140 mmHg or more, diastolic blood pressure 90 mmHg or more, use of antihypertensive medications, or self-report; diabetes was defined as fasting glucose more than 125 mg/dL, use of insulin or oral hypoglycemic medications, or self-report. AGES-Reykjavik = Age, Gene/Environment Susceptibility-Reykjavik Study; CHS = Cardiovascular Health Study; CVD = cardiovascular disease.
As a proportion of all deaths for those in the survival to age 90 years or older group.
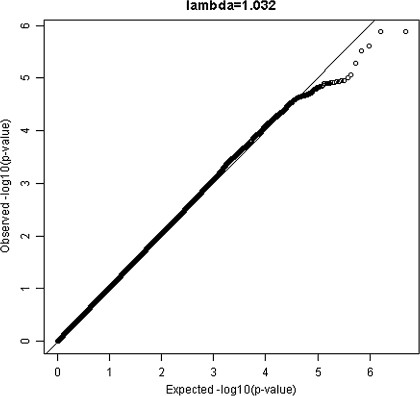
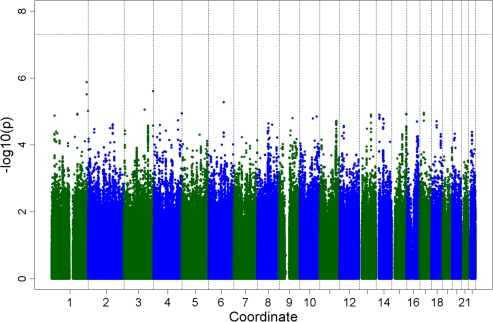
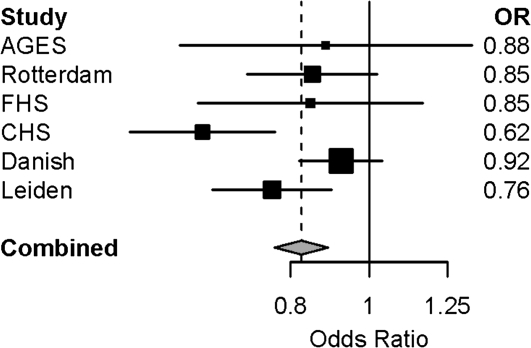
The genomic control inflation factor lambda (λGC) for each cohort was less than 1.05 (45). After meta-analysis, overall inflation of the meta-analysis p values was minor (λGC = 1.034; Figure 1). None of the SNP–longevity associations achieved the prespecified level of genome-wide significance of p < 5 × 10−8 (Figures 1 and 2). There were 273 SNP associations with meta-analysis p < 10−4, and of these, 7 SNP associations had p < 10−5 (Supplementary Table 2). Under the null hypothesis that there are no associations in the genome, we would expect 0.0001 × ∼2.3 million = ∼230 hits. Table 2 shows the top 24 independent SNPs associated with longevity along with the number of supporting SNPs (additional SNPs with linkage disequilibrium r2 > .80 and p < 10−4). Thus, for example, there were 19 supporting SNPs on chromosome 15 in or near the longevity assurance homologue 3 (LASS3) gene, with the strongest association (OR = 0.79, p = 1.2 × 10−5) noted for rs8029244. The study-specific ORs for the 24 SNP associations shown in Table 2 were in the same direction and were of similar magnitude across the four cohorts (Figure 3; Supplementary Table 3).
Figure 1.
Quantile–quantile plot for the 2,287,520 single-nucleotide polymorphisms in the meta-analysis of survival to age 90 years or older.
Figure 2.
Plot of genome-wide association study for longevity meta-analysis (persons surviving to age ≥90 years, n = 1,836, and comparison group, n = 1,955) showing the −log10 (p values) based on the fixed-effects meta-analysis by chromosome. Line indicates threshold for genome-wide significance of 5 × 10−8.
Table 2.
Top 24 SNP Associations Ranked by p Value for Meta-analysis of Four Cohorts for Survival to Age 90 Years or Older (n = 1,836) Compared With Survival to Age 55–80 Years (n = 1,955)* and Second Discovery Stage Meta-analysis Results
| Hit Number | Marker Name | Chromosome | Position | Closest Reference Gene | Distance (bp) From Closest Gene or Function | Alleles |
Study Effect Direction | Number of Supporting SNPs | Effective Sample Size (proportion of total) | Second Discovery Stage Meta-analysis–CHARGE Plus Leiden and Denmark Cohorts |
||||||
| Minor | Major | MAF | OR | p Value | Study Effect Direction Leiden/Denmark | OR | p Value | |||||||||
| 1 | rs4443878 | 1 | 238971041 | RGS7 | 34,398 | T | C | .04 | 0.41 | 1.3 × 10−6 | −−−− | 10 | 2,435 | ++ | 0.83 | .068 |
| 2 | rs9825185 | 3 | 196415923 | C3orf21 | Intron | C | A | .13 | 1.44 | 2.5 × 10−6 | ++++ | 24 | 0.64 | −− | 1.10 | .045 |
| 3 | rs954551 | 6 | 102886028 | GRIK2 | 262,554 | G | A | .25 | 0.77 | 5.3 × 10−6 | −−−− | 55 | 0.88 | † | — | — |
| 4 | rs7624691 | 3 | 138345769 | IL20RB | 133,159 | C | T | .43 | 0.80 | 8.8 × 10−6 | −−−− | 67 | 0.92 | ++ | 0.95 | .092 |
| 5 | rs10888267 | 1 | 246126046 | OR2W3 | Missense | C | T | .45 | 1.25 | 9.7 × 10−6 | ++++ | 2 | 0.92 | † | — | — |
| 6 | rs9972933 | 17 | 28589381 | ACCN1 | Intron | T | C | .23 | 0.77 | 1.1 × 10−5 | −−−− | 30 | 0.91 | +− | 0.89 | .003 |
| 7 | rs2739532 | 4 | 190944424 | C | G | .27 | 1.48 | 1.1 × 10−5 | ?+?+ | 9 | 0.99 | ‡ | — | — | ||
| 8 | rs8029244 | 15 | 98826098 | LASS3 | Intron | A | G | .49 | 0.79 | 1.2 × 10−5 | −−−− | 108 | 0.37 | +− | 0.90 | .002 |
| 9 | rs16850255 | 1 | 175085164 | PAPPA2 | 6,573 | C | T | .21 | 0.75 | 1.2 × 10−5 | −−−− | 81 | 0.86 | ++ | 0.92 | .041 |
| 10 | rs1543505 | 14 | 22432468 | REM2 | 5,739 | G | A | .28 | 1.27 | 1.3 × 10−5 | ++++ | 23 | 0.83 | −+ | 1.12 | .001 |
| 11 | rs7321904 | 13 | 80681190 | SPRY2 | 868,103 | T | C | .07 | 0.64 | 1.3 × 10−5 | −−−− | 40 | 0.93 | ++ | 0.92 | .179 |
| 12 | rs17401847 | 1 | 20111053 | OTUD3 | 971 | G | A | .15 | 1.38 | 1.4 × 10−5 | ++++ | 26 | 0.90 | −+ | 1.12 | .015 |
| 13 | rs3124736 | 10 | 115487102 | CASP7 | 6,450 | A | G | .03 | 2.30 | 1.4 × 10−5 | +++? | 0 | 0.89 | ‡ | — | — |
| 14 | rs690232 | 9 | 92422258 | DIRAS2 | Intron | A | G | .30 | 1.27 | 1.6 × 10−5 | +++− | 68 | 0.64 | † | — | — |
| 15 | rs9664222 | 10 | 89328613 | MINPP1 | 25,489 | A | C | .21 | 0.77 | 1.6 × 10−5 | −−−− | 55 | 0.93 | −− | 0.82 | 6.77 × 10−7 |
| 16 | rs11157721 | 14 | 49586464 | LOC196913 | 33,655 | T | C | .39 | 0.79 | 1.7 × 10−5 | −−−− | 53 | 0.86 | +− | 0.90 | .002 |
| 17 | rs4690810 | 4 | 166500130 | SC4MOL | 16,456 | C | T | .35 | 0.79 | 1.9 × 10−5 | −−−− | 81 | 0.85 | ++ | 0.93 | .044 |
| 18 | rs11605096 | 11 | 113047320 | TMPRSS5 | 16,162 | A | C | .12 | 0.71 | 1.9 × 10−5 | −−−− | 94 | 0.93 | † | — | — |
| 19 | rs16972414 | 18 | 35709920 | PIK3C3 | 2,079,276 | G | A | .30 | 0.79 | 2.0 × 10−5 | −−−− | 94 | 0.93 | † | — | — |
| 20 | rs12935091 | 16 | 70082709 | ZNF19 | 1,954 | G | A | .07 | 0.62 | 2.0 × 10−5 | −−−− | 46 | 0.94 | −+ | 0.80 | .002 |
| 21 | rs210332 | 14 | 53262222 | BMP4 | 223,982 | C | T | .19 | 1.33 | 2.3 × 10−5 | ++++ | 2 | 0.65 | † | — | — |
| 22 | rs17369174 | 8 | 76128602 | CRISPLD1 | 19,256 | G | T | .10 | 0.69 | 2.3 × 10−5 | −−−− | 83 | 0.79 | +− | 0.86 | .014 |
| 23 | rs6721003 | 2 | 166931758 | SCN7A | 38,026 | A | G | .45 | 1.23 | 2.4 × 10−5 | ++++ | 55 | 0.91 | −+ | 1.09 | .006 |
| 24 | rs4734457 | 8 | 101573533 | ANKRD46 | 28,644 | A | C | .10 | 1.75 | 2.5 × 10−5 | ++++ | 0 | 0.99 | +− | 1.10 | .098 |
Notes: p Values are for the inverse variance–weighted meta-analysis. Distances to genes are given in base pairs. Position is for NCBI Build 36. ORs are for each additional minor allele. Number of supporting SNPs: The number of SNPs within 500 kb of the top SNP that are in LD with the top SNP in the HapMap CEU release 22 (r2 ≥ .10) and have association p < .05. For imputed and direction, study-specific information is presented in the order: Age, Gene/Environment Susceptibility-Reykjavik Study, Rotterdam Study, FHS, CHS, Leiden, and Denmark. Direction: + = minor allele increases odds of survival more than 90; − = minor allele decreases odds of survival; ? = not tested. CEU = Centre d'Etude du Polymorphisme Humain European Ancestry; CHARGE = Cohorts for Heart and Aging Research in Genomic Epidemiology; CHS = Cardiovascular Health Study; FHS = Framingham Heart Study; LD = linkage disequilibrium; MAF = minor allele frequency; OR = odds ratio; SNPs = single-nucleotide polymorphisms.
For information on all SNP associations with p < 10−4, see Supplementary Table 2.
Genotyping requested, not completed.
Genotyping not requested.
Figure 3.
Study-specific odds ratios (ORs) and 95% confidence intervals for MINPP1 (rs9664222) longevity association.Note: AGES-Reykjavik = Age, Gene/Environment Susceptibility-Reykjavik Study; CHS = Cardiovascular Health Study; FHS = Framingham Heart Study.
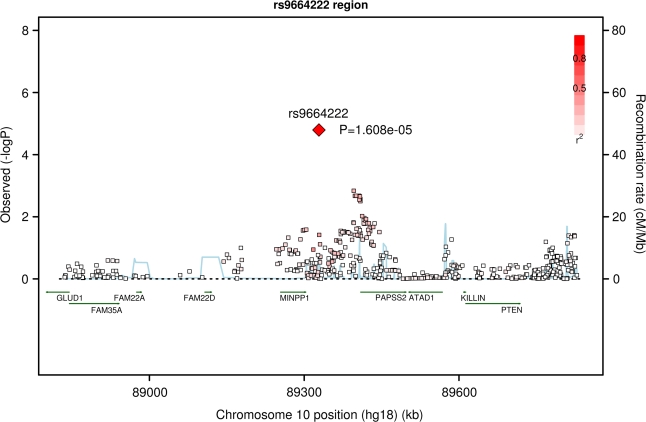
Of the 24 strongest independent regions shown in Table 2, the 22 SNPs tested in all four CHARGE cohorts were selected for further evaluation, and 16 were successfully genotyped in the second stage cohorts. Only 1 of the 16 SNPs had a smaller p value after including the replication studies in a joint meta-analysis, with the p value decreasing about 10-fold, from 1.61 × 10−5 to 6.77 × 10−7 and corresponding OR of 0.82. This SNP, rs9664222, is ∼25 kb from the MINPP1 gene (Figure 4). In the CHARGE analysis, the minor allele was associated with a lower odds of survival past age 90 (OR = 0.77). The Leiden study yielded a similar effect estimate (OR = 0.76, p = .0014), whereas the Danish study showed a nonsignificant trend in the same direction (OR = 0.92, p = .19). Findings for the other SNPs were inconsistent in direction of association such that the meta-analysis p values increased with inclusion of the second stage cohorts (Supplementary Table 3). Pathway analysis did not reveal significant findings in the top associations, though some groupings were biologically plausible.
Figure 4.
Regional plot for rs9664222 near MINPP1.
DISCUSSION
The CHARGE Consortium collaboration allowed us to conduct a meta-analysis of GWAS for longevity in a sample of long-lived individuals and a corresponding comparison group derived from the same longitudinal community-based cohort studies. Although none of the SNP associations for longevity in the first discovery phase achieved prespecified level of genome-wide significance, a polymorphism associated with the MINPP1 genes was among the strongest associations observed in our sample, with effect sizes that were similar within the four cohorts. The finding related to the MINPP1 gene was strengthened after including two additional cohorts in a second discovery phase but did not reach genome-wide significance. Among the top 10 associations in the initial meta-analysis, additional SNP associations of potential interest in longevity include SNPs in or near LASS3, ACCN1, IL20RB, and PAPPA2. These SNPs are near genes that have not previously been reported to be associated with longevity in human populations but are interesting because these genes are conserved in basic biologic pathways.
The MINPP1 gene codes multiple inositol polyphosphate phosphatases, which are compartmentalized to the endoplasmic reticulum lumen. MINPP1-deficient mice have no obvious defects, though targeted deletion in vitro is associated with slowed cellular proliferation (46). There is no evidence that this SNP is functional; furthermore, its distance from the gene shows that it is not in strong linkage disequilibrium with SNPs in MINPP1 (47). However, it is well known that important regulatory elements are found outside of genes. This SNP is within 50 kb of two copy number variants. The finding of an SNP near a gene regulating proliferation is intriguing because of the higher rate of cancer death in the comparison group.
The initial finding in the LASS3 gene region was of interest because of the historical association of its homologue with longevity in yeast (46). The LASS gene family contains a group of highly conserved genes that are found in all eukaryotic species. LASS isoforms are mammalian homologues of the yeast longevity assurance gene 1, which encodes a protein that regulates life span (48). The strongest association was noted for rs8029244; this SNP is in the intronic enhancer region of the LASS3 gene. LASS3 is a member of the ceramide synthase family, which is important in sphingolipid metabolism, cell differentiation, cell cycling, and apoptosis (46). LASS3 may be involved in sphingolipid synthesis or its regulation (49).
IL20RB, interleukin 20 receptor beta IL-20, plays a role in skin inflammation and the development of hematopoietic cells (50) and is of interest because of the strong associations of inflammation with the aging process (51). IL-20 is a pleiotropic cytokine with potent inflammatory, angiogenic, and chemoattractive characteristics and is involved in inflammatory diseases, such as psoriasis, atherosclerosis, and rheumatoid arthritis (50). The ACCN1 gene encodes amiloride-sensitive sodium channels with two hydrophobic transmembrane regions and a large extracellular loop, which has many cysteine residues with conserved spacing (52,53). The member encoded by this gene may play a role in neurotransmission. ACCN1 was found to be associated with multiple sclerosis (54). Pregnancy-associated plasma protein A2 (PAPPA2) is a metalloproteinase regulating local insulin-like growth factor pathway action (55). Genetic deletion extends life span in the mouse by 30%–40% (56) and is characterized by delay in thymic involution (57) and low rates of tumor incidence (56). Although the associations reported here did not reach the a priori specified level of significance, the findings are important to report so that they can be replicated in studies without whole-genome genotyping and compared with future studies, such as in centenarian studies and family studies of longevity. Effect size estimates noted here support the likelihood that longevity is a complex process, in that there were no variants with large effects, supporting the hypothesis that there may be many genes with small effects that contribute to longevity.
The strengths of this study include the community-based prospective design and the long-term follow-up of these cohorts. In all cases, vital status was confirmed using death certificates and hospital records. Another strength was our ability to use controls that were equally well characterized and were drawn from within the same cohorts. The number of long-lived individuals reported here is very large relative to other studies in the literature, allowing greater ability to identify SNPs with small effects. The cohorts were relatively homogeneous with respect to ancestry, limited to Caucasians of European decent. Our top associations were homogeneous across cohorts. Screening for latent population substructure also supported ethnic homogeneity. Thus, the findings reported are less likely to be due to population stratification.
There are important aspects of the study that need to be kept in mind when interpreting the results. The differences in causes of death in the longevous individuals versus the comparison groups are expected as death from cancer tends to occur earlier in life than death from heart disease or dementia. Many of the long-lived people are still alive and we do not yet know what their ultimate cause of death will be, but it is likely that cancer will be underrepresented among persons achieving longevity. Power remains a limitation. Thus, future GWAS aiming to identify variants for this phenotype will have to consider small effect sizes and target a sample size larger than our nearly 2,000 long-lived persons. DNA collection in cohort studies is a recent enough phenomenon that relatively few cohort members who had DNA collected have had the opportunity to survive to age 90 years. Continuous study of these and other similarly designed cohorts will allow us to extend this study to larger numbers and to older ages.
In our case comparison analysis, we attempted to account for birth cohort, but the overlap between birth year of the comparison group and of the long-lived participants was limited. Further follow-up of these cohorts is needed to increase our ability to examine potential birth cohort effects. The study design of the cohorts examined in the second stage was different from the initial four-study CHARGE meta-analysis in that the comparison groups were derived from younger participants, living and deceased, who were not from the same cohort as the individuals achieving longevity. Certainly, there are important environmental factors that would be necessary for the fulfillment of the genetic potential for longevity. Heterogeneity in environmental exposures and gene–environment interactions require further study. Finally, these results cannot be extended to populations of other ancestry.
In conclusion, this meta-analysis of GWAS data for longevity from four large cohorts and two additional cohorts has implicated several genes involved in conserved basic mechanisms of cellular function. Analysis of more extreme survival phenotypes such as centenarians, additional follow-up to increase sample size in these cohorts for this phenotype, or evaluation of more specific phenotypes such as disease-free survival may support and refine these initial findings.
FUNDING
The CHS research reported in this article was supported by contract numbers N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, and N01-HC-45133 and grant numbers U01 HL080295 and R01 HL087652 from the NHLBI and R01 AG023629 from the NIA, with additional contribution from the National Institute of Neurological Disorders and Stroke. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm. DNA handling and genotyping was supported in part by National Center for Research Resources grant M01-RR00425 to the Cedars-Sinai General Clinical Research Center Genotyping core and National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The FHS phenotype–genotype analyses were supported by the NIA grant numbers R01AG029451, R01AG028321, R01AG033193, and R01 AG031287. The FHS of the NHLBI of the National Institutes of Health (NIH) and Boston University School of Medicine were supported by the NHLBI's FHS contract number N01-HC-25195 and its contract with Affymetrix, Inc. for genotyping services (contract number N02-HL-6-4278). Analyses reflect intellectual input and resource development from the FHS investigators participating in the SNP Health Association Resource project. A portion of this research was conducted using the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. D.P.K.’s effort was supported by a grant from the National Institute of Arthritis, Musculoskeletal and Skin Diseases and the NIA (R01 AR/AG 41398). The RS is supported by the Erasmus Medical Center and Erasmus University Rotterdam; the Netherlands Organisation for Scientific Research (NWO); the Netherlands Organisation for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly; the Ministry of Education, Culture and Science; the Ministry of Health, Welfare and Sports; and the European Commission (DG XII). The genetic analyses were supported by the Netherlands Organization for Scientific Research grant number 175.01.2005.011. This study was supported by the Netherlands Genomics Initiative-Sponsored Netherlands Consortium for Healthy Aging (nr 050-060-810). The work of S.W. was supported by Netspar. The AGES-Reykjavik is funded by NIH contract number N01-AG-12100, the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament). Genotyping was conducted at the NIA Intramural Research Program Laboratory of Neurogenetics. The Leiden Longevity Study was supported by the Innovation Oriented research Program on Genomics, the Centre for Medical Systems Biology, and the Netherlands Consortium for Healthy Aging in the framework of the Netherlands Genomics Initiative, the Netherlands Organization for Scientific Research, the Netherlands Organization for Health Research and Development (ZonMw), the Danish National Programme for Research Infrastructure 2007 from the Danish Agency for Science Technology and Innovation, and Unilever PLC, UK. The Longevity Consortium, funded by the NIA, grant number U19 AG023122, provided administrative resources to CHARGE investigators for this phenotype as well as scientific opportunity funds to conduct follow-up genotyping. The Danish 1905 Cohort Survey was supported by grants from the Danish National Research Foundation and NIH/NIA grant P01 AG08761. The Danish Aging Research Center is supported by a grant from the Velux Foundation. The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by NHLBI contract numbers N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022, R01HL087641, R01HL59367, and R01HL086694; National Human Genome Research Institute contract number U01HG004402; and NIH contract number HHSN268200625226C. Infrastructure was partly supported by grant number UL1RR025005, a component of the NIH and NIH Roadmap for Medical Research.
CONFLICT OF INTEREST
The content is solely the responsibility of the authors and does not necessarily represent the views of the NIA, NHLBI, the National Institute of Neurological Disorders and Stroke, or NIH.
SUPPLEMENTARY MATERIAL
Supplementary material can be found at: http://biomed.gerontologyjournals.org/
Acknowledgments
We are indebted to the participants and staff in the AGES-Reykjavik, the CHS, the FHS, and the RS for their important contributions. We acknowledge the NHLBI, which has made the Framingham SNP Health Association Resource project possible.
References
- 1.Perls T, Shea-Drinkwater M, Bowen-Flynn J, et al. Exceptional familial clustering for extreme longevity in humans. J Am Geriatr Soc. 2000;48(11):1483–1485. [PubMed] [Google Scholar]
- 2.Perls T, Kohler IV, Andersen S, et al. Survival of parents and siblings of supercentenarians. J Gerontol A Biol Sci Med Sci. 2007;62(9):1028–1034. doi: 10.1093/gerona/62.9.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perls T, Terry D. Genetics of exceptional longevity. Exp Gerontol. 2003;38(7):725–730. doi: 10.1016/s0531-5565(03)00098-6. [DOI] [PubMed] [Google Scholar]
- 4.Hjelmborg JV, Iachine I, Skytthe A, et al. Genetic influence on human lifespan and longevity. Hum Genet. 2006;119(3):312–321. doi: 10.1007/s00439-006-0144-y. [DOI] [PubMed] [Google Scholar]
- 5.Herskind AM, McGue M, Holm NV, Sorensen TI, Harvald B, Vaupel JW. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870-1900. Hum Genet. 1996;97(3):319–323. doi: 10.1007/BF02185763. [DOI] [PubMed] [Google Scholar]
- 6.Iachine IA, Holm NV, Harris JR, et al. How heritable is individual susceptibility to death? The results of an analysis of survival data on Danish, Swedish and Finnish twins. Twin Res. 1998;1(4):196–205. doi: 10.1375/136905298320566168. [DOI] [PubMed] [Google Scholar]
- 7.Schoenmaker M, de Craen AJM, de Meijer PHEM, et al. Evidence of genetic enrichment for exceptional survival using a family approach: the Leiden Longevity Study. Eur J Hum Genet. 2005;14(1):79–84. doi: 10.1038/sj.ejhg.5201508. [DOI] [PubMed] [Google Scholar]
- 8.Butler RN, Austad SN, Barzilai N, et al. Longevity genes: from primitive organisms to humans. J Gerontol A Biol Sci Med Sci. 2003;58(7):581–584. doi: 10.1093/gerona/58.7.b581. [DOI] [PubMed] [Google Scholar]
- 9.Browner WS, Kahn AJ, Ziv E, et al. The genetics of human longevity. Am J Med. 2004;117(11):851–860. doi: 10.1016/j.amjmed.2004.06.033. [DOI] [PubMed] [Google Scholar]
- 10.Franceschi C, Olivieri F, Marchegiani F, et al. Genes involved in immune response/inflammation, IGF1/insulin pathway and response to oxidative stress play a major role in the genetics of human longevity: the lesson of centenarians. Mech Ageing Dev. 2005;126(2):351–361. doi: 10.1016/j.mad.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 11.van Heemst D, Beekman M, Mooijaart SP, et al. Reduced insulin/IGF-1 signalling and human longevity. Aging Cell. 2005;4(2):79–85. doi: 10.1111/j.1474-9728.2005.00148.x. [DOI] [PubMed] [Google Scholar]
- 12.Holzenberger M, Dupont J, Ducos B, et al. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421(6919):182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- 13.Matsuoka S, Ballif BA, Smogorzewska A, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316(5828):1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 14.Reiner AP, Diehr P, Browner WS, et al. Common promoter polymorphisms of inflammation and thrombosis genes and longevity in older adults: the Cardiovascular Health Study. Atherosclerosis. 2005;181(1):175–183. doi: 10.1016/j.atherosclerosis.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 15.Reiner AP, Carlson CS, Jenny NS, et al. USF1 gene variants, cardiovascular risk, and mortality in European-Americans. Analysis of two U.S. cohort studies. Arterioscler Thromb Vasc Biol. 2007;27:2736–2742. doi: 10.1161/ATVBAHA.107.154559. [DOI] [PubMed] [Google Scholar]
- 16.Walston JD, Fallin MD, Cushman M, et al. IL-6 gene variation is associated with IL-6 and C-reactive protein levels but not cardiovascular outcomes in the Cardiovascular Health Study. Hum Genet. 2007;122:485–494. doi: 10.1007/s00439-007-0428-x. [DOI] [PubMed] [Google Scholar]
- 17.Arking DE, Atzmon G, Arking A, Barzilai N, Dietz HC. Association between a functional variant of the KLOTHO gene and high-density lipoprotein cholesterol, blood pressure, stroke, and longevity. Circ Res. 2005;96(4):412–418. doi: 10.1161/01.RES.0000157171.04054.30. [DOI] [PubMed] [Google Scholar]
- 18.Atzmon G, Rincon M, Schechter CB, et al. Lipoprotein genotype and conserved pathway for exceptional longevity in humans. PLoS Biol. 2006;4(4):e113. doi: 10.1371/journal.pbio.0040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barzilai N, Atzmon G, Schechter C, et al. Unique lipoprotein phenotype and genotype associated with exceptional longevity. JAMA. 2003;290(15):2030–2040. doi: 10.1001/jama.290.15.2030. [DOI] [PubMed] [Google Scholar]
- 20.Puca AA, Daly MJ, Brewster SJ, et al. A genome-wide scan for linkage to human exceptional longevity identifies a locus on chromosome 4. Proc Natl Acad Sci U S A. 2001;98(18):10505–10508. doi: 10.1073/pnas.181337598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reed T, Dick DM, Uniacke SK, Foroud T, Nichols WC. Genome-wide scan for a healthy aging phenotype provides support for a locus near D4S1564 promoting healthy aging. J Gerontol A Biol Sci Med Sci. 2004;59(3):227–232. doi: 10.1093/gerona/59.3.b227. [DOI] [PubMed] [Google Scholar]
- 22.Lunetta KL, D’Agostino RB, Sr, Karasik D, et al. Genetic correlates of longevity and selected age-related phenotypes: a genome-wide association study in the Framingham Study. BMC Med Genet. 2007;8(suppl 1):S13. doi: 10.1186/1471-2350-8-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39(5):631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 26.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39(5):645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 27.Thomas G, Jacobs KB, Yeager M, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40(3):310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 28.Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 29.Helgadottir A, Thorleifsson G, Manolescu A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 30.Psaty BM, O'Donnell CJ, Gudnason V, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium: design of prospective meta-analyses of genome-wide association studies from five cohorts. Circ: Cardiovasc Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris TB, Launer LJ, Eiriksdottir G, et al. Age, Gene/Environment Susceptibility-Reykjavik Study: multidisciplinary applied phenomics. Am J Epidemiol. 2007;165(9):1076–1087. doi: 10.1093/aje/kwk115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fried LP, Borhani NO, Enright P, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1(3):263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 33.Dawber TR, Meadors GF, Moore FE., Jr. Epidemiological approaches to heart disease: the Framingham Study. Am J Public Health. 1951;41(3):279–281. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dawber TR, Kannel W, Lyell L. An approach to longitudinal studies in a community: the Framingham Study. Ann N Y Acad Sci. 1963;107:539–556. doi: 10.1111/j.1749-6632.1963.tb13299.x. [DOI] [PubMed] [Google Scholar]
- 35.Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The Framingham Offspring Study. Design and preliminary data. Prev Med. 1975;4(4):518–525. doi: 10.1016/0091-7435(75)90037-7. [DOI] [PubMed] [Google Scholar]
- 36.Kannel W, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families. The Framingham offspring study. Am J Epidemiol. 1979;110(3):281–290. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 37.Hofman A, Breteler MM, van Duijn CM, et al. The Rotterdam Study: objectives and design update. Eur J Epidemiol. 2007;22(11):819–829. doi: 10.1007/s10654-007-9199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hofman A, Grobbee DE, de Jong PT, van den Ouweland FA. Determinants of disease and disability in the elderly: the Rotterdam Elderly Study. Eur J Epidemiol. 1991;7(4):403–422. doi: 10.1007/BF00145007. [DOI] [PubMed] [Google Scholar]
- 39.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 40.van Gent CM, van der Voort HA, de Bruyn AM, Klein F. Cholesterol determinations. A comparative study of methods with special reference to enzymatic procedures. Clin Chim Acta. 1977;75(2):243–251. doi: 10.1016/0009-8981(77)90195-4. [DOI] [PubMed] [Google Scholar]
- 41.Westendorp RGJ, van Heemst D, Rozing M, et al. Nonagenarian siblings and their offspring display lower risk of mortality and morbidity than sporadic Nonagenarians: the Leiden Longevity Study. J Am Geriatr Soc. 2009;57:1634–1637. doi: 10.1111/j.1532-5415.2009.02381.x. [DOI] [PubMed] [Google Scholar]
- 42.Nybo H, Petersen HC, Gaist D, et al. Predictors of mortality in 2,249 nonagenarians—the Danish 1905-Cohort Survey. J Am Geriatr Soc. 2003;51(10):1365–1373. doi: 10.1046/j.1532-5415.2003.51453.x. [DOI] [PubMed] [Google Scholar]
- 43.Oeth P, Beaulieu M, Park C, et al. iPLEX assay: increased plexing efficiency and flexibility for Mass ARRAY system through single base primer extension with mass-modified terminators. www.sequenom.com. http://jmgroup.pl/kawaska/download/iPLEX%20Application%20note.pdf. Accessed November 10, 2006. Application note: 1–12. [Google Scholar]
- 44.de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Devlin B, Roeder K. Genomic control for association studies. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 46.Chi H, Yang X, Kingsley PD, et al. Targeted deletion of Minpp1 provides new insight into the activity of multiple inositol polyphosphate phosphatase in vivo. Mol Cell Biol. 2000;20(17):6496–6507. doi: 10.1128/mcb.20.17.6496-6507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ioannidis JPA, Thomas G, Daly MJ. Validating, augmenting and refining genome-wide association signals. Nat Rev Genet. 2009;10(5):318–329. doi: 10.1038/nrg2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teufel A, Maass T, Galle PR, Malik N. The longevity assurance homologue of yeast lag1 (Lass) gene family (review) Int J Mol Med. 2009;23(2):135–140. [PubMed] [Google Scholar]
- 49.Mizutani Y, Kihara A, Igarashi Y. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem J. 2005;390(Pt 1):263–271. doi: 10.1042/BJ20050291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei CC, Hsu YH, Li HH, et al. IL-20: biological functions and clinical implications. J Biomed Sci. 2006;13(5):601–612. doi: 10.1007/s11373-006-9087-5. [DOI] [PubMed] [Google Scholar]
- 51.van den Biggelaar AHJ, Huizinga TWJ, de Craen AJM, et al. Impaired innate immunity predicts frailty in old age. The Leiden 85-plus study. Exper Gerontol. 2004;39(9):1407–1414. doi: 10.1016/j.exger.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 52.Saugstad JA, Roberts JA, Dong J, Zeitouni S, Evans RJ. Analysis of the membrane topology of the acid-sensing ion channel 2a. J Biol Chem. 2004;279(53):55514–55519. doi: 10.1074/jbc.M411849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waldmann R, Champigny G, Voilley N, Lauritzen I, Lazdunski M. The mammalian degenerin MDEG, an amiloride-sensitive cation channel activated by mutations causing neurodegeneration in Caenorhabditis elegans. J Biol Chem. 1996;271(18):10433–10436. doi: 10.1074/jbc.271.18.10433. [DOI] [PubMed] [Google Scholar]
- 54.Bernardinelli L, Murgia SB, Bitti PP, et al. Association between the ACCN1 gene and multiple sclerosis in Central East Sardinia. PLoS One. 2007;2(5):e480. doi: 10.1371/journal.pone.0000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Page NM, Butlin DJ, Lomthaisong K, Lowry PJ. The characterization of pregnancy associated plasma protein-E and the identification of an alternative splice variant 3. Placenta. 2001;22(8–9):681–687. doi: 10.1053/plac.2001.0709. [DOI] [PubMed] [Google Scholar]
- 56.Conover CA, Bale LK. Loss of pregnancy-associated plasma protein A extends lifespan in mice 21. Aging Cell. 2007;6(5):727–729. doi: 10.1111/j.1474-9726.2007.00328.x. [DOI] [PubMed] [Google Scholar]
- 57.Vallejo AN, Michel JJ, Bale LK, Lemster BH, Borghesi L, Conover CA. Resistance to age-dependent thymic atrophy in long-lived mice that are deficient in pregnancy-associated plasma protein A. Proc Natl Acad Sci. 2009;106(27):11252–11257. doi: 10.1073/pnas.0807025106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.