Abstract
The Göttingen minipig is an excellent model for studying effects of dietary high-fat intake on obesity. In this study, we analyzed the expression level of microRNA-122 (miRNA-122) and its target mRNA, CAT1, in intact young male minipigs fed either high-cholesterol or standard diet for 11 wk. MiRNA-122 and CAT1 are known to be important regulators of lipid metabolism. The weight of the young minipigs was monitored once a week during the feeding period; measurements of total cholesterol, triglycerides, high-density lipoproteins, and low-density lipoproteins were recorded at 4 time points (8, 14, 16, and 19 wk of age) in fasting animals during the feeding scheme. Body weight, total cholesterol, and high-density lipoproteins were higher in pigs fed the high-cholesterol compared with the standard diet. In contrast, the level of triglycerides was lower in pigs on the high-cholesterol diet than those receiving the standard diet. Pigs fed high-cholesterol also had lower miRNA-122 levels than did those fed the standard diet. These results suggest that in our minipigs, the increase in weight and cholesterol levels resulting from subchronic (11 wk) feeding of a high-cholesterol diet is correlated with a decrease in the expression of miRNA-122, confirming the implication of this microRNA in obesity. Gene expression levels of CAT1 did not differ between groups.
Abbreviations: CAT1, cationic amino acid transporter 1; HCD, high-cholesterol diet; miRNA, microRNA; MSD, minipig standard diet; qPCR, qualitative PCR
Obesity has reached epidemic proportions globally and has become a serious health problem that causes many severe diseases. Obesity and overweight pose major risks for serious diet-related chronic diseases, including type 2 diabetes, cardiovascular diseases, and stroke. Obesity is a complex disease that can be caused by both genetic and environmental factors. Although genes are important in determining a person's susceptibility to weight gain, energy balance is determined by calorie intake and physical activity. Many genes have been associated with obesity-related traits,22 and lately it has been revealed that microRNAs (miRNAs) also may play a role in obesity. MicroRNAs are small noncoding RNAs that regulate gene expression at posttranscriptional level. Some miRNAs have been associated with lipid metabolism:36 miRNA-33525 miRNA-143,8 miRNA-122,7 and miRNA-2723 among others. Moreover, lipid accumulation in adipocytes leads to obesity and therefore defects in lipid metabolism are correlated with pathologic development of obesity. However, the study of miRNA is still in its infancy in the obesity field, and high priority is given to establishing a useful animal model.
The Göttingen minipig breed is an excellent model for studying the effect of dietary high-fat intake on obesity, glucose homeostasis, and susceptibility to diabetes. Göttingen minipigs fed a high-fat diet increase in body weight, fat content, and cholesterol levels compared with their counterparts that receive a standard diet.2,15,20,31,33 In particular, miRNA-122 is a 22-bp highly evolutionarily conserved mature miRNA10 that derives from a liver-specific noncoding RNA precursor transcribed from the hrc gene.1 It is one of the most studied miRNAs, and miRNA-122 initially was discovered during a cloning study of tissue-specific small RNAs in mice,19 in which miRNA-122 accounted for 72% of all cloned miRNAs in liver. Studies in mice have shown that the level of miRNA-122 increases from 12.5 d after gestation and reaches a plateau just before birth, followed by a slow increase to as much as 70% of the total population of miRNAs in adult liver.1 In pigs, miRNA-122 was identified recently through sequencing of a small-RNA library from liver. 30
Several important functions assigned to miRNA-122 include acting as an enhancer of replication of hepatitis C viral RNA during infection,16,17,27 as a tumor suppressor regulating hepatocarcinogenesis,34 and as a biomarker for early signs of drug-induced liver injury.35 However, its main role is related to lipid metabolism and liver homeostasis. Inhibition of miRNA-122 in obese mice resulted in a remarkable reduction of cholesterol levels in plasma and a decrease in hepatic fatty-acid metabolism, making miRNA-122 an attractive therapeutic target for metabolic and cardiovascular diseases.7,10
Various miRNA-122 knockdown studies using different antisense oligonucleotide approaches revealed derepression of several liver mRNAs, suggesting that miRNA-122 regulates the expression of many mRNAs in liver, including glycogen synthase 1, cationic amino acid transporter 1 (CAT1) and aldolase A.6,7,9,18,21 Most of the identified targets showed only moderate derepression, implying that miRNA-122 might fine-tune several gene-regulatory networks in liver.6 Moreover, miRNA-122 has been shown to interact directly with CAT1 through posttranscriptional regulation of the CAT1 transcript at the level of mRNA degradation.1,17 Six target sites for the miRNA-122 have been predicted in the 3′ untranslated region and 2 sites in the 5′ untranslated region of the human CAT1 transcript. Interestingly, during mouse liver development, CAT1 mRNA decreases in almost inverse correlation with miRNA-122, a pattern that supports the prediction that CAT1 is a direct target of miRNA-122.1 CAT1 is the first cloned amino-acid transporter, and its important metabolic functions make this factor essential for cell survival under stress conditions. This requirement has been confirmed in CAT1 knockout mice, which develop severe anemia and die within 12 h after birth.28 The CAT1 gene is expressed ubiquitously in adult tissues, with low levels in liver; however, it is highly expressed in developmental liver.24 Together these results suggested CAT1 as the most likely miRNA-122 target for us to investigate.
In the present study, the expression levels of miRNA-122 and its target CAT1 were correlated with biochemical and physical measurements of risk factors for obesity in Göttingen minipigs.
Materials and Methods
Biological material.
Twelve intact barrier-bred young male Göttingen minipigs (Sus scrofa; Ellegaard Göttingen Minipigs, Dalmose, Denmark; for a health status description, see www.minipigs.dk) were used in the present study. The experimental feeding started when pigs were 8 wk old and ended at 19 wk of age. The 12 minipigs were assigned randomly to receive a standard diet (MSD group; Minipig Maintenance, Altromin 9020, Christian Petersen, Gentofte, Denmark; http://www.altromin.de/) or a high-cholesterol diet (HCD group; Porcine Western Diet 824100, Special Diets Services, Wilham, UK). The high-cholesterol diet was a modified minipig chow diet with 2% cholesterol, 22.77% crude fat (monounsaturated fatty acids 0.55% and polyunsaturated fatty acids 6.86%), and 3.5% fiber. The metabolizable energy of the standard chow was 10.5 MJ/Kg and that of the high-cholesterol diet was 19.3 MJ/Kg. All the animals were fed restricted rations of 1744 KJ × (body weight)0.52 twice daily to maintain both groups on equal weight curves according to their age; water was available ad libitum. The minipigs were housed under controlled conditions (temperature, 18 to 22 °C; relative humidity, 30% to 70%) and were allowed to acclimate at the research facilities for 1 wk before starting the experiment. The 19-wk-old minipigs were fasted 12 h prior to anesthesia and euthanasia.
The minipigs used in this study were treated in accordance to the Animal Experimentation Act of Denmark, which is in accordance with the Council of Europe Convention 123. The study was licensed by the National Animal Experimentation Board according to Danish law.
Body weight and biochemical measurements.
Body weight was measured weekly (Figure 1), and venous blood samples were obtained from fasted animals at ages 8, 14, 16, and 19 wk. Serum levels of total cholesterol, triglycerides, high-density lipoproteins (HDL), and low-density lipoproteins (LDL) were determined. Samples were centrifuged (1800 × g, 10 min), and serum stored at –80 °C until analysis. Total cholesterol, triglycerides, HDL and LDL were measured on an automated analyzer (Pentra 400, Horiba ABX, Les Ulis, France).
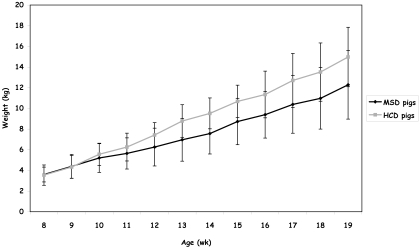
Figure 1.
Age (wk) and weight (kg) of the mininipigs used in the feeding study. Body weight was measured weekly in MSD pigs (n = 5) and HCD pigs (n = 7). Data are the mean ± SD for each group of pigs at each time point. Slope: MSD group, 0.76; HCD group, 1.03 (P = 0.0498, Student t test). Two-sided ANOVA revealed a significant (P = 0.00014) difference in body weight between groups. A Tukey test showed that there was no difference in mean body weight between the groups until week 11. From week 12, HCD minipigs weighed more (P < 0.05) than MSD animals.
Tissue collection and RNA extraction.
Liver samples were collected immediately after euthanasia. Tissue samples were cut in small pieces, snap-frozen in liquid nitrogen, and stored at –80 °C until use. Total RNA was extracted in duplicate for each animal (2 biologic replicates per animal) by using 50 to 100 mg tissue and TRI Reagent (Molecular Research Center, Cincinnati, OH) according to the manufacturer's recommendations. RNA yield was quantified spectrophotometrically (NanoDrop 1000, ThermoScientific, Wilmington, DE). RNA quality was evaluated by visual inspection of the rRNA 28S and 18S bands on an agarose gel and by measuring the ‘RNA integrity number’ (model 2100 Bioanalyzer with the Eukaryote Total RNA NanoChip, Agilent Technologies, Waldbronn, Germany). Average RNA integrity numbers (maximum, 10) were 8.56 for the MSD group and 8.65 for the HCD group.
cDNA synthesis.
For detection of miRNA-122 transcripts, cDNA was synthesized by using a commercial method (Exiqon, Vedbaek, Denmark). Shortly, 100 ng total RNA was polyA-tailed by using poly(A) polymerase and reverse-transcribed for 1 h at 42 °C. Enzymes were inactivated by incubation for 5 min at 95 °C. Prior to use in qualitative PCR (qPCR), all cDNAs were diluted 1:10 in RNAse-free TE buffer.
For detection of CAT1 transcripts, 1 µg DNAse-treated total RNA was reverse-transcribed (ImpromII, Promega, Madison, WI) by using a 1:3 mixture of oligo-dT and random hexamers according to the manufacturer's recommendations. Prior to use in qPCR, cDNA was diluted 1:8 in RNAse-free TE buffer.
qPCR.
Primers enriched for locked nucleic acids were used to improve the sensitivity and specificity of miRNA detection. Primers for miRNA-122, miR-16 (for normalization), and let-7a (for normalization) were provided by Exiqon (Vedbaek, Denmark). Primers were designed for the CAT1 gene (cat1.exon8 5′ CCA TGC CGC GAG TTA TCT ATG 3′ and cat1.exon9 5′ GTC AAA GAG GAA GGC CAT GA 3′) by using GenBank sequence AY371320 and the Primer3 program.32 For the reference genes riboprotein L4 (RPL4), hydroxymethylbilane synthase (HMBS), and TATA-binding protein (TBP), primer sequences described previously26 were used.
Briefly, 5 µL QuantiFast SYBR Green PCR Master Mix (Qiagen, Hilden, Germany) was mixed with 500 nmol of each primer and 2 µL diluted cDNA or water or standard in PCR plates (AB Gene, ThermoScientific, Waltham, MA) and underwent PCR amplification (model Mx3000P, Stratagene, La Jolla, CA). The cycling conditions were: denaturation at 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s, 60 °C for 30 s. Fluorescence was measured automatically during amplification, and one 3-segment cycle (95 °C for 1 min, 55 °C for 30 s, 95 °C for 30 s) was performed to obtain the melting curve for the product. The baseline adjustment method of the thermocycler (Mx3000, Stratagene) software was used to determine the cycle threshold in each reaction.
Statistical analysis.
Physiologic data are presented as mean ± SEM. Because weight gain showed a strong linear relation with weeks of diet (average r2 = 0.98 ± 0.02), the weight gain for each animal was calculated as the slope of the linear regression curve of the weight as a function of time of feeding. The results for the MSD group were compared with the results for the HCD group by using a one-sided Student t test for unpaired samples. Body weights for MSD and HCD groups also were compared by 2-way ANOVA followed by Tukey test. Differences were considered significant when P values were lower than 0.05.
Biochemical data are presented as mean ± SEM. Because there was no linear relationship between the lipid measurements and time of feeding, data sets for weeks 8 and 19 were compared by using a 2-sided Student t test for unpaired samples. Differences were considered significant when P values were lower than 0.05.
Regarding statistical analysis of the qPCR data, cycle threshold values of the MSD samples and HCD samples were normalized by using 2 reference miRNAs (let-7a and miRNA-16) for the miRNA-122 study and by using 3 reference genes (RPL4, HBMS, and TBP) for the CAT1 study. Normalized values for the MSD and HCD groups were compared by using an algorithm (Relative Expression Software Tool, version 38429) for which the mathematical model is based on the PCR efficiency of each gene investigated and the mean deviation in cycle threshold between control (MSD) and sample (HCD) groups. The resulting expression ratios were tested for significance by using a randomization test, which is based on the assumption that the diets were allocated randomly; the test does not imply normal distribution of the samples. Differences were considered significant when P values were lower than 0.05.
Results
Body weight.
The weight of the minipigs followed the recommended weight curves according to age in both diet groups. Nevertheless, the animals that received a high-cholesterol diet showed a body weight increase of 24% compared with the pigs on standard chow (Figure 1). At the end of the feeding scheme (at age 19 wk), the minipigs in the HCD group weighed 15 ± 2.9 kg and those in the MSD group weighed 12.3 ± 3.3 kg. On average the pigs in the MSD control group gained 0.76 ± 0.23 kg/wk, whereas HSD pigs gained 1.03 ± 0.25 kg/wk, indicating that the animals fed a high-cholesterol diet gained weight faster (P = 0.0498, Student t test) than did the control group. Moreover, 2-sided ANOVA revealed a significant (P = 0.00014) difference in body weight between the 2 groups. A Tukey test showed that the mean body weight in the 2 groups remained similar until week 11. However, from week 12, HCD animals weighed more (P < 0.05) than did MSD pigs.
Biochemical measurements.
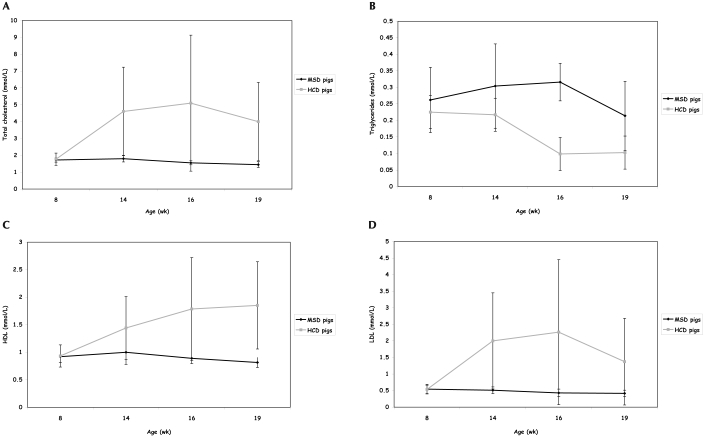
Serum total cholesterol increased substantially (P = 0.042) in the HCD group from 1.77 ± 0.36 mmol/L at week 8 to 3.99 ± 2.3 mmol/L at week 19. In contrast, total cholesterol in MSD pigs decreased (P = 0.018) from 1.72 ± 0.12 mmol/L at week 8 to 1.45 ± 0.16 mmol/L at week 19 (Figure 2 A).
Figure 2.
Lipid profiles (mean ± SD; mmol/L) and age (wk) of the minipigs used in the feeding study. Venous blood samples were obtained from fasted animals (MSD group, n = 5; HCD group, n = 7) at 4 time points. Values at 8 and 19 wk were compared to yield the reported P values. (A) Total cholesterol (HCD group, P = 0.042; MSD group, P = 0.018). (B) Triglycerides (HCD group, P = 0.0016; MSD group, P = 0.48). (C) HDL (HCD group, P = 0.019; MSD group, P = 0.13). (D) LDL (HCD group, P = 0.15; MSD group, P = 0.14).
Serum triglycerides fluctuated in the MSD group from 0.262 ± 0.098 mmol/L at week 8 to 0.21 ± 0.10 mmol/L at week 19 (P = 0.48). In the HCD group, triglycerides decreased (P = 0.0016) from 0.225 ± 0.03 mmol/L at week 8 to 0.10 ± 0.10 mmol/L at week 19 (Figure 2 B).
The serum HDL level in the MSD group was 0.92 ± 0.10 mmol/L at week 8 and 0.81 ± 0.09 at week 19 (P = 0.13). In contrast, in the HCD group the HDL level increased (P = 0.019) from 0.933 ± 0.20 mmol/L at week 8 to 1.85 ± 0.8 mmol/L at week 19 (Figure 2 C). In the MSD group, serum LDL was 0.544 ± 0.14 mmol/L at week 8 and 0.416 ± 0.09 mmol/L at week 19 (P = 0.14), whereas the LDL level in HCD pigs was 0.535 ± 0.13 mmol/L at week 8 and 1.37 ± 1.3 mmol/L at week 19 (P = 0.15; Figure 2D).
qPCR.
To study the expression of miRNA-122, we compared the qPCR results from the 5 MSD pigs with those of the 7 HCD pigs. We found that miRNA-122 was 1.4 times less abundant in the HCD pigs compared with the MSD pigs (P = 0.0015; Table 1). In contrast, the level of the CAT1 transcript did not differ between the groups.
Table 1.
Cycle threshold values (mean ± SD) from qPCR
| Cycle threshold value |
||
| Group | miRNA-122 | CAT1 |
| MSD (n = 5) | 22.50 ± 0.14 | 28.42 ± 0.72 |
| HCD (n = 7) | 23.13 ± 0.37 | 28.27 ± 0.73 |
Data reflect 2 biologic replicates per animal.
Discussion
The Göttingen minipig is an established useful model for studying the effect of dietary high-fat intake on obesity, glucose homeostasis and susceptibility to diabetes.2,15,20,31,33 In the present study, we used Göttingen minipigs to investigate the effect of high-cholesterol diet on the expression of miRNA-122 and its targeted mRNA, CAT1, by using qPCR. The qPCR results were correlated with biochemical and physical measurement of risk factors for obesity (that is, body weight and total cholesterol, triglycerides, HDL, and LDL levels).
The weight of the minipigs followed the recommended age-corrected weight-curves in both diet groups. Nevertheless, the pigs fed a high-cholesterol diet gained weight faster than did the control group, but the between-group differences in body weight were significant only after week 11.
The minipigs were group-housed according to the diet they were fed. We chose this arrangement to increase animal wellbeing and for practical reasons. As cohoused minipigs get older, a hierarchy develops within the group, such that the dominant pigs usually eat more and grow faster than subordinates. This situation led to variation in weight within each group and likely explains the high standard deviation in both groups. Individually housing the pigs would have given us better control over feeding, thereby decreasing the standard deviations, this option was not possible in the present study.
The decrease in triglycerides level in the HCD group that we found in the present study is in accordance with previous findings in male minipigs.14,20 In contrast, another study15 found that triglycerides were increased in female minipigs that received a high-fat, high-energy diet (0.24 ± 0.03 mmol/L) compared with that in the group fed a low-fat, low-energy diet (0.13 ± 0.04 mmol/L). Furthermore, 2 independent studies in Ossabaw miniature swine4,5 also showed increased triglycerides levels in both female and male pigs. In fact, an increase in triglycerides in response to atherogenic diet more closely mimics the disease in humans.
Although high-cholesterol diet previously was reported to induce an increase in the LDL:HDL ratio3,4,5,13,14 this ratio did not vary significantly (P = 0.35) over time in the HCD pigs in the present study. This discrepancy can be explained by differences in diet composition, the method of feeding (that is, restrictively or ad libitum), amount of consumption (which can vary due to the hierarchy within the group), the breed of pigs used (minipigs and Landrace pigs,13 male Sinclair miniature pigs,3 and Ossabaw miniature pigs4,5), and the sex of the animals used.2,14
The qPCR technique is a sensitive way to measure changes in gene expression. We found that although miRNA-122 was downregulated in the HCD group, subchronic feeding of the high-cholesterol diet did not alter CAT1 transcript levels. A subchronic feeding scheme likely will not reveal all consequences of a high-cholesterol diet, and our study suggests a delay between effects on miRNA-122 response and the CAT1 response.
miRNA-122 is an important regulator of liver metabolism.10 Studies of antisense targeting against miRNA-122 in a diet-induced obesity mouse model resulted in decreased plasma cholesterol levels.6,7,18 Moreover, silencing miRNA-122 increased the expression of several genes. For example, a cDNA array experiment7 identified 108 upregulated genes (including CAT1), even though it was not designed to find direct miRNA-122 targets but to characterize the downstream effects of miRNA-122 inhibition. Of all the genes identified as targets of miRNA-122, CAT1 seemed to be the most promising candidate for us to investigate because of its implication in liver metabolism1,24 and because of its prediction as the most likely target for miRNA-122.1,21
Unlike miRNA-122, CAT1 is expressed almost ubiquitously, with the exception of the adult liver.1 In addition, miRNA-122 has a negative effect on the stability of CAT1 mRNA. The amount of mRNA for CAT1 increased from 1.5- to 3.5-fold after inhibition of miRNA-122.1 Furthermore, CAT1 and miRNA-122 show inversed patterns of expression during liver development.10
In the present study we saw a diet-induced reduction of miRNA-122 expression. Furthermore, the expression levels of CAT1 were unchanged after 11 wk of feeding a high-cholesterol diet but seemed to have a tendency to increase after long-term high-calorie diet (results not shown). If miRNA-122 is repressed (or downregulated), it cannot destabilize CAT1 mRNA; therefore, mRNA CAT1 levels increase, and its protein product can mediate the bidirectional transport of cationic amino acids, allowing cells to resume growth.12
In several studies, 6,7,9,18 silencing of miRNA-122 decreases plasma cholesterol independent of dose, that is, increased inhibition of miRNA-122 does not decrease the cholesterol level further.7 The decrease in the plasma cholesterol level reflects reduction of both the HDL and LDL fractions. Furthermore, we noted that downregulation of miRNA-122 is significant after 11 wk of high-cholesterol diet, but the expression of CAT1 does not increase. In the present study, the response to the decrease in miRNA-122, which would be an increase in the CAT1 mRNA level, is not established simultaneously. Therefore, even though it has been postulated that miRNAs are “fast molecular switches” or “fine-tuners” of gene expression,11 the response seems to have a time lag. One possible explanation is that CAT1 is not the primary target of miRNA-122. The fact that silencing miRNA-122 reduces the level of cholesterol suggests that downregulation of miRNA-122 is a defense mechanism to prevent high levels of cholesterol.
From the present study we conclude that the increase in weight and cholesterol levels resulting from subchronic (11 wk) feeding of a high-cholesterol diet to minipigs is correlated with a decrease in the expression of miRNA-122. In addition, the levels of CAT1 mRNA are unchanged after 11 wk of high-cholesterol diet. Exposing a larger number of animals to long-term (more than 11 wk) feeding of high-cholesterol diet might provide further insight into the role of the CAT1 gene in diet-induced obesity in minipigs.
Acknowledgments
We thank Minna Jakobsen for technical assistance. This work was supported by The Danish Research Council (FTP).
References
- 1.Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, Zaret KS, Taylor JM. 2004. miRNA-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high-affinity cationic amino-acid transporter CAT1. RNA Biol 1:106–113 [DOI] [PubMed] [Google Scholar]
- 2.Christoffersen BO, Grand N, Golozoubova V, Svendsen O, Raun K. 2007. Gender-associated differences in metabolic syndrome-related parameters in Göttingen minipigs. Comp Med 57:493–504 [PubMed] [Google Scholar]
- 3.Dixon JL, Stoops JD, Parker JL, Laughlin MH, Weisman GA, Sturek M. 1999. Dyslipidemia and vascular dysfunction in diabetic pigs fed an atherogenic diet. Arterioscler Thromb Vasc Biol 19:2981–2992 [DOI] [PubMed] [Google Scholar]
- 4.Dyson MC, Alloosh M, Vuchetich JP, Mokelke EA, Sturek M. 2006. Components of metabolic syndrome and coronary artery disease in female Ossabaw swine fed excess atherogenic diet. Comp Med 56:35–45 [PubMed] [Google Scholar]
- 5.Edwards JM, Neeb ZP, Alloosh MA, Long X, Bratz IN, Peller CR, Byrd JP, Kumar S, Obukhov AG, Sturek M. 2010. Exercise training decreases store-operated Ca2+ entry associated with metabolic syndrome and coronary atherosclerosis. Cardiovasc Res 85:631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elmén J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjärn M, Hansen JB, Hansen HF, Straarup EM, McCullagh K, Kearney P, Kauppinen S. 2008. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to upregulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res 36:1153–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. 2006. MiR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 3:87–98 [DOI] [PubMed] [Google Scholar]
- 8.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, Bennett CF, Lollo B, Griffey R. 2004. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem 279:52361–52365 [DOI] [PubMed] [Google Scholar]
- 9.Fabani MM, Gait MJ. 2008. miRNA-122 targeting with LNA/2’-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA 14:336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Girard M, Jacquemin E, Munnich A, Lyonnet S, Henrion-Caude A. 2008. MiR-122, a paradigm for the role of microRNAs in the liver. J Hepatol 48:648–656 [DOI] [PubMed] [Google Scholar]
- 11.Hammell CM. 2008. The microRNA–argonaute complex: a platform for mRNA modulation. RNA Biol 5:123–127 [DOI] [PubMed] [Google Scholar]
- 12.Hatzoglou M, Fernandez J, Yaman I, Closs E. 2004. Regulation of cationic amino acid transport: the story of the CAT1 transporter. Annu Rev Nutr 24:377–399 [DOI] [PubMed] [Google Scholar]
- 13.Jacobsson L. 1986. Comparison of experimental hypercholesterolemia and atherosclerosis in Göttingen minipigs and Swedish domestic swine. Atherosclerosis 59:205–213 [DOI] [PubMed] [Google Scholar]
- 14.Jacobsson L. 1989. Comparison of experimental hypercholesterolemia and atherosclerosis in male and female minipigs of the Göttingen strain. Artery 16:105–117 [PubMed] [Google Scholar]
- 15.Johansen T, Hansen HS, Richelsen B, Malmlöf R. 2001. The obese Göttingen minipig as a model of the metabolic syndrome: dietary effects on obesity, insulin sensitivity, and growth hormone profile. Comp Med 51:150–155 [PubMed] [Google Scholar]
- 16.Jopling CL. 2008. Regulation of hepatitis C virus by microRNA-122. Biochem Soc Trans 36:1220–1223 [DOI] [PubMed] [Google Scholar]
- 17.Jopling CL, Norman KL, Sarnow P. 2006. Positive and negative modulation of viral and cellular mRNAs by liver-specific microRNA miRNA-122. Cold Spring Harb Symp Quant Biol 71:369–376 [DOI] [PubMed] [Google Scholar]
- 18.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. 2005. Silencing of microRNAs in vivo with ‘antagomirs.’. Nature 438:685–689 [DOI] [PubMed] [Google Scholar]
- 19.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. 2002. Identification of tissue-specific microRNAs from mouse. Curr Biol 12:735–739 [DOI] [PubMed] [Google Scholar]
- 20.Larsen MO, Rolin B, Wilken M, Carr RD, Svendsen O. 2002. High-fat high-energy feeding impairs fasting glucose and increases fasting insulin levels in the Göttingen minipig: results from a pilot study. Ann N Y Acad Sci 967:414–423 [DOI] [PubMed] [Google Scholar]
- 21.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. 2003. Prediction of mammalian microRNA targets. Cell 115:787–798 [DOI] [PubMed] [Google Scholar]
- 22.Li S, Loos RJ. 2008. Progress in the genetics of common obesity: size matters. Curr Opin Lipidol 19:113–121 [DOI] [PubMed] [Google Scholar]
- 23.Lin Q, Gao Z, Alarcon RM, Ye J, Yun Z. 2009. A role of miR-27 in the regulation of adipogenesis. FEBS J 276:2348–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacLeod CL. 1996. Regulation of cationic amino acid transporter (CAT) gene expression. Biochem Soc Trans 24:846–852 [DOI] [PubMed] [Google Scholar]
- 25.Nakanishi N, Nakagawa Y, Tokushige N, Aoki N, Matsuzaka T, Ishii K, Yahagi N, Kobayashi K, Yatoh S, Takahashi A, Suzuki H, Urayama O, Yamada N, Shimano H. 2009. The upregulation of microRNA-335 is associated with lipid metabolism in liver and white adipose tissue of genetically obese mice. Biochem Biophys Res Commun 385:492–496 [DOI] [PubMed] [Google Scholar]
- 26.Nygard AB, Jørgensen CB, Cirera S, Fredholm M. 2007. Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol Biol 8:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan QW, Henry SD, Scholte BJ, Tilanus HW, Janssen HL, van der Laan LJ. 2007. New therapeutic opportunities for hepatitis C based on small RNA. World J Gastroenterol 13:4431–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perkins CP, Mar V, Shutter JR, del Castillo J, Danilenko DM, Medlock ES, Ponting IL, Graham M, Stark KL, Zuo Y, Cunningham JM, Bosselman RA. 1997. Anemia and perinatal death result from loss of the murine ecotropic retrovirus receptor mCAT1. Genes Dev 11:914–925 [DOI] [PubMed] [Google Scholar]
- 29.Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for groupwise comparison andstatistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reddy AM, Zheng Y, Jagadeeswaran G, Macmil SL, Graham WB, Roe BA, Desilva U, Zhang W, Sunkar R. 2009. Cloning, characterization and expression analysis of porcine microRNAs. BMC Genomics 10:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ritskes-Hoitinga J, Bollen P. 1997. Nutrition of Göttingen minipigs: facts, assumptions, and mysteries. Pharmacol Toxicol 80 Suppl 2:5–9 [DOI] [PubMed] [Google Scholar]
- 32.Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386 [DOI] [PubMed] [Google Scholar]
- 33.Spurlock ME, Gabler NK. 2008. The development of porcine models of obesity and the metabolic syndrome. J Nutr 138:397–402 [DOI] [PubMed] [Google Scholar]
- 34.Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, Lin CD, Liao YL, Wang JL, Chau YP, Hsu MT, Hsiao M, Huang HD, Tsou AP. 2009. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology 49:1571–1582 [DOI] [PubMed] [Google Scholar]
- 35.Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Hood LE, Galas DJ. 2009. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA Mar 106: 4402–4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilfred BR, Wang WX, Nelson PT. 2007. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab 91:209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]