Abstract
A recently developed proteomic strategy, the “GG-azide”-labeling approach, is described for the detection and proteomic analysis of geranylgeranylated proteins. This approach involves metabolic incorporation of a synthetic azido-geranylgeranyl analog and chemoselective derivatization of azido-geranylgeranyl-modified proteins by the “click” chemistry, using a tetramethylrhodamine-alkyne. The resulting conjugated proteins can be separated by 1-D or 2-D and pH fractionation, and detected by fluorescence imaging. This method is compatible with downstream LC-MS/MS analysis. Proteomic analysis of conjugated proteins by this approach identified several known geranylgeranylated proteins as well as Rap2c, a novel member of the Ras family. Furthermore, prenylation of progerin in mouse embryonic fibroblast cells was examined using this approach, demonstrating that this strategy can be used to study prenylation of specific proteins. The “GG-azide”-labeling approach provides a new tool for the detection and proteomic analysis of geranylgeranylated proteins, and it can readily be extended to other post-translational modifications.
Keywords: 2-D, Azide, Click chemistry, Protein geranylgeranylation, Rap2c
1 Introduction
Protein prenylation is a post-translational modification of proteins involving the addition of isoprenyl lipids [1–5]. Most prenylated proteins contain the C-terminal CaaX motif (C is cysteine, A is an aliphatic amino acid, and X is a variable residue that dictates the prenyl group that is added). A 15-carbon farnesyl group is added by protein farnesyltransferase (FTase) when X is serine, methionine, glutamine, cysteine, or alanine. Farnesylated proteins include Ras proteins, Rheb proteins, nuclear lamins, and Hdj2. When X is leucine or phenylalanine, a 20-carbon geranylgeranyl (GG) group is added by protein geranylgeranyltransferase type I (GGTase-I). Rho family proteins such as RhoA, Cdc42, and Rac, as well as the γ-subunit of heterotrimeric G-proteins, are geranylgeranylated [6]. Rab proteins involved in protein transport across the secretory and endocytosis pathways are geranylgeranylated by Rab geranylgeranyltransferase [7, 8]. These proteins usually end with CC or CXC at the C termini, and both cysteine residues are geranylgeranylated.
Recent studies have highlighted the physiological importance of protein geranylgeranylation. Characterization of GGTase-I-deficient cells showed proliferation inhibition and accumulation of p21CIP1/WAF1, pointing to the significance of GGTase-I in cell proliferation and cell cycle progression [9]. Conditional knockout of GGTase-I results in the inhibition of lung tumor growth and increases survival [9]. Recent studies have shown that a number of geranylgeranylated proteins play important roles in tumorigenesis and metastasis. In addition to RhoA and Cdc42 proteins, RalA and RalB were found to be activated downstream of Ras in most pancreatic cancer cells harboring an oncogenic K-ras [10–12]. In addition, Dlc1, a RhoA GTPase-activating protein, was found to be a major class of tumor suppressor [13, 14]. Inhibition of protein geranylgeranylation is a promising approach for developing anticancer drugs. Inhibitors of GGTase-I are currently undergoing preclinical studies and have been shown to disrupt oncogenic and tumor survival pathways, inhibit proliferation and anchorage-independent growth, and induce apoptosis [15–19].
Here, we report a recently developed strategy, the “GG-azide”-labeling approach, for the detection and proteomic analysis of geranylgeranylated proteins based on metabolic incorporation of a synthetic azido-GG analog and chemoselective reaction between azido-geranylgeranyl-modified proteins and a TAMRA-alkyne. TAMRA-labeled, geranylgeranylated samples can be separated by 1-D or 2-D and pH fractionation, and detected with fluorescence imaging. This method can be combined with LC-MS/MS for proteomic analysis of geranylgeranylated proteins. The “GG-azide”-labeling strategy is an extension of the tagging-via-substrate approach we previously developed for the detection and proteomic analysis of farnesylated proteins [20]. Tagging-via-substrate technology involves metabolic incorporation of a synthetic azido-farnesyl analog and the Staudinger conjugation reaction between azido-farnesyl-modified proteins and a biotinylated phosphine capture reagent [21, 22]. The bioti-nylated, azido-farnesyl-modified proteins are detected by Western blotting analysis using horseradish peroxidase-conjugated streptavidin. The protein conjugates can also be affinity-purified by streptavidin-conjugated agarose beads.
In this study, proteomic analysis of the TAMRA-labeled proteins led to the identification of several known geranylgeranylated proteins as well as Rap2c, a novel member of the Ras family. This new experimental approach will be useful for the identification of geranylgeranylated proteins and studies designed to ascertain the biological significance of protein geranylgeranylation. The “GG-azide”-labeling approach can readily be extended to other protein modifications, such as farnesylation and palmitoylation.
2 Materials and methods
2.1 Materials
Azide GG Alcohol (N3GGOH), Click-iT® Tetramethylrhodamine (TAMRA) Glycoprotein Dection Kit (C33370), SYPRO® Ruby protein gel stain, 4–12% NuPAGE® bis-Tris Mini gels, Lithium dodecyl sulfate (LDS) sample buffer, MOPS SDS running buffer, Zoom® IEF fractionator, Zoom® Disks (pH 3.0, 4.6, 5.4, 6.2, 7.0, and 10), IEF anode and cathode buffers, Zoom® Focusing buffers, Zoom® narrow pH gradient IPG strips, Zoom® carrier ampholytes, urea, thiourea, CHAPS, DTT, EZQ® Protein Quantitation Kit, and trypsin were obtained from Invitrogen (Carlsbad, CA, USA). N,N-dimethylacrylamide and tributylphosphine were from Sigma-Aldrich (St. Louis, MO, USA). DMEM was from Mediatech; fetal bovine serum (FBS) was from Hyclone, and Protease Inhibitor Cocktail was from Roche Applied Sciences (Indianapolis, IN, USA).
2.2 Metabolic labeling with azido GG alcohol
MCF-7 and COS-7 cells were grown in DMEM/10% FBS, and Jurkat cells were grown in RPMI 1640/10% FBS. Cells were grown to 50% confluency and labeled with 50 µM azido-GG alcohol for 24–48 h. The cells were washed five times with cold Dulbecco’s PBS (Mediatech). Cells were harvested in 1% SDS/100mM Tris-HCl, pH 8, 1 × Protease Inhibitor Cocktail, and then sonicated three times for 10 s each, vortexed for 5 min and centrifuged at 10 000 × g at 4°C for 15 min. The cell debris pellet was discarded, and the protein-containing supernatant was precipitated with methanol/chloroform/water. Labeling with azido farnesyl alcohol and lovastatin treatment were performed as described in [20].
2.3 Detection of azido-geranylgeranylated proteins
The protein pellet was solubilized in 1% SDS/100mM Tris-HCl, pH 8. The lysate (50 µg) was labeled with TAMRA-alkyne in the presence of Cu(I) for 1 h at room temperature as per the Click-iT® TAMRA Glycoprotein Detection Kit instructions (Invitrogen). Labeled samples were precipitated using methanol/chloroform/water. To detect azido-geranylgeranylated proteins, the protein pellet was resolubilized in 1 × SDS sample butter and size-fractionated by SDS-PAGE. The azido-geranylgeranyled-modified proteins were detected with a Typhoon 9410 scanner.
2.4 pH Fractionation of TAMRA-labeled Lysates
The precipitated TAMRA-labeled MCF-7 lysates (2 mg) were resuspended in 3.4 mL 7 M urea, 2 M thiourea, 2% CHAPS, 1% Zwittergent 3–10, 60 mM DTT, 0.1% Zoom® Focusing buffer pH 3–7 and 0.1% Zoom focusing buffer pH 7–12. The samples were centrifuged at 3000 rpm for 10 min, and 650 µL of supernatant was loaded into each of the five assembled chambers of the Zoom® IEF fractionator (Invitrogen) and fractionated according to the Zoom® IEF fractionator instructions. The five narrow pH fractions were precipitated using methanol/chloroform/water. The precipitated fractions were resuspended in 50 µL 100 mM Tris, 1% SDS, pH 8, and protein amounts were determined using the EZQ protein quantitation kit (Invitrogen). For 1-D PAGE analysis 7 µg of pH 3–4.6 fractions and 20 µg of pH 4.6–5.4, 5.4–6.2, 6.2–7, and 7–10 fractions were diluted into LDS sample buffer plus DTT (50 mM final) and resolved on a 1 mm NuPAGE® 4–12% bis-Tris gel with MOPS running buffer. TAMRA signal was detected using a FX Pro Plus fluorescence scanner (Bio-Rad) with 532 nm excitation and a 555 nm long pass emission filter. The gel was then fixed in an aqueous solution of 50% methanol, 7% acetic acid overnight, and then stained for total proteins using SYPRO® Ruby protein gel stain (30 min using microwave heating) followed by a 30-min destain in 10% methanol, 7% acetic acid, and a water rinse. The gels were imaged a second time on the FX Pro Plus scanner using 473 nm excitation and a 555 nm long pass emission filter.
2.5 2-D SDS-PAGE
The resuspended narrow pH range fractions in 100 mM Tris, 1% SDS, pH 8 were then reduced and alkylated with tributyl phosphine (200 mM) and N,N-dimethylacrylamide (0.5%) by heating at 65°C for 10 min, followed by rotation end-over-end at RT for 20 min. The samples were precipitated with methanol/chloroform/water and resuspended in 180 µL 7 M urea, 2M thiourea, 2% CHAPS, 2% ASB-14, 66 mM DTT, 1% ampholytes (pH 4–6 ampholytes for pH 4.6–5.4 fractions, pH 5–7 ampholytes for pH 5.4–6.2 fractions, pH 6–8 ampholytes for pH 6.1–7.0 fractions, and pH 6–8 and 9–11 ampholytes for pH 7–10 fractions). The samples were centrifuged at 20 000 rpm for 3 min, and 170 µL (approximately 200 µg) supernatant was loaded onto narrow pH range IPG strips specific for each fraction (pH 4.5–5.5 strips for pH 4.6–5.4 fractions, pH 5.3–6.3 strips for pH 5.4–6.2 fractions, pH 6.1–7.1 strips for pH 6.1–7.0 fractions, and pH 6–10 strips for pH 7–10 fractions). The IPG strips were rehydrated for 90 min and then focused for 20 min at 200 V, 25 min at 450 V, 20 min at 700 V, and 105 min at 2000 V, after which they were incubated in 1 × LDS sample buffer plus 50 mM DTT, and resolved on a 1 mm NuPAGE® 4–12% bis-Tris gel with MOPS running buffer. TAMRA signal was detected and the gels were stained for total proteins using SYPRO® Ruby protein gel stain and imaged again as detailed above. The TAMRA and SYPRO® Ruby protein stain images were overlayed and spots were excised from the SYPRO® Ruby-stained gels corresponding to the locations of the TAMRA-labeled proteins.
2.6 Protein identification using MS
For protein identification, spots were excised from the SYPRO® Ruby-stained gels corresponding to the locations of the TAMRA-labeled proteins and digested overnight using trypsin, and tryptic peptides were then isolated as reported previously [23]. The resulting peptides were subjected to LC-MS/MS analysis at the Vincent Coates Foundation Mass Spectrometry Laboratory at Stanford University (http://mass-spec.stanford.edu). Nanoreversed phase HPLC was performed using an Eksigent 2-D nanoLC (Eksigent, Dublin, CA, USA) with buffer A consisting of 0.1% formic acid in water and buffer B 0.1% formic acid in ACN. A fused silica column self-packed with duragel C18 (Peeke, Redwood City, CA, USA) matrix was used with a linear gradient from 5% B to 40% B over 60 min at a flow rate of 450nL/min. The nanoHPLC was interfaced with an Advion Nanomate (Ithaca, NY, USA) for nanoESI into the mass spectrometer. The mass spectrometer was a LCQ Deca XP Plus (Thermo Scientific), which was set in data-dependent acquisition mode to perform MS/MS on the top three most intense ions with a dynamic exclusion setting of two. The DTA files were extracted from the raw data using the bioworks browser (Thermo Scientific).
The extracted DTA files were analyzed using two search engines: MASCOT (version MASCOT, Matrix Science, London, UK) and X! Tandem (version 2007.01.01.1, www.thegpm.org). MASCOT was used to search the National Center for Biotechnology Information Non-redundant (NCBInr)_070510 database, which was selected for Homo sapiens. X! Tandem was used to search the NCBInr_060405 database. Both NCBInr databases were used with the following search parameters: a fragment ion mass tolerance of 1.5 Da (monoisotopic) and a parent ion tolerance of 1.5 Da (monoisotopic). Acrylamide adduct of cysteine was specified in MASCOT and X! Tandem as a fixed modification, whereas oxidation of methionine was specified in both search engines as a variable modification.
To validate MS/MS-based peptide and protein identifications, Scaffold (version Scaffold_2_00_03, Proteome Software, Portland, OR, USA) was used. Peptide identifications were accepted only when they exceeded peptide thresholds of the search engines. For MASCOT, peptide identifications required that the ion scores must be greater than both the associated identity scores and 41. For X! Tandem, peptide identifications required —log (expected scores) scores to be greater than 2. To ensure confident protein identifications, proteins must contain at least three identified peptides.
2.7 Geranylgeranylation of a mutant form of prelamin A (progerin) assessed by the “GG-azide”-labeling approach
Mice expressing a mutant Lmna allele (LmnaHG/+), yielding the truncated prelamin A (progerin) found in Hutchinson-Gilford progeria syndrome) were created by gene-targeting and described previously [24, 25]. Progerin contains a CaaX motif and is normally farnesylated. Recently, Davies et al. [26] used gene targeting to create a mutant Lmna allele (LmnaggHG) that yields a geranylgeranylated form of progerin. Mouse embryonic fibroblasts (MEFs) heterozygous for each mutant allele (LmnaHG/+ and LmnaggHG/+) were incubated with 50 µM azido-GG alcohol for 48 h and harvested in 1% SDS and 50 mM Tris, pH 8. Protein extracts were prepared from the fibroblasts prepared by sonication, chloroform-methanol precipitation, and resuspension in 1% SDS and 50 mM Tris, pH 8. Proteins that had incorporated azido GG alcohol were labeled with TAMRA-alkyne as per the Click-iT® TAMRA Glycoprotein Detection Kit instructions (Invitrogen) [26]. Proteins in cell extracts were then fractionated on 4–12% gradient polyacrylamide bis-Tris gels (Invitrogen), and labeled proteins were visualized with a Typhoon 9410 Variable Mode Imager (GE Healthcare, Piscataway, NJ, USA). After imaging, the separated proteins were transferred to nitrocellulose for Western blotting with an antibody against antilamin A/C (sc-6215, Santa Cruz Biotechnology, Santa Cruz, CA, USA) [26].
3 Results
3.1 Detection of geranylgeranylated proteins by the “GG-azide”-labeling approach
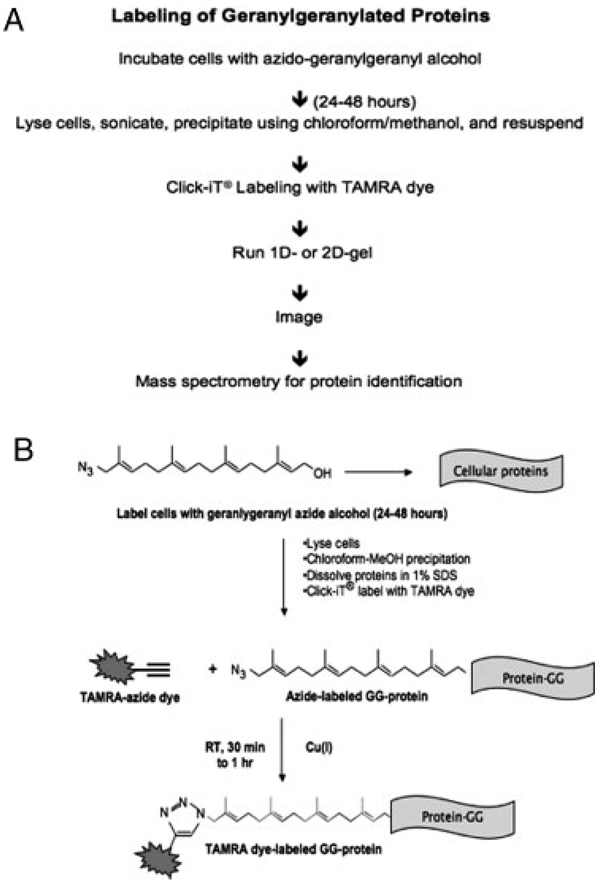
The “GG-azide”-labeling approach utilizes the Cu(I)-catalyzed chemoselective cycloaddition reaction between an azide and an alkyne to detect geranylgeranylated proteins modified with azido groups. Geranylgeranylated proteins are metabolically labeled by feeding cells with azido-geranylgeranyl alcohol (N3-GG-OH), which is converted to a diphosphate derivative that acts as a replacement for geranylgeranyl diphosphate, a substrate for protein GGTases. Cells are then collected and lysed. GG-azide-modified proteins in the lysate undergo chemoselective derivatization via the “click” reaction with fluorescent (TAMRA)-alkyne. Following electrophoresis, TAMRA-labeled samples can be detected by fluorescence imaging and are compatible with downstream LC-MS/MS and MALDI MS analysis (Fig. 1).
Figure 1.
Metabolic labeling and “click” tagging of geranylgeranylated proteins using the Cu(I)-catalyzed azide–alkyne cycloaddition reaction. (A) Overview of the “GG-azide”-labeling approach for the labeling and detection of geranylgeranylated proteins. (B) Schematic representation of the “click” reaction.
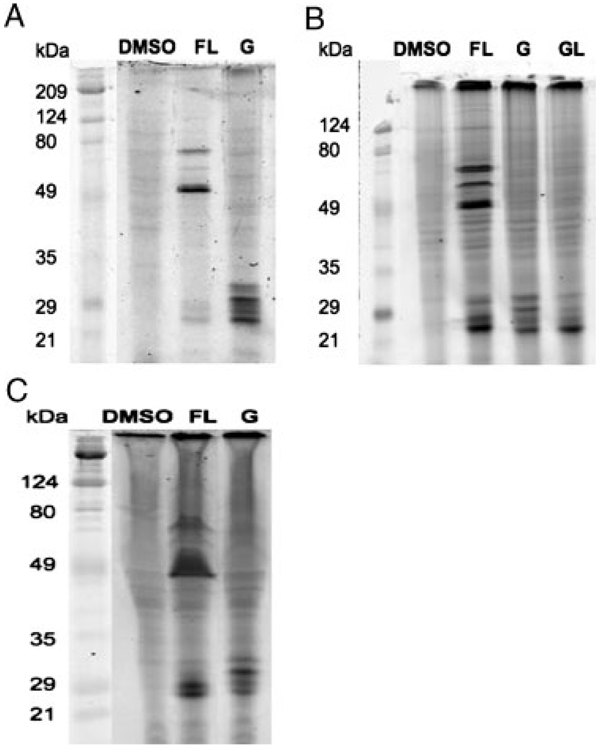
To test whether cultured mammalian cells will process the exogenous azido-GG substrate for protein geranylgeranylation, we performed a metabolic labeling experiment of Jurkat cells with N3-GG-OH or DMSO, followed by the “click” reaction with fluorescent alkyne and gel electrophoresis. When N3-GG modification was visualized by TAMRA-labeled protein detection, the TAMRA label was attached to proteins only in the cells that had been incubated in the presence of N3-GG-OH (Fig. 2A, lane G). Interestingly, geranylgeranylated proteins were detected predominantly in a region between 20 and 35 kDa. Faint bands were observed in regions above the 20–35 kDa region, suggesting that geranylgeranylated proteins of higher molecular weight exist.
Figure 2.
Azide-GG-OH is a substrate for protein geranylgeranylation in vivo. (A) Labeling and detection of geranylgeranylated proteins in Jurkat cells. Jurkat cells were metabolically labeled with DMSO, N3-GG-OH (G) or N3-F-OH in the presence of lovastatin (FL, where F stands for N3-F-OH) for 48 h. (B) and (C) Labeling and detection of geranylgeranylated proteins in other cell lines. Metabolic labeling experiments of COS-7 (B) and MCF-7 (C) were performed using N3-GG-OH (G), N3-F-OH (F) in the presence or absence of lovastatin (L) as indicated.
Metabolic labeling experiments with N3-GG-OH were also performed using COS-7 cells. When N3-GG modification was visualized by TAMRA-labeled protein detection, only the TAMRA label was found attached to proteins when the cells were incubated in the presence of N3-GG-OH, but not in the presence of DMSO (Fig. 2B, compare lanes DMSO and G). TAMRA-labeled proteins were predominantly detected between ~20 and 35 kDa. The profiles of geranylgeranylated proteins were also assessed in MCF-7 cells (Fig. 2C, lane G). Similar to Jurkat and COS-7 cells, geranylgeranylated proteins were predominantly detected between ~20 and 35 kDa in MCF-7 cells. When the metabolic labeling experiment of MCF-7 cells was performed with N3-GG-OH in the presence of excess geranylgeraniol (GG-OH), TAMRA-labeled proteins (20–30kDa) were not observed, suggesting that labeling observed was due to geranylgeranylated proteins (data not shown).
To compare the profile of geranylgeranylated proteins observed with that of farnesylated proteins, metabolic labeling experiments with azido-farnesyl alcohol (N3-F-OH) were performed in Jurkat (Fig. 2A), COS-7 (Fig. 2B), and MCF-7 (Fig. 2C) cells. As previously reported, N3-F-OH is converted to farnesyl diphosphate (FPP) and used as a substrate by protein FTase [20]. The labeling experiment was performed in the presence of both N3-F-OH and lovastatin, an inhibitor of HMG-CoA reductase. Lovastatin blocks mevalonate synthesis, depleting a pool of endogenous FPP and thereby enhances the detection of azide-labeled farnesylated proteins [20, 27, 28]. In the presence of N3-F-OH alone, the bands were too weak to be detected (data not shown), suggesting that N3-F-OH was diluted by endogenous FPP. In the case of geranylgeranylated proteins, pretreating cells with lovastatin resulted in little improvement in labeling (Fig. 2B, compare lanes G and GL). In the presence of both N3-F-OH and lovastatin, N3-F modification was observed (Fig. 2A–C, lane FL). These experiments revealed that the profile of farnesylated proteins was distinct from that of geranylgeranylated proteins (Fig. 2A–C, compare lanes G and FL). For instance, three major farnesylated bands were detected at positions corresponding to 49-, 60-, and 80-kDa proteins. In addition, a number of bands were detected in the region between 20 and 30 kDa. In contrast, when the cells were labeled with N3-GG-OH, labeling was concentrated in a region between 20 and 30 kDa (Fig. 2A – C).
3.2 pH Fractionation of TAMRA-labeled, geranylgeranylated proteins in MCF-7 cells
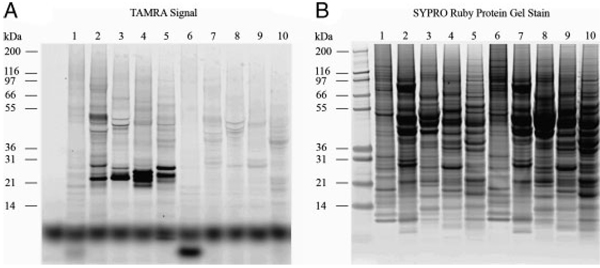
To uncover geranylgeranylated proteins present in lower abundance in cells and to achieve higher resolution of separation, TAMRA-labeled, geranylgeranylated proteins were separated by pH fractionation (IEF), a separation method by isoelectric point. N3-GG-labeled lysates were fractionated by the Zoom® IEF fractionator into five different pH fractions (3–4.6, 4.6–5.4, 5.4–6.2, 6.2–7.0, and 7–10) and resolved by 1-D gel electrophoresis. As shown in Fig. 3B, each fraction contained a different set of proteins as detected by SYPRO® Ruby post-stain. Different bands of TAMRA-labeled, geranylgeranylated proteins were detected in different pH fractions (Fig. 3A). Interestingly, we observed higher molecular weight geranylgeranylated bands at ~50kDa in the pH 4.6–5.4 fraction.
Figure 3.
pH Fractionation of TAMRA-labeled, geranylgeranylated proteins in MCF-7 cells followed by 1-D SDS-PAGE. TAMRA-labeled samples were subjected to pH fractionation on the Zoom® IEF fractionator into five different pH fractions and then resolved by 1-D SDS-PAGE. (lane 1: DMSO, pH 3–4.6; lane 2: N3-GG-OH, pH 4.6–5.4; lane 3: N3-GG-OH, pH 5.4–6.2; lane 4: N3-GG-OH, pH 6.2–7; lane 5: N3-GG-OH, pH 7–10; lane 6: N3-GG-, pH 3–4.6; lane 7: DMSO, pH 4.6–5.4; lane 8: DMSO, pH 5.4–6.2; lane 9: DMSO, pH 6.2–7; lane 10: DMSO, pH 7–10.
3.3 2-D and MS identification of azide-GG-modified proteins in MCF-7 cells
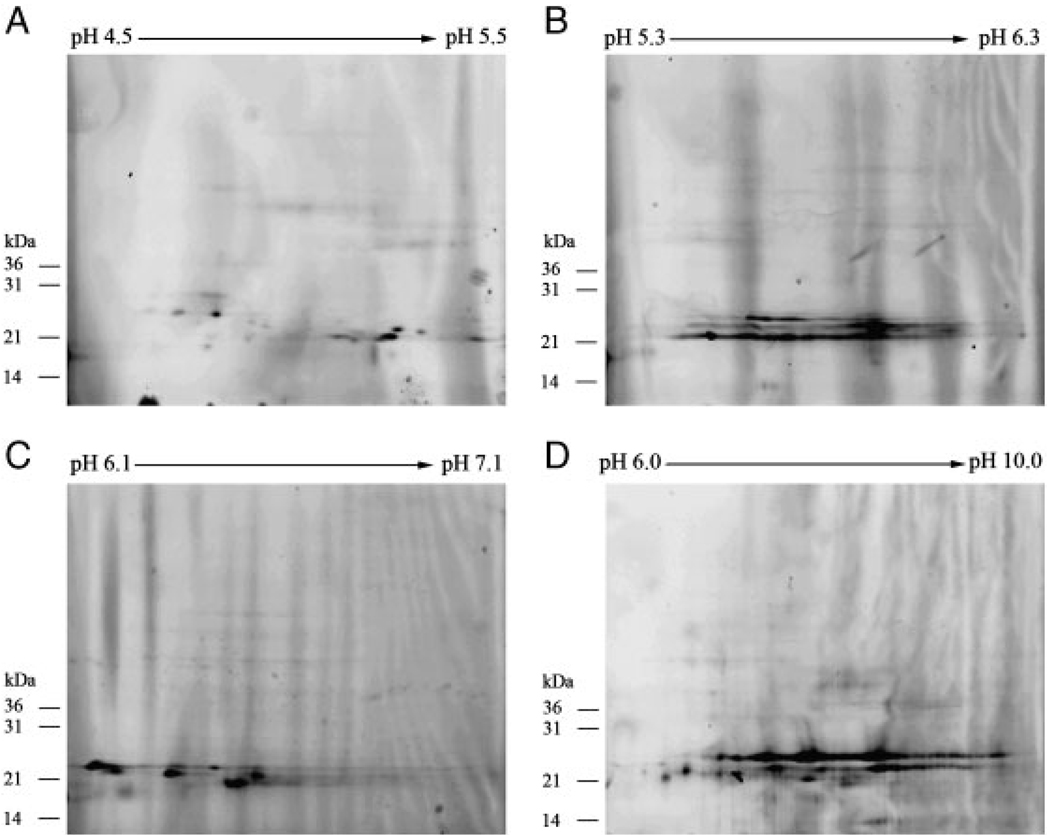
To separate geranylgeranylated proteins, we combined pH fractionation with 2-D. TAMRA-labeled lysates were fractionated by Zoom® IEF fractionator and subjected to narrow pH range 2-D SDS-PAGE. Geranylgeranylated proteins were identified by TAMRA-labeled protein detection (Fig. 4). This achieved further separation of geranylgeranylated proteins.
Figure 4.
pH Fractionation of TAMRA-labeled, geranylgeranylated proteins in MCF-7 cells followed by 2-D SDS-PAGE. TAMRA-labeled samples were pH fractionated by the Zoom® IEF fractionator into different pH fractions, followed by narrow pH range 2-D SDS-PAGE. TAMRA-labeled, geranylgeranylated proteins were visualized by TAMRA-labeled protein detection.
To identify geranylgeranylated proteins, spots representing geranylgeranylated proteins were excised from SYPRO® Ruby-stained gels (Supporting Information Fig. S1). Excised spots (26 in total, A–Z) were digested with trypsin, and the resulting tryptic peptides were subjected to LC-MS/MS. Geranylgeranylated proteins were identified by peptide sequence with the search programs MASCOT and X! Tandem. A protein database search (NCBInr) with the MS data led to the identification of several previously known geranylgeranylated proteins, including members of the Rab protein family. Supporting Information Table S1 lists of all the proteins identified for each spot. The list of TAMRA-labeled, geranylgeranylated proteins identified by this experiment is summarized in Table 1. Interestingly, Rap2c, a member of the Ras family, was identified as a geranylgeranylated protein [29]. In addition, our results suggest that N3-GG-OH was incorporated into proteins containing the CaaX motif as well as proteins that terminate with CC or CXC (Table 1). The identification of a number of known geranylgeranylated proteins confirms the ability of the GG-azide technology for efficient isolation and identification of geranylgeranylated proteins.
Table 1.
Identification of TAMRA-labeled, geranylgeranylated proteins in MCF-7 cellsa)
| Protein name | CAAX, CC, or CXC motif |
Protein molecular weight (AMU) |
% Coverage of protein in excised gel spots |
|---|---|---|---|
| RAB7, member of RAS oncogene family | SCSC | 23 472.00 | 28 |
| RAB11B | CQNL | 24 417.90 | 25 |
| RAB14, member of RAS oncogene family | GCGC | 23 909.60 | 25 |
| RAB1A, member of RAS oncogene family | GGCC | 22 660.40 | 25 |
| RAB3D, member of RAS oncogene family | SCSC | 24 249.50 | 26 |
| Ras-related small GTP-binding protein RAB5 | CCSN | 23 549.90 | 17 |
| RAB6A, member of RAS oncogene family | GCSC | 23 531.40 | 24 |
| RAB2, member of RAS oncogene family | GGCC | 23 528.20 | 25 |
| RAP2C, member of RAS oncogene family | CVVQ | 20 754.40 | 40 |
| RAB18, member of RAS oncogene family | CSVL | 22 959.50 | 26 |
Excised spots (26 in total) from Sypro Ruby-stained gels were digested with trypsin, and the resulting tryptic peptides were subjected to MS. TAMRA-labeled, geranylgeranylated proteins were identified by peptide sequencing with the search programs MASCOT and X! Tandem.
3.4 Application of “GG-azide”-labeling approach: examination of prenylation of nuclear lamin A
The isoprenylation of prelamin A has recently received significant interest [24, 25]. Hutchinson–Gilford progeria syndrome is a genetic disorder characterized by disease phenotypes that resemble premature aging [30]. The disease is caused by the synthesis of a mutant prelamin A, commonly called progerin, that contains an internal 50 amino acid deletion. Progerin retains its carboxyl-terminal CaaX motif and is normally farnesylated. The farnesyl lipid anchor is believed to be critical for the pathogenesis of disease [24, 25]. To explore the significance of the isoprenylation of progerin, mice expressing a mutant version of progerin expected to be geranylgeranylated were created [26]. MEF cells from these mice represented valuable reagents for testing “GG-azide”-labeling techniques [25].
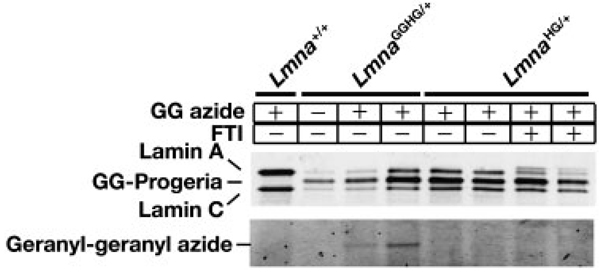
Fibroblasts from the wild-type mice (Lmna+/+), mice expressing the geranylgeranylated version of progerin (LmnaggHG/+), and mice expressing farnsylated progerin (LmnaHG/+) were examined in metabolic labeling experiments (Fig. 5). Lamin A and lamin C are expressed in wild-type fibroblasts and can be easily detected by Western blot with an antibody against lamin A/C. An additional band, progerin, intermediate in size between lamin A and lamin C, is found in the LmnaggHG/+ and LmnaHG/+ fibroblasts. Metabolic labeling with N3-GG-OH revealed that the progerin in LmnaggHG/+ fibroblasts was geranylgeranylated. As expected, there was no labeling of the progerin in LmnaHG/+ fibroblasts. These results show that the “GG-azide”-labeling approach can be used effectively to analyze geranylgeranylated proteins in knock-in mice.
Figure 5.
Examination of prenylation of progerin, a mutant form of prelamin A. Fibroblasts from mice expressing geranylgeranylated progerin (LmnaggHG/+), farnesylated progerin (LmnaHG/+), or mature lamin A (Lmna+/+) were metabolically labeled with N3-GG-OH for 48h. Geranylgeranylated, TAMRA-labeled proteins were visualized with a fluorescence imager. Separated proteins were also detected by Western blotting using an antibody against lamin A/C.
4 Discussion
Here, we describe a novel approach, called “GG-azide”-labeling approach, for the detection and proteomic analysis of geranylgeranylated proteins based on metabolic incorporation of a synthetic azido-GG analog and a chemoselective reaction between azido-geranylgeranyl-modified proteins. The N3-GG-OH was able to enter cells efficiently and become incorporated into geranylgeranylated proteins, and the fluorescent-labeled samples, obtained after the “click” reaction, can be separated by 1-D or 2-D and pH fractionation. Our experimental approach is simple and reduces the time required for visualizing geranylgeranylated proteins from days or months with autoradiographic/ fluorographic methods to minutes with fluorescence imaging. It is also very sensitive and avoids the use of radioactive substrates. Following 2-D, the modified proteins can be analyzed by LC-MS/MS, making it possible to use the “GG-azide”-labeling approach to identify novel geranylgeranylated proteins. In the previous work, we demonstrated the application of MALDI-TOF peptide fingerprinting for the identification of click-labeled serum glycoproteins isolated from metabolically labeled HER2/neu mice [31].
Previously, crystallographic analysis of FTase and GGTase-I complexed with substrate peptides has defined rules of protein substrate selectivity, and those rules were used to classify known substrates of the prenyltransferases (55 in total) and generate a list of potential FTase and GGTase-I substrates (65 in total) [32]. More recently, the Prenylation Prediction Suite (PrepS), an amino acid sequence-based predictor, has been used to predict potential substrates of FTase [33, 34]. However, it is possible that not all proteins containing the consensus motifs are prenylated; therefore, it is necessary to experimentally validate potential substrates of prenyltransferases. Previously reported methods for in vitro or in vivo analysis of potential substrates of protein prenylation involve transcription/translation of a cloned construct and protein prenylation in the presence of 3H-labeled lipid anchor precursors followed by auto-radiography [35–37] or thin layer chromatography scanning [38]. Recently, Nguyen et al., reported the synthesis and characterization of biotin-geranylpyrophosphate (BGPP) to detect prenylated proteins in cells [39, 40]. BGPP is an unnatural phosphoisoprenoid that can be incorporated into Rab GTPases by Rab geranylgeranyltransferase. In addition, using a structure-guided protein engineering approach, Nguyen et al., designed mutants of FTase and GGTase-I that are capable of using BGPP as a substrate [40].
Using the “GG-azide”-labeling approach, major bands of geranylgeranylated proteins were detected in a region between 20 and 30 kDa with minor bands in higher molecular weight range. This may suggest that the majority of geranylgeranylated proteins represent those of the Ras superfamily G-proteins including the Rho family and the Rab family proteins. In contrast, we found four to five major bands of farnesylated proteins in a region between 50 and 70 kDa. Some of these likely represent nuclear lamins, as lamin A and lamin C appear as a band of 72 and 62 kDa, respectively, by SDS-PAGE [41]. In addition, we detected bands of farnesylated proteins in the range of 20–30 kDa. These presumably reflect those of the Ras superfamily G-proteins.
We have used three different mammalian cell lines, T-cell line Jurkat, monkey cell line COS-7 and human breast cancer cell line MCF-7, to demonstrate labeling of geranylgeranylated proteins. We have also obtained labeling of geranylgeranylated proteins in another breast cancer cell line MDA-MB231 (unpublished observation). Thus, the method is applicable to the analysis of geranylgeranylated proteins in a variety of cell lines. It is interesting to point out that the overall profile of geranylgeranylated proteins is similar among different cell lines including transformed and untransformed cells. It has been reported that the profile of protein prenylation in FRTL-5 thyroid cells is altered by transformation with v-K-ras [42]. So far, we have not seen major differences in the patterns of geranylgeranylated proteins between transformed and untransformed cells. More extensive analysis is needed to investigate this point. In addition, “GG-azide” labeling was performed with MEFs from wild-type mice (Lmna+/+) and knock-in mice expressing either geranylgeranylated progerin (LmnaggHG/+) or farnesylated progerin (LmnaHG/+). Metabolic labeling indicated that the progerin in LmnaggHG/+ MEFs was indeed geranylgeranylated, whereas no labeling of progerin was detected in LmnaHG/+ MEFs. Our results demonstrate that the azide-labeling approach can be used to study the prenylation of lamins and other specific proteins of interest.
In our study, we identified Rap2C, which is a novel member of the Ras family, as a newly identified geranylgeranylated protein. Previous studies have shown that overexpression of Rap2C in HEK293T cells activated the transcriptional activities of serum response element [43]. When overexpressed in cultured mammalian cells, Rap2C was found to localize to the plasma membrane [29]. Interestingly, using an amino acid sequence-based predictor of protein prenylation called PrePS, Maurer-Stroh et al., have predicted that Rap2C is likely a target of inhibitors of FTase; however, their prediction was not experimentally verified [34]. The discrepancy between results obtained using the “GG-azide”-labeling approach and those obtained using the PrePS indicates that although amino acid sequences can help identify the presence of a variety of proteins ending with the CAAX motif, not all proteins containing a CAAX motif are farnesylated or geranylgeranylated as predicted. To further investigate whether Rap2C is indeed a substrate of protein geranylgeranylation, metabolic labeling experiments using N3-GG-OH can be performed by following treatment with inhibitors of GGTase. Furthermore, it will be interesting to investigate the biological significance of prenylation for the functions of Rap2C.
Our identification of Rap2c as a novel geranylgeranylated protein suggests that the “GG-azide” technology is sufficiently sensitive to identify novel substrates of protein geranylgeranylation. As discussed in Section 1, recent studies have pointed to the importance of protein gera-nylgeranylation in cancer. Activation or overexpression of geranylgeranylated proteins such as RhoA and RalA has been detected in human cancers. Geranylgeranyltransferase inhibitors cause cell cycle arrest and inhibition of tumor growth in animal model systems. However, proteins that are targeted by geranylgeranyltransferase inhibitors to induce such effects are still being investigated. Our “GG-azide”-labeling approach will be useful for identification of geranylgeranylated proteins and examination of changes in modification caused by GGTase inhibitors that could be potential anticancer agents.
Acknowledgments
The authors thank the Vincent Coates Foundation Mass Spectrometry Laboratory at Stanford University (SUMS). They also thank Chris Adams at SUMS for advice and help with MS/ MS data analysis. This work is supported by NIH grant CA41996 (F.T.) and NIH grants AR050200 (S.G.Y.), HL76839 (S.G.Y.), HL86683 (S.G.Y.), GM66152 (S.G.Y.), a March of Dimes grant 6-FY2007-1012 (S.G.Y.), and an Ellison Medical Foundation Senior Scholar Award (S.G.Y.). LC is supported by the Ruth L. Kirschstein National Research Science Award (GM07185).
Abbreviations
- BGPP
biotin-geranylpyrophosphate
- FBS
fetal bovine serum
- FPP
farnesyl diphosphate
- FTase
farnesyltransferase
- GG
geranylgeranyl
- GGTase-I
geranylgeranyltransferase-I
- LDS
lithium dodecyl sulfate
- MEFs
mouse embryonic fibroblasts
- NCBInr
National Center for Biotechnology Information Non-redundant
- N3-F-OH
azido-farnesyl alcohol
- N3-GG-OH
azido-geranylgeranyl alcohol
- PrePS
Prenylation Prediction Suite
- TAMRA
tetramethylrhodamine
Footnotes
The authors have declared no conflict of interest.
References
- 1.Tamanoi F, Sigman DS, editors. The Enzymes. Vol. 21. San Diego: Academic Press; 2001. [Google Scholar]
- 2.Zhang FL, Casey PJ. Annu. Rev. Biochem. 1996;65:241–270. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 3.Glomset JA, Gelb MH, Farnsworth CC. Trends Biochem. Sci. 1990;15:139–142. doi: 10.1016/0968-0004(90)90213-u. [DOI] [PubMed] [Google Scholar]
- 4.Gelb MH, Brunsveld L, Hrycyna CA, Michaelis S, Tamanoi F, Van Voorhis WC, Waldmann H. Nat. Chem. Biol. 2006;2:518–528. doi: 10.1038/nchembio818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox AD, Der CJ. Curr. Opin. Cell Biol. 1992;4:1008–1106. doi: 10.1016/0955-0674(92)90133-w. [DOI] [PubMed] [Google Scholar]
- 6.Taylor JS, Reid TS, Terry KL, Casey PJ, Beese LS. EMBO J. 2003;22:5963–5974. doi: 10.1093/emboj/cdg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leung KF, Baron R, Seabra MC. J. Lipid Res. 2006;47:467–475. doi: 10.1194/jlr.R500017-JLR200. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Seabra MC, Deisenhofer J. Structure. 2000;8:241–251. doi: 10.1016/s0969-2126(00)00102-7. [DOI] [PubMed] [Google Scholar]
- 9.Sjogren AK, Andersson KM, Liu M, Cutts BA, Karlsson C, Wahlstrom AM, Dalin M, et al. J. Clin. Invest. 2007;117:1294–1304. doi: 10.1172/JCI30868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim KH, Baines AT, Fiordalisi JJ, Shipitsin M, Feig LA, Cox AD, Der CJ, Counter CM. Cancer Cell. 2005;7:533–545. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 11.Lim KH, O’Hayer K, Adam SJ, Kendall SD, Campbell PM, Der CJ, Counter CM. Curr. Biol. 2006;16:2385–2394. doi: 10.1016/j.cub.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 12.Falsetti SM, Wang DA, Peng H, Carrico D, Cox AD, Der CJ, Hamilton AD, Sebti SM. Mol. Cell Biol. 2007;27:8003–8014. doi: 10.1128/MCB.00057-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue W, Krasnitz A, Lucito R, Sordella R, Vanaelst L, Cordon-Cardo C, Singer S, et al. Genes Dev. 2008;22:1439–1444. doi: 10.1101/gad.1672608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lahoz A, Hall A. Genes Dev. 2008;22:1724–1730. doi: 10.1101/gad.1691408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dan HC, Jiang K, Coppola D, Hamilton A, Nicosia SV, Sebti SM, Cheng JQ. Oncogene. 2004;23:706–715. doi: 10.1038/sj.onc.1207171. [DOI] [PubMed] [Google Scholar]
- 16.Miquel K, Pradines J, Sun J, Qian Y, Hamilton AD, Sebti SM, Favre G. Cancer Res. 1997;57:1846–1850. [PubMed] [Google Scholar]
- 17.Sun J, Blaskovich MA, Knowles D, Qian Y, Ohkanda J, Bailey RD, Hamilton AD, Sebti SM. Cancer Res. 1999;59:4919–4926. [PubMed] [Google Scholar]
- 18.Sun J, Ohkanda J, Coppola D, Yin H, Kothare M, Busciglio B, Hamilton AD, Sebti SM. Cancer Res. 2003;63:8922–8929. [PubMed] [Google Scholar]
- 19.Lu J, Chan L, Fiji HD, Dahl R, Kwon O, Tamanoi F. Mol. Cancer Ther. 2009;8:1218–1226. doi: 10.1158/1535-7163.MCT-08-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kho Y, Kim SC, Jiang C, Barma D, Kwon SW, Cheng J, Jaunbergs J, et al. Proc. Natl. Acad. Sci. USA. 2004;101:12479–12484. doi: 10.1073/pnas.0403413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saxon E, Luchansky SJ, Hang HC, Yu C, Lee SC, Bertozzi CR. J. Am. Chem. Soc. 2002;124:14893–14902. doi: 10.1021/ja027748x. [DOI] [PubMed] [Google Scholar]
- 22.Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Proc. Natl. Acad. Sci. USA. 2002;99:19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. Nat. Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 24.Yang SH, Bergo MO, Toth JI, Qiao X, Hu Y, Sandovl S, Meta M, et al. Proc. Natl. Acad. Sci. USA. 2005;102:10291–10296. doi: 10.1073/pnas.0504641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang SH, Meta M, Qiao X, Frost D, Bauch J, Coffinier C, Majumdar S, et al. J. Clin. Invest. 2006;116:2115–2121. doi: 10.1172/JCI28968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies BSJ, Yang SH, Farber E, Lee R, Buck SB, Andres DA, Spielmann HP, et al. J. Lipid Res. 2009;50:126–134. doi: 10.1194/jlr.M800424-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinensky M, Beck LA, Leonard S, Evans R. J. Biol. Chem. 1990;265:19937–19941. [PubMed] [Google Scholar]
- 28.Kim R, Rine J, Kim SH. Mol. Cell. Biol. 1990;10:5945–5949. doi: 10.1128/mcb.10.11.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paganini S, Guidetti GF, Catricalà S, Trionfini P, Panelli S, Balduini C, Torti M. Biochimie. 2006;88:285–295. doi: 10.1016/j.biochi.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 30.Debusk FL. J. Pediatr. 1972;80:697–724. doi: 10.1016/s0022-3476(72)80229-4. [DOI] [PubMed] [Google Scholar]
- 31.Agnew B, Nyberg T, Hart C, Lakshmanaswamy R. 55th ASMC Conference; 2007. Abstract TP274. [Google Scholar]
- 32.Reid TS, Terry KL, Casey PJ, Beese LS. J. Mol. Biol. 2004;343:417–433. doi: 10.1016/j.jmb.2004.08.056. [DOI] [PubMed] [Google Scholar]
- 33.Maurer-Stroh S, Eisenhaber F. Genome Biol. 2005;6:R55. doi: 10.1186/gb-2005-6-6-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maurer-Stroh S, Koranda M, Benetka W, Schneider G, Sirota FL, Eisenhaber F. PLoS Comput. Biol. 2007;3:e66. doi: 10.1371/journal.pcbi.0030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hancock JF. Methods Enzymol. 1995;255:60–65. doi: 10.1016/s0076-6879(95)55009-7. [DOI] [PubMed] [Google Scholar]
- 36.Wilson AL, Maltese WA. Methods Enzymol. 1995;250:79–91. doi: 10.1016/0076-6879(95)50064-2. [DOI] [PubMed] [Google Scholar]
- 37.Wang DA, Sebti SM. J. Biol. Chem. 2005;280:19243–19249. doi: 10.1074/jbc.M411472200. [DOI] [PubMed] [Google Scholar]
- 38.Benetka W, Koranda M, Maurer-Stroh S, Pittner F, Eisenhaber F. Biomed. Chromatogr. Biochem. 2006;7:6. doi: 10.1186/1471-2091-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen UT, Cramer J, Gomis J, Reents R, Gutierrez-Rodriguez M, Goody RS, Alexandrov K, Waldmann H. Chembiochem. 2007;8:408–423. doi: 10.1002/cbic.200600440. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen UT, Guo Z, Delon C, Wu Y, Deraeve C, Fränzel B, Bon RS, et al. Nat. Chem. Biol. 2009;5:227–235. doi: 10.1038/nchembio.149. [DOI] [PubMed] [Google Scholar]
- 41.Laliberté JF, Dagenais A, Filion M, Bibor-Hardy V, Simard R, Royal A. J. Cell Biol. 1984;98:980–985. doi: 10.1083/jcb.98.3.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laezza C, Di Marzo V, Bifulco M. Proc. Natl. Acad. Sci. USA. 1998;95:13646–13651. doi: 10.1073/pnas.95.23.13646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo Z, Yuan J, Tang W, Chen X, Gu X, Luo K, Wang Y, et al. Mol. Biol. Rep. 2007;34:137–144. doi: 10.1007/s11033-006-9023-9. [DOI] [PubMed] [Google Scholar]