Abstract
When naive CD4+ Th cells encounter cognate pathogen-derived Ags they expand and develop the capacity to express the appropriate effector cytokines for neutralizing the pathogen. Central to this differentiation process are epigenetic modifications within the effector cytokine genes that allow accessibility to the transcriptional machinery. In contrast, when mature self-reactive CD4 cells encounter their cognate epitopes in the periphery they generally undergo a process of tolerization in which they become hyporesponsive/anergic to antigenic stimulation. In the current study, we used a TCR transgenic adoptive transfer system to demonstrate that in a dose-dependent manner parenchymal self-Ag programs cognate naive CD4 cells to acetylate histones bound to the promoter region of the Ifng gene (which encodes the signature Th1 effector cytokine) during peripheral tolerization. Although the Ifng gene gains transcriptional competence, these tolerized CD4 cells fail to express substantial amounts of IFN-γ in response to antigenic stimulation apparently because a blockage in TCR-mediated signaling also develops. Nevertheless, responsiveness to antigenic stimulation is partially restored when self-Ag-tolerized CD4 cells are retransferred into mice infected with a virus expressing the same Ag. Additionally, there is preferential boosting in the ability of these CD4 cells to express IFN-γ relative to other cytokines with expression that also becomes impaired. Taken together, these results suggest that epigenetic modification of the Ifng locus during peripheral CD4 cell tolerization might allow for preferential expression of IFN-γ during recovery from tolerance.
Inflammation induced by infection supports the activation of naive T cells specific for Ags expressed by the infectious agents. These activated T cells then differentiate into effector T cells that express the appropriate effector molecules for neutralizing the pathogen. During this differentiation process, epigenetic modifications are induced in regions of the chromatin that contain genes encoding certain effector molecules, such as IFN-γ in the case of Th1 effector CD4+ T cells. This change in chromatin structure opens these regions so that RNA polymerase and supporting transcription factors can bind to critical cis-acting DNA sequences and thereby promote gene transcription (1–7). In contrast, when naive T cells encounter cognate self-Ags under steady-state conditions they undergo a distinct tolerogenic differentiation program that results in non-responsiveness to antigenic stimulation and thus an inability to express effector functions (8–11).
Although immunogenic and tolerogenic T cell differentiation programs result in diametrically opposed outcomes, there are some common elements. Thus, when naive TCR transgenic clonotypic CD4 cells are induced to undergo either Th1 differentiation or tolerization following adoptive transfer into recipient mice that express cognate vaccinia viral Ag or parenchymal self-Ag, respectively, they exhibit similar proliferative responses upon initially encountering Ag (12, 13). Naive CD4 cells undergoing tolerization also transiently gain the capacity to provide help to CD8 cells (14, 15). The ability of CD4 cells undergoing tolerization to express the Th1 signature cytokine IFN-γ, however, remains substantially lower than the ability in virally primed counterparts throughout their respective differentiation programs (13, 15).
In the current study, we examined the response of naive CD4 cells undergoing self-Ag-induced tolerization at the molecular level. Surprisingly, we found that tolerized CD4 cells exhibit certain features that are normally associated with Th1 effectors. Thus, in direct relation to the amount of self-Ag expression, tolerized CD4 cells contain acetylated histones bound to the promoter region of the Ifng gene and are able to express moderate levels of IFN-γ when stimulated with agents that bypass TCR-proximal signaling steps. Additionally, tolerized CD4 cells express moderate amounts of the Th1 master-regulatory factor T-bet (16) and IL-12Rβ2, both of which are expressed in Th1 cells and contribute to Th1 differentiation and function (7, 17). Although the Ifng locus gains a degree of transcriptional competence during tolerization, it appears that a blockage in TCR-induced signaling substantially dampens Ag-induced IFN-γ expression.
The ability of CD4 cells to open their Ifng locus during peripheral tolerization raised the possibility that they might be able to readily express IFN-γ if they could recover from tolerance, as can occur following removal of the initial source of tolerizing Ag (18, 19). This might be important during certain tumor immunotherapy scenarios such as when tumor vaccines are administered following standard therapies that achieve states of minimal residual disease in which expression of the targeted tumor Ags is diminished to levels insufficient to maintain cognate T cells in a tolerant state (20, 21). Thus, the ability of the Ifng locus to open during peripheral tolerization might subsequently allow such tumor-specific CD4 cells that have undergone tolerance reversal to express IFN-γ more rapidly in response to vaccination.
To directly assess the potential of peripherally tolerized CD4 cells to express IFN-γ in response to immunization after the tolerizing Ag has been removed, naive CD4 cells were first adoptively transferred into recipient mice expressing cognate self-Ag to induce tolerance. Cells were subsequently retransferred into secondary recipients that had been infected with a vaccinia virus that expresses the same Ag. Notably, these virally recovered CD4 cells exhibited an improved ability to express several cytokines including IL-2 and TNF-α, but IFN-γ expression was boosted to the greatest extent.
Materials and Methods
Mice and adoptive transfer
Naive clonotypic CD4 cells that are specific for an I-Ed-restricted epitope deriving from influenza (PR8 strain) hemagglutinin (HA)3 (110SFERFEIFPKE120) were prepared from the CD8-depleted lymph nodes (LN) of 6.5 TCR transgenic mice (22) on the B10.D2 (H-2d) background that express the Thy1.1+ congenic marker. These clonotypic CD4 cells were labeled with the fluorescent tracking dye CFSE immediately before adoptive transfer into the indicated recipient mice as previously described (13). A total of 2 × 106 clonotypic CD4 cells were transferred unless otherwise indicated. The 6.5 clonotypic adoptive transfer recipients on the B10.D2 Thy1.2+ congenic background included C3-HA transgenic mice that express HA under the control of the rat C3(1) promoter, which directs HA expression in a variety of parenchymal tissues at either a low amount of HA (self-HAlow) (23) or a high amount (self-HAhigh) (12). Recipients also included nontransgenic mice infected with a recombinant vaccinia virus that expresses HA (viral-HA) as previously described (13). TEa TCR transgenic mice on the B6 (H-2b) Thy1.1+ background, provided by Dr. A. Rudensky (University of Washington School of Medicine, St. Louis, MO) express a clonotypic TCR that is specific for an I-Ed-derived (Eα) peptide (52AS-FEAQGALANIAVDKA68) presented by I-Ab (24). Naive TEa clonotypic CD4 cells were prepared from CD8-depleted LN of TEa TCR transgenic mice. Cells were labeled with CFSE before adoptive transfer into the indicated B6 Thy1.2+ recipients and challenged with an i.p. bolus of 250 µg of soluble Eα peptide. Adoptive retransfer experiments were performed by pooling spleen samples from the indicated primary adoptive transfer recipients, relabeling them with CFSE, and retransferring aliquots containing 1 × 106 clonotypic CD4 cells into the indicated secondary recipients as previously described (25, 26). The Animal Care and Use Committee of the University of Connecticut Health Center approved all mouse protocols used in this study.
Flow cytometry
The functional response of 6.5 and TEa clonotypic CD4 cells were analyzed by FACS following recovery from spleens of the indicated adoptive transfer recipients as previously described (13, 26, 27). In short, clonotypic CD4 cells (identified as CD4+Thy1.1+) were analyzed for frequency and dilution of CFSE directly ex vivo. Intracellular cytokine staining was performed by stimulating the indicated spleen samples with either the appropriate peptide (100 µg/ml) or PMA plus ionomycin in the presence of brefeldin A for 5 h, and subsequently staining with anti-Thy1.1, anti-CD4, and the appropriate anti-cytokine mAbs following fixation and permeabilization. Cytokine expression profiles were generated by gating on CD4+Thy1.1+ cells that exhibited diluted CFSE fluorescence (refer to Fig. 1A). Total cytokine expression is expressed in arbitrary units, calculated by multiplying the percentage of cytokine-expressing clonotypic CD4 cells (determined using isotype staining controls) by the corresponding mean fluorescence intensity value as previously described (13). The same FACS analysis criteria were used in both single and double (i.e., retransfer) adoptive transfer experiments.
FIGURE 1.
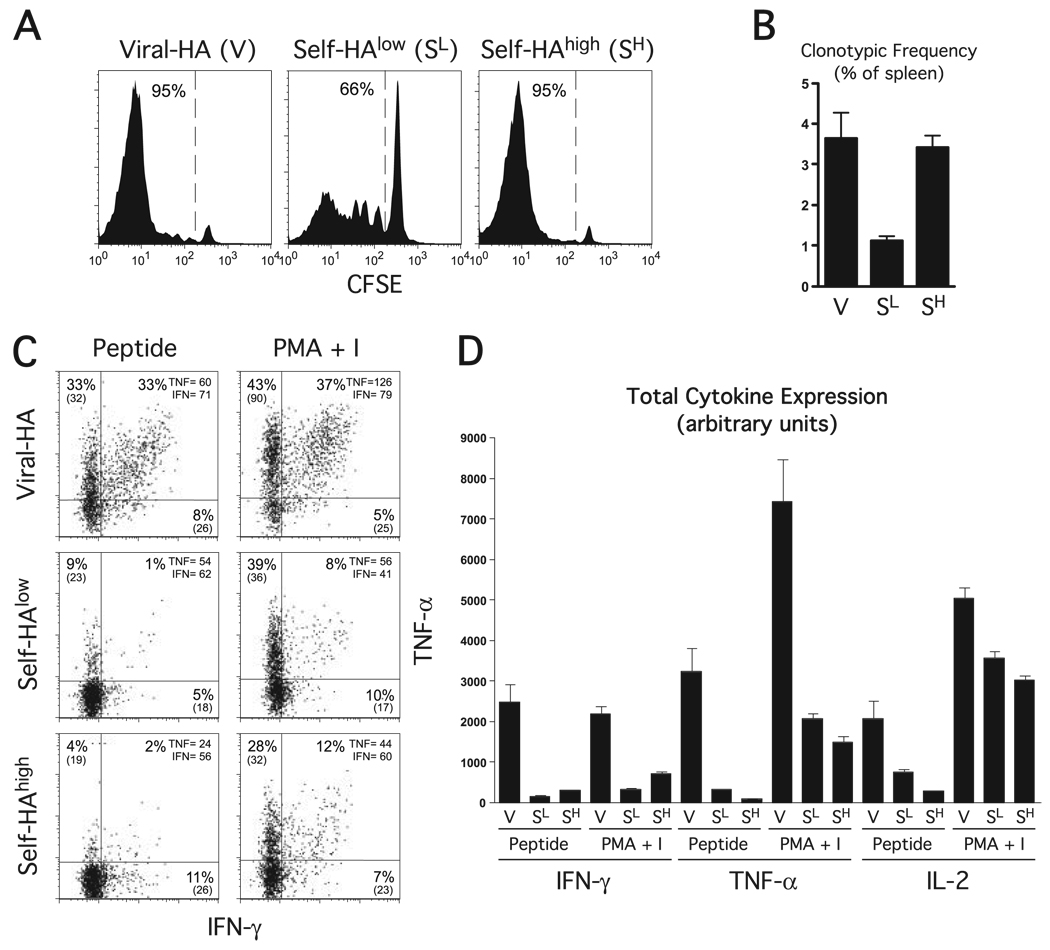
Functional response and properties of naive clonotypic CD4 cells exposed to cognate viral or self-Ag. CFSE-labeled naive Thy1.1 + HA-specific clonotypic CD4 cells were adoptive transferred into Thy1.2+ viral-HA (V), self-HAlow (SL), or self-HAhigh (SH) recipients and recovered from spleens 6 days later for analysis. A, In vivo proliferation of clonotypic CD4 cells. Representative CFSE dilution histograms with the percentage of clonotypic CD4 cells displaying diluted CFSE fluorescence indicated. B, The frequency of clonotypic CD4 cells is expressed as a percentage within the total splenocyte population. C, The ability of clonotypic CD4 cells exposed to cognate viral and self-Ag to express cytokines in response to in vitro restimulation. Plots representative of intracellular IFN-γ vs TNF-α expression following in vitro restimulation with HA peptide or PMA plus ionomycin (PMA + I), with the percentage of clonotypic CD4 cells expressing each cytokine and the corresponding mean fluorescence intensity (MFI) value indicated. D, Quantitation of cytokine expression shown in C. Total intracellular IFN-γ, TNF-α, and IL-2 is expressed in arbitrary units, and was calculated by multiplying the percentage of cytokine-expressing clonotypic CD4 cells by the corresponding mean fluorescence intensity. Data are expressed as the mean ± SEM for n= 4 recipients per group.
Isolation of clonotypic CD4 cells, RT-PCR, and chromatin immunoprecipitation (ChIP)
Clonotypic CD4 cells were isolated from recipient spleens either by positively selecting Thy1.1+ cells using MACS columns as previously described (27) (for a portion of the self-HAhigh and viral-HA samples) or by FACS sorting CD4+Thy1.1+ CFSE-diluted cells using a FACSVantage SE cell sorter (BD Biosciences) (for all of the self-HAlow samples and the remaining self-HAhigh and viral-HA samples). Each self-HAlow sample contained spleens pooled from three separate mice, whereas self-HAhigh and viral-HA samples each contained a spleen from a single animal. RT-PCR and ChIP data obtained from self-HAhigh and viral-HA counterparts isolated by the different methods were comparable (data not shown). Clonotypic CD4 cell samples were analyzed directly ex vivo (i.e., without in vitro restimulation), and each sample was divided and analyzed by real-time PCR-based RT-PCR and ChIP assays to measure T-bet and IL-12Rβ2 mRNA and histone H3 acetylation at the Ifng and Tnfa promoters, respectively, using our previously established protocols and PCR primers (27). In short, after isolating RNA and reverse transcribing cDNA, T-bet and IL-12Rβ2 mRNAs were quantified by first normalizing for input sample amounts by calculating the difference in threshold cycle, CT, values relative to HPRT message, and then using the ΔΔ threshold cycle method to calculate the ratio between experimental samples and a representative naive CD4 cell sample. Similarly, after precipitating sheared genomic DNA-chromatin complexes with anti-acetylated histone H3, the extent of histone H3 acetylation at the Ifng and Tnfa promoters was determined by first normalizing to the CD3ε enhancer (Δ threshold cycle), and then calculating the ratio between individual experimental samples and a representative naive CD4 cell sample (ΔΔCT).
Results
Parenchymal self-Ag can induce histone acetylation within the Ifng promoter region in naive CD4 cells undergoing peripheral tolerization
We have previously developed a TCR transgenic adoptive transfer system to study CD4 cell peripheral tolerization to parenchymal self-Ag vs Th1 differentiation to viral Ag (12, 13). Thus, naive clonotypic CD4 cells specific for an I-Ed-restricted epitope of influenza HA obtained from 6.5 TCR transgenic donor mice (22) on the B10.D2 (H-2d) Thy1.1+ background are labeled with the fluorescent tracking dye CFSE and adoptively transferred into B10.D2 Thy1.2+ recipient mice, and the functional response of these clonotypic CD4 cells is subsequently monitored via FACS using the congenic Thy1.1 marker. We previously found that when naive clonotypic CD4 cells are transferred into C3-HA transgenic recipients that express HA as a generic parenchymal self-Ag at either low (self-HAlow, 142 founder line) or high (self-HAhigh, 137 founder line) amounts they initially undergo a vigorous proliferative response in secondary lymphoid organs as indicated by the dilution of CFSE fluorescence (albeit proliferation is more robust in self-HAhigh recipients). Despite this initial proliferative phase, clonotypic CD4 cells recovered from both self-HAlow and self-HAhigh recipients are impaired in their ability to undergo further proliferation and to express a variety of cytokines in response to antigenic restimulation, and are therefore referred to as tolerized or functionally impaired. In contrast, when naive clonotypic CD4 cells are adoptively transferred into nontransgenic viral-HA recipients, they proliferate vigorously and differentiate into Th1 effectors that are capable of expressing IFN-γ, TNF-α, and IL-2 (12, 13, 26). Consistent with these previous findings, in the current study, when clonotypic CD4 cells were recovered from spleens 6 days following adoptive transfer into either viral-HA or self-HAhigh recipients, proliferation (i.e., CFSE dilution) (Fig. 1A) and accumulation (Fig. 1B) were similarly robust. Proliferation and accumulation was less pronounced in self-HAlow recipients. Also consistent with our previous studies (12, 13, 26), the virally primed clonotypic CD4 cells displayed a classic Th1 effector phenotype marked by the ability to express intracellular IFN-γ, TNF-α, and IL-2 following in vitro restimulation with HA peptide, whereas the self-HAhigh tolerized clonotypic CD4 cells expressed severely reduced levels of all three cytokines (Fig. 1, C and D). Interestingly, although clonotypic CD4 cells recovered from self-HAlow recipients also displayed a tolerized phenotype marked by low cytokine expression potentials, the pattern was distinct from self-HAhigh recipients. Thus, although IFN-γ expression potential was substantially lower in both self-HAlow (p = 0.02 by unpaired two-tailed t test) and self-HAhigh (p = 0.003) recipient groups compared with viral-HA recipients, it was 2-fold greater in self-HAhigh than in self-HAlow recipients (p = 0.02). Conversely, TNF-α (p = 0.0001) and IL-2 (p = 0.0006) expression potentials were at least 2-fold higher in self-HAlow recipients compared with self-HAhigh recipients. Ruling out the possibility that these different patterns of peptide-induced intracellular cytokine expression in the clonotypic CD4 cells recovered from the three recipient groups were due to differences in APC activities, splenic APCs isolated from each recipient group exhibited comparable abilities to stimulate cytokine expression in clonotypic CD4 cells isolated from all three sample groups (Fig. 2). Taken together, these data indicate that greater levels of self-Ag induce a more profound inability to express TNF-α and IL-2, whereas IFN-γ expressing potential is most profoundly impaired following exposure to lower levels of self-Ag.
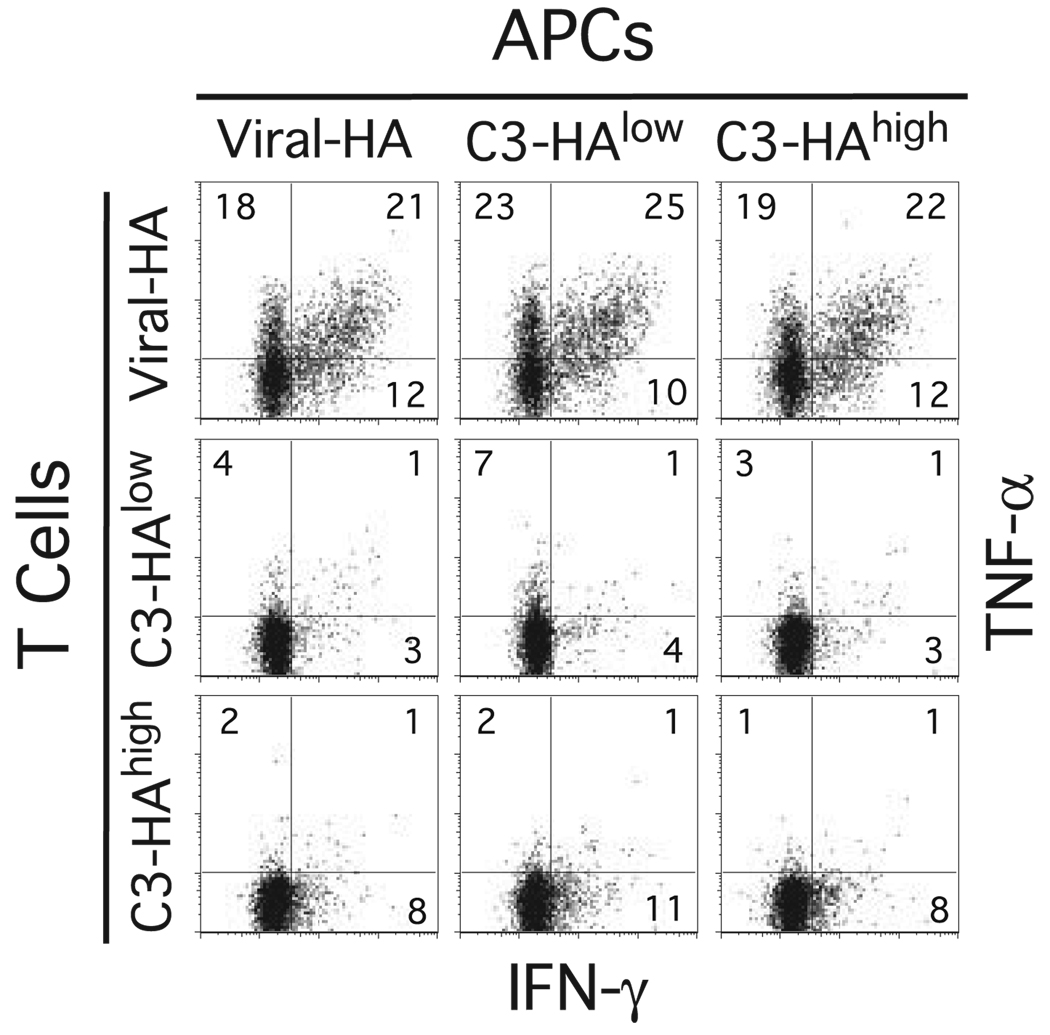
FIGURE 2.
Splenic APCs recovered from both viral-HA and self-HA adoptive transfer recipients are equivalent in their abilities to stimulate clonotypic CD4 cells to express intracellular cytokines in vitro. Spleens recovered from viral-HA, self-HAlow, and self-HAhigh recipients 6 days after receiving adoptive transfers of naive clonotypic CD4 cells were fractionated into T cell (depleted of APCs using magnetic beads) and APC (depleted of T cells using magnetic beads) fractions, remixed in the nine possible APC to T cell combinations at a 1:1 ratio, and incubated with HA peptide followed by intracellular cytokine staining. Data shown are representative of two independent experiments.
To begin analyzing the intrinsic defects responsible for impaired cytokine expression potential in self-Ag-tolerized clonotypic CD4 cells, we asked whether cytokine expression could be rescued through bypassing TCR-proximal signaling steps using PMA plus ionomycin, which directly activate protein kinase C and releases intracellular Ca2+, respectively. PMA plus ionomycin rescued IL-2 expression in tolerized CD4 cells recovered from both self-HAlow and self-HAhigh recipients almost to the level observed in virally primed counterparts, indicating that impaired IL-2 expression potential is mostly conferred via a blockage in signaling steps that are proximal to the TCR. PMA plus ionomycin also rescued TNF-α expression in both tolerized clonotypic CD4 cell populations, albeit only to about one-fourth the level of virally primed effectors, indicating that both TCR-proximal and –distal signaling defects operate to diminish TNF-α expression potential in tolerized CD4 cells (Fig. 1, C and D). Notably, PMA plus ionomycin elicited increased IFN-γ expression in self-HAhigh-tolerized CD4 cells relative to expression observed in peptide restimulated counterparts (p = 0.0005) that reached about one-third the level observed in viral-HA counterparts. Because naive clonotypic CD4 cells are unable to express appreciable levels of IFN-γ in response to PMA plus ionomycin stimulation (27), and because the chromatin structure of the Ifng gene locus exists in a condensed configuration that is inaccessible to the transcriptional machinery (7, 17, 28), this result suggested that self-Ag causes the Ifng gene locus in cognate naive CD4 cells to undergo some degree of chromatin/epigenetic modification, but that these tolerized cells are nevertheless unable to express substantial levels of IFN-γ in response to antigenic stimulation due to a blockage in signaling steps that are proximal to the TCR.
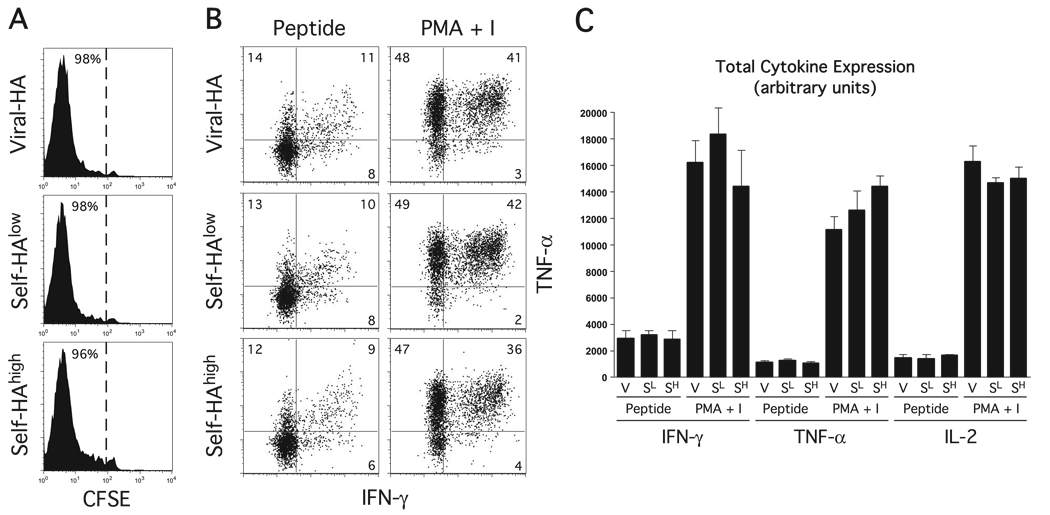
PMA plus ionomycin also stimulated greater IFN-γ expression in self-HAlow-tolerized clonotypic CD4 cells compared with stimulation with peptide-pulsed APCs (p = 0.0001), although the level remained ~2.5-fold lower compared with self-HAhigh counterparts (p = 0.002). Because the self-HAlow and self-HAhigh transgenic mice contain the same transgene expression cassette and display a similar tissue distribution of HA expression, but differ in the amounts of HA protein that they express by ~1000-fold (12), we suspected that the enhanced IFN-γ expression potential in clonotypic CD4 cells recovered from self-HAhigh relative to self-HAlow recipients resulted from greater in vivo stimulation of the clonotypic HA-specific TCR. We could not rule out the possibility, however, that the clonotypic CD4 cell responses in these two mice were also influenced by some other immunological parameter, perhaps resulting from the different genomic sites of transgene integration. To assess this possibility, the response of a naive clonotypic CD4 cell population responding to a tolerogenic form of Ag that is unrelated to HA was compared in these two transgenic founder lines. More specifically, self-HAlow and self-HAhigh transgenic mice backcrossed from the original B10.D2 (H-2d, HA-restricting) genetic background to the B6 (H-2b, HA-nonrestricting) background (which is similar to B10.D2 excepting the MHC locus) received adoptive transfers of naive CFSE-labeled TEa TCR transgenic clonotypic CD4 cells that recognize an I-Ed-derived peptide presented by I-Ab (24), and were subsequently challenged with a bolus of cognate soluble Eα peptide. Viral-HA-infected nontransgenic B6 recipients were also included as a control. In all three recipient groups, the TEa CD4 cells underwent comparable proliferative responses (Fig. 3A) and developed similar capacities to express IFN-γ, TNF-α (Fig. 3, B and C), and IL-2 (Fig. 3C). The observation that these HA-unrelated TEa responses did not differ between self-HAlow and self-HAhigh transgenic recipients suggests that these two transgenic founder lines do not differ in their intrinsic CD4 cell priming/tolerization machinery, further suggesting that the difference in HA-specific clonotypic CD4 cell cytokine expression potential that develops in self-HAlow vs self-HAhigh recipients (Fig. 1) is the consequence of differences in the amounts of HA Ag expression.
FIGURE 3.
Self-HAlow and self-HAhigh transgenic mice do not differ in their intrinsic CD4 cell priming/tolerization capacity. Naive CFSE-labeled TEa clonotypic CD4 cells on the B6 (H-2b) Thy1.1+ background that are specific for an I-Ed-derived peptide (Eα) presented by I-Ab were adoptively transferred into self-HAlow, self-HAhigh, or viral-HA recipients that had been backcrossed to the B6 Thy1.2+ background. These adoptive transfer recipients were then challenged with a bolus of soluble Eα peptide, and recovered from spleens 6 days later for analysis. FACS plots from n= 3 recipients per group represent CFSE dilution (A) and intracellular IFN-γ vs TNF-α expression following in vitro restimulation with Eα peptide or PMA plus ionomycin (PMA + I) (B). C, Quantification of cytokine expression in B is total intracellular cytokines expressed in arbitrary units and calculated as described in Fig. 1.
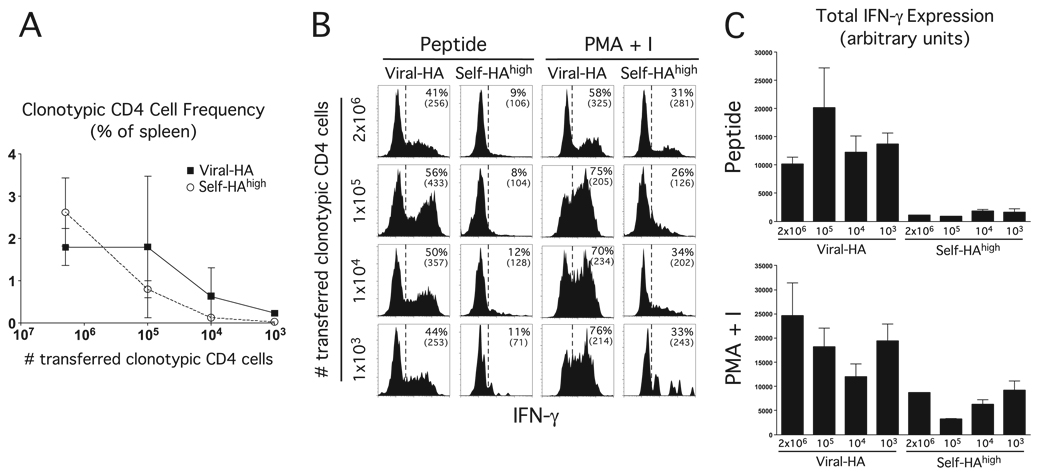
It has recently been shown in various TCR transgenic adoptive transfer systems that the functional outcome of antigenic priming can be influenced by the number of transferred clonotypic T cells (29–31). Given our unexpected finding that when over a million naive clonotypic CD4 cells are induced to undergo peripheral tolerization following adoptive transfer into self-HAhigh recipients, they develop some potential to express IFN-γ, we wished to determine whether the same outcome would occur when the number of clonotypic CD4 cells more closely resembles physiological conditions. Thus, a titrated number of naive clonotypic CD4 cells were transferred into self-HAhigh or viral-HA controls, and subsequently recovered from spleens 6 days later to assess their potential to express IFN-γ following in vitro restimulation with HA peptide vs PMA plus ionomycin (Fig. 4). The number of transferred clonotypic CD4 cells included: 2 × 106 (the number used in the previous experiments), 1 × 105, 1 × 104, and 1 × 103 (a number previously shown (31) to approximate a physiological precursor frequency). In viral-HA recipients, the frequency of expanded clonotypic CD4 cells remained stable when the number of transferred naive cells was reduced from 2 × 106 to 1 × 105 (~2% of the total spleen), but progressively decreased thereafter, reaching a frequency of ~0.2% when 1 × 103 naive clonotypic CD4 cells were transferred. The frequency of expanded clonotypic CD4 cells in self-HAhigh recipients was more severely affected by reducing the input number of naive cells, although expanded cells were still detectable (~0.03%) when 1 × 103 naive clonotypic CD4 cells were transferred (Fig. 4A). The ability of these expanded clonotypic CD4 cells to express IFN-γ, however, was not appreciably affected by the input number of naive cells (Fig. 4, B and C). Most notably, clonotypic CD4 cells recovered from self-HAhigh recipients that had received 1 × 103 naive input cells were able to express the same level of IFN-γ following restimulation with PMA plus ionomycin compared with self-HAhigh recipients that had received 2 × 106 naive input cells (p = 0.8). This result suggests that the development of IFN-γ expression potential in tolerized clonotypic CD4 cells under our standard experimental conditions does recapitulate physiological conditions.
FIGURE 4.
The ability of clonotypic CD4 cells exposed to cognate self-Ag to express IFN-γ is not influenced by the number of adoptively transferred naive clonotypic CD4 cells. The indicated number of naive clonotypic CD4 cells were adoptively transferred into viral-HA or self-HAhigh recipients, and recovered from spleens 6 days later for analysis. A, Frequency of clonotypic CD4 cells is expressed as the percentage of total splenocytes. FACS plots (B) and quantification (C) representative of intracellular IFN-γ expression following in vitro restimulation with HA peptide or PMA plus ionomycin from n= 3 recipients per group.
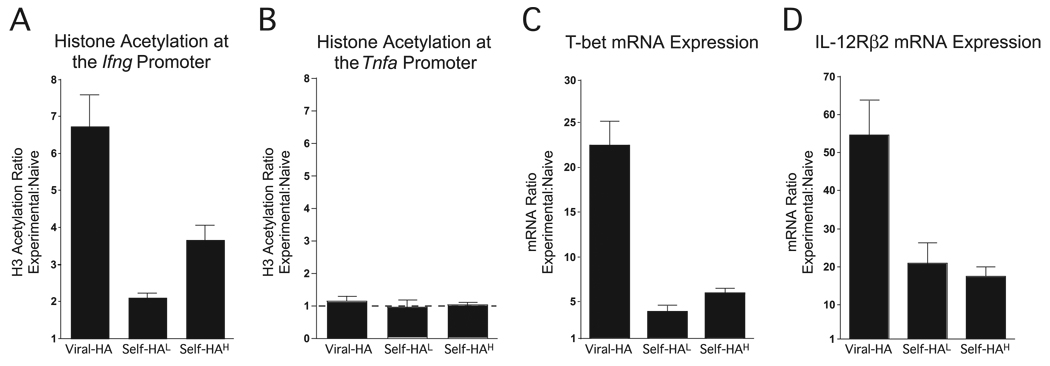
The development of competence to express IFN-γ in CD4 cells involves epigenetic modification of the Ifng gene locus that allows accessibility to the transcriptional machinery, an early and well-correlated modification being acetylation of histone H3 in the proximal promoter region (7, 28). Thus, to directly confirm that the ability of self-Ag-tolerized clonotypic CD4 cells to express IFN-γ in response to PMA plus ionomycin stimulation does in fact indicate that the chromatin structure of the Ifng locus can open during tolerization, the extent of histone H3 acetylation at the proximal promoter was measured using a real-time PCR-based ChIP assay (27) (Fig. 5A). As shown in our previous work (27), Th1 effector clonotypic CD4 cells recovered from viral-HA recipients exhibited a 6.5-fold increase in the extent of histone H3 acetylation at the Ifng proximal promoter relative to naive controls (where the Ifng promoter is bound to nonacetylated histones (7, 28)). Substantial histone H3 acetylation was also observed in clonotypic CD4 cells recovered from self-HAhigh recipients, and although the magnitude was only 3.5-fold greater than naive cells, the relative differences in histone acetylation at the Ifng promoter and the ability to express IFN-γ in response to PMA plus ionomycin stimulation were roughly similar when comparing viral-HA vs self-HAhigh recipients (compare Fig. 5A to Fig. 1D). Histone acetylation at the Ifng promoter was also evident in clonotypic CD4 cells recovered from self-HAlow recipients, although it was only 2-fold greater than naive cells (Fig. 5A), once again correlating with the weaker ability of these CD4 cells to express IFN-γ in response to PMA plus ionomycin stimulation (refer to Fig. 1D).
FIGURE 5.
CD4 cells that have undergone peripheral tolerization in response to cognate self-Ag display acetylated histone H3 bound to the Ifng promoter region and also exhibit other characteristics normally associated with Th1 effectors. Clonotypic CD4 cell samples isolated from viral-HA (n = 9 recipients), self-HAlow (self-HAL, n = 3), each determination deriving from three pooled recipient samples), and self-HAhigh (self-HAH, n = 11 recipients) were divided and analyzed by quantitative real-time PCR-based ChIP to measure histone H3 acetylation at the Ifng (A) and Tnfa (B) proximal promoters. Quantitative real-time RT-PCR to measure T-bet (C) and IL-12Rβ2 (D) mRNAs. Data are expressed as a ratio relative to naive clonotypic CD4 cells.
We have previously shown that histone H3 acetylation at the Tnfa promoter is comparable in naive, Th1 effector and tolerized Th1 effector clonotypic CD4 cells (27). In the current study, we found that H3 acetylation at the Tnfa promoter also did not differ between clonotypic CD4 cells recovered from virally primed Th1 effector and self-HAlow or self-HAhigh-tolerized clonotypic CD4 cells (Fig. 5B). Taken together, these results suggest that while in tolerized CD4 cells the extent of opening in the Ifng locus influences their ability to express IFN-γ, epigenetic modification of the Tnfa locus might not play a role in regulating TNF-α expression during tolerization.
Because opening of the Ifng locus is normally associated with Th1 differentiation, we wished to assess whether tolerized CD4 cells exhibit other similarities to Th1 effectors. Both self-HAlow and self-HAhigh-tolerized clonotypic CD4 cells expressed increased amounts (relative to naive CD4 cells) of mRNA encoding for the Th1 master regulatory factor T-bet (16), albeit T-bet mRNA in tolerized CD4 cells did not reach the level observed in virally primed Th1 effectors (Fig. 5C). Tolerized CD4 cells also expressed appreciable mRNA levels corresponding to another Th1 differentiation factor, IL-12Rβ2 (17, 32), although once again IL-12Rβ2 mRNA levels in tolerized CD4 cells were intermediate to those observed in naive and virally primed Th1 effector counterparts (Fig. 5D). Taken together, these data indicate that tolerized CD4 cells can exhibit multiple similarities to bona fide Th1 effectors.
Opening of the Ifng locus during tolerization is associated with preferential boosting of IFN-γ expression in response to subsequent immunization
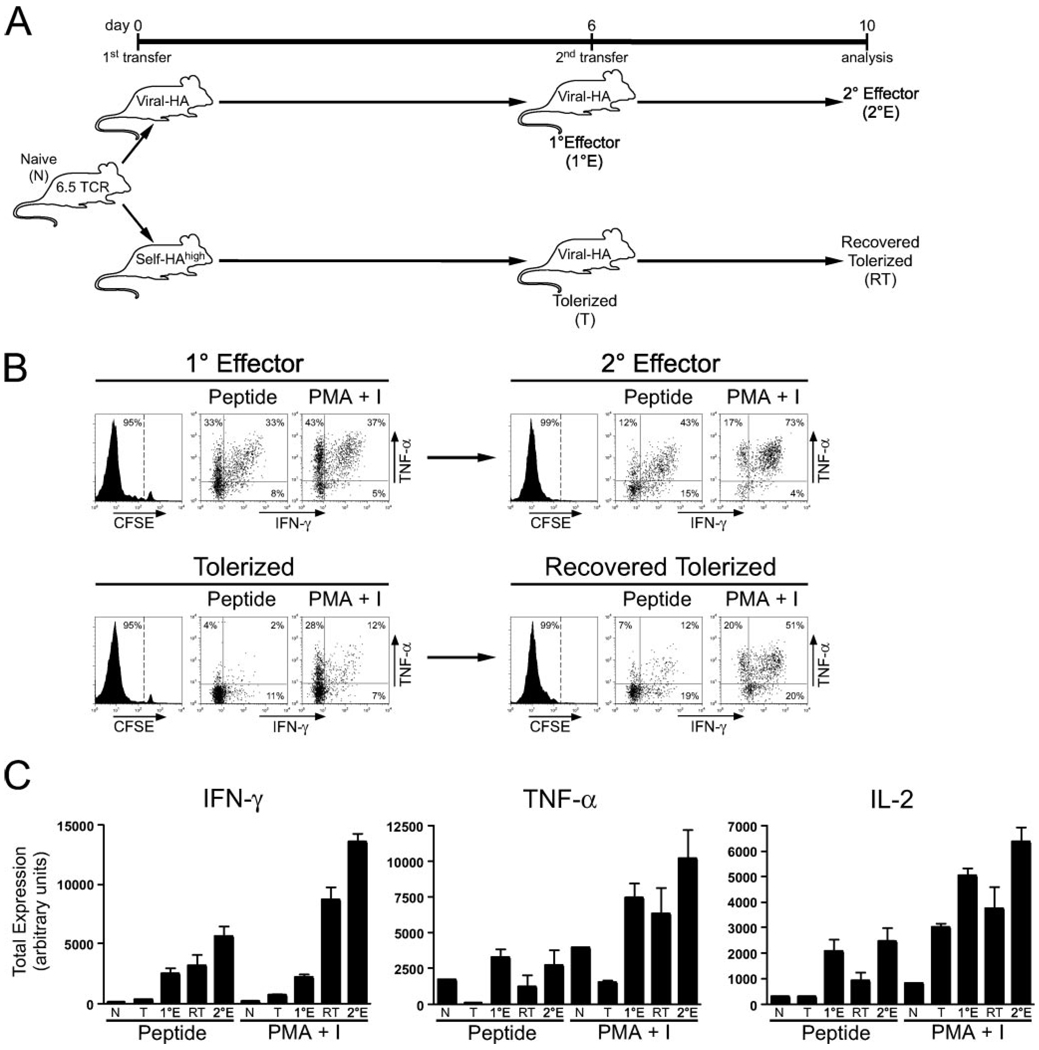
Our observation that tolerized CD4 cells contain acetylated histones bound to the Ifng promoter region was rather interesting, particularly because these cells remain substantially impaired in their ability to express this cytokine in response to antigenic stimulation apparently because a blockage in TCR-mediated signaling also develops. Because it has been reported that tolerized CD4 cells can regain responsiveness to TCR stimulation following removal of the initial source of tolerizing Ag (18, 19), we reasoned that opening of the Ifng locus during peripheral tolerization might subsequently allow such functionally recovered tolerized CD4 cells to express IFN-γ more rapidly if they are challenged with an immunogenic form of the same Ag. To test this possibility, naive clonotypic CD4 cells were first transferred into C3-HAhigh and viral-HA recipients to induced tolerance and Th1 differentiation, respectively, and then recovered from spleens and relabeled with CFSE before retransfer into secondary recipients that had been infected with viral-HA (Fig. 6).
FIGURE 6.
Viral priming preferentially boosts the ability of tolerized CD4 cells to express IFN-γ. Viral-HA-primed clonotypic CD4 cells (primary effectors or 1°E) and self-HAhigh-tolerized (T) clonotypic CD4 cells (both pooled from the samples shown in Fig. 1) were relabeled with CFSE and retransferred into viral-HA-infected secondary recipients (1 × 106 clonotypic CD4 cells per secondary recipient) to generate secondary effector (2°E) and recovered tolerized (RT) clonotypic CD4 cells, respectively. A, Diagram of the retransfer experimental schema. B, Plots of representative CFSE dilution and intracellular IFN-γ vs TNF-α expression. C, Total intracellular IFN-γ, TNF-α, and IL-2 expression is expressed in arbitrary units and was calculated as described in Fig. 1. Note that data corresponding to 1°E and tolerized clonotypic CD4 cells are the same as in Fig. 1 for n= 4 recipients per group.
Consistent with our previous findings (27), when clonotypic CD4 cells initially primed with viral-HA were retransferred into a second set viral-HA-infected recipients, they underwent a second round of vigorous CFSE-dilution and further improved their ability to express IFN-γ in response to both HA peptide (p = 0.02) and PMA plus ionomycin (p = 0.0001) restimulation, suggesting the possibility that their Ifng locus underwent further epigenetic modification. Notably, although the clonotypic CD4 cells that were initially transferred into C3-HAhigh recipients were no longer undergoing cell cycle progression (only 7.6% ± 1.3 (mean ± SEM) were undergoing blastogenesis) despite the continual expression of the transgenic Ag that initiated the transient proliferative response, following retransfer into viral-HA secondary recipients these tolerized CD4 cells did in fact undergo comparable CFSE dilution (Fig. 6B) and accumulation (p = 1.0) as the control secondary effectors that had encountered viral-HA for the second time, indicating that at least some degree of functional recovery had taken place. Viral priming also improved the ability of the tolerized CD4 cells to express IL-2 (p = 0.04) and TNF-α (p = 0.15) in response to peptide stimulation, although these levels remained ~2–3-fold lower compared with control primary and secondary effectors (in all cases p > 0.05) (Fig. 6, A and B). Given that impaired IL-2 expression in tolerized CD4 cells is mediated mostly via a blockage in TCR-proximal signaling (Fig. 1), and that IL-2 expression elicited by peptide stimulation in these virally recovered tolerized CD4 cells only reached 50% the levels observed in controls that had not been exposed to self-Ag (Fig. 6C), these data suggested that that the TCR-proximal signaling blockage had only been partially repaired. Nevertheless, HA peptide-stimulated IFN-γ expression in the virally recovered tolerized CD4 cells was comparable to control primary effectors (p = 0.5). Additionally, PMA plus ionomycin stimulation elicited much greater IFN-γ expression in the recovered tolerized CD4 cells than in primary effectors (p = 0.0008), suggesting the possibility that viral priming further augmented epigenetic modifications within the Ifng locus that were initiated during tolerization. That the recovered tolerized CD4 cells were able to express greater levels of IFN-γ than the primary effectors in response to PMA plus ionomycin, but not peptide, stimulation confirmed that the TCR-proximal signaling blockage had not been fully repaired.
Taken together, the finding that Ag-induced IFN-γ, but not IL-2 and TNF-α, expression potential was boosted by viral priming to the level of control primary effectors despite the lingering partial blockage in TCR-proximal signaling is consistent with the notion that epigenetic modification of the Ifng locus during peripheral tolerization allows for preferential expression of IFN-γ during viral-mediated functional recovery.
Discussion
Naive CD4+ Th cells that are primed by pathogen-derived Ags are programmed to differentiate into effectors that are capable of producing the appropriate effector cytokines for neutralizing the pathogen. This capability in turn is regulated through epigenetic modification of the effector cytokine gene loci from a condensed to an open configuration that is competent for transcription (1, 7, 28). In contrast, mature self-reactive T cells generally undergo a process of peripheral tolerization in which they become unable to express effector cytokines (33, 34). Our current result that the Ifng gene locus opens in CD4 cells that are undergoing tolerization in response to cognate self-Ag was therefore somewhat unexpected.
The extent to which the Ifng gene opens in tolerized CD4 cells increases in relation to the amount of cognate self-Ag. This may relate in part to the fact that greater levels of self-Ag induce more robust proliferation, and that epigenetic modification within the Ifng locus is linked to cell cycle progression (6). Even though tolerized CD4 cells contain acetylated histones bound to the Ifng promoter region, they remain unable to express substantial amounts of IFN-γ in response to antigenic stimulation apparently because a blockage in TCR-mediated signaling also develops. We have also previously shown that in response to immunization with cognate peptide complexed to the heat shock protein gp96 naive CD4 cells neither become tolerant nor develop the capacity to express either IFN-γ or the Th2 signature effector cytokine IL-4 (35). It therefore appears that there is a degree of independence in the pathways that regulate epigenetic modification of the Ifng locus and the decision to become tolerant (as defined by a blockage in TCR-mediated signaling (36, 37)). The reason for this flexibility may in part relate to the ability of tolerized CD4 cells to regain antigenic responsiveness following the removal of the initial source of tolerizing Ag (18, 19). Thus, the ability of the Ifng locus to undergo epigenetic modification during peripheral tolerization might allow such functionally recovered CD4 cells to express IFN-γ more rapidly once they have been primed in response to an immunogenic form of the same Ag. Consistent with this possibility, in the current study we found that CD4 cells that initially underwent peripheral tolerization in response to cognate self-Ag preferentially gained the ability to express IFN-γ (in comparison to TNF-α and IL-2) when the tolerizing self-Ag was immediately replaced with the same Ag expressed in the context of a virus. TNF-α and IL-2 expression were only partially rescued by this protocol, apparently because the blockage in TCR-proximal signaling had not been fully repaired. It might have been possible that TCR-proximal signaling would have been more fully restored by providing a longer recovery interval between removal of the tolerizing self-Ag and challenge with viral-Ag. Nevertheless, the finding that even without a recovery period IFN-γ expression was boosted to a level observed in virally primed controls despite the lingering impairment in TCR-proximal signaling efficiency strongly suggests that opening of the Ifng locus initiated during tolerization contributes to this effect.
Opening of the Ifng locus during peripheral CD4 cell tolerization might be important during certain tumor immunotherapy scenarios. Thus, CD4 cells specific for tumor-associated Ags can be susceptible to peripheral tolerization (20, 38), but could potentially recover function following standard treatments that induce states of minimal residual disease that would be associated with reduced loads of tolerogenic tumor Ag. If the Ifng locus opened in tumor-reactive CD4 cells undergoing tolerization, vaccines administered following such standard treatments could potentially enable these cells to more rapidly develop the capacity to express IFN-γ (which has potent antitumor activity in numerous systems (39–42)). Consistent with this possibility, we have recently shown in a transgenic prostate cancer model that tumor-specific CD4 cells exposed to tolerogenic prostate-tumor Ag exhibit an enhanced ability to differentiate into IFN-γ-expressing effectors in response to vaccination when androgen ablation therapy (which reduces the tolerogenic Ag load) is administered first (20).
Acknowledgment
We thank Dr. Alexander Rudensky for the TEa TCR transgenic mice.
Footnotes
This work was supported by Grants AI057441 and CA109339 (to A.J.A.) from the National Institutes of Health.
Abbreviations used in this paper: HA, hemagglutinin; ChIP, chromatin immunoprecipitation; LN, lymph node; viral-HA, recombinant vaccinia virus expressing HA.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Murphy KM, Reiner SL. Decision making in the immune system: The lineage decisions of helper T cells. Nat. Rev. Immunol. 2002;2:933–944. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- 2.Glimcher LH, Townsend MJ, Sullivan BM, Lord GM. Recent developments in the transcriptional regulation of cytolytic effector cells. Nat. Rev. Immunol. 2004;4:900–911. doi: 10.1038/nri1490. [DOI] [PubMed] [Google Scholar]
- 3.Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity. 2006;24:677–688. doi: 10.1016/j.immuni.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Pulendran B. Variegation of the immune response with dendritic cells and pathogen recognition receptors. J. Immunol. 2005;174:2457–2465. doi: 10.4049/jimmunol.174.5.2457. [DOI] [PubMed] [Google Scholar]
- 5.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 6.Bird JJ, Brown DR, Mullen AC, Moskowitz NH, Mahowald MA, Sider JR, Gajewski TF, Wang CR, Reiner SL. Helper T cell differentiation is controlled by the cell cycle. Immunity. 1998;9:229–237. doi: 10.1016/s1074-7613(00)80605-6. [DOI] [PubMed] [Google Scholar]
- 7.Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. TH cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat. Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 8.Janeway CA., Jr Approaching the asymptote? evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 1989;54:1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Matzinger P. Tolerance, danger, and the extended family. Annu. Rev. Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- 10.Jenkins MK, Khoruts A, Ingulli E, Mueller DL, McSorley SJ, Reinhardt RL, Itano A, Pape KA. In vivo activation of antigen-specific CD4 T cells. Annu. Rev. Immunol. 2001;19:23–45. doi: 10.1146/annurev.immunol.19.1.23. [DOI] [PubMed] [Google Scholar]
- 11.Steinman RM, Nussenzweig MC. Avoiding horror autotoxicus: the importance of dendritic cells in peripheral T cell tolerance. Proc. Natl. Acad. Sci. USA. 2002;99:351–358. doi: 10.1073/pnas.231606698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adler AJ, Huang CT, Yochum GS, Marsh DW, Pardoll DM. In vivo CD4+ T cell tolerance induction versus priming is independent of the rate and number of cell divisions. J. Immunol. 2000;164:649–655. doi: 10.4049/jimmunol.164.2.649. [DOI] [PubMed] [Google Scholar]
- 13.Higgins AD, Mihalyo MA, McGary PW, Adler AJ. CD4 cell priming and tolerization are differentially programmed by APCs upon initial engagement. J. Immunol. 2002;168:5573–5581. doi: 10.4049/jimmunol.168.11.5573. [DOI] [PubMed] [Google Scholar]
- 14.Huang CT, Huso DL, Lu Z, Wang T, Zhou G, Kennedy EP, Drake CG, Morgan DJ, Sherman LA, Higgins AD, Pardoll DM, Adler AJ. CD4+ T cells pass through an effector phase during the process of in vivo tolerance Induction. J. Immunol. 2003;170:3945–3953. doi: 10.4049/jimmunol.170.8.3945. [DOI] [PubMed] [Google Scholar]
- 15.Zhou G, Lu Z, McCadden JD, Levitsky HI, Marson AL. Reciprocal Changes in tumor antigenicity and antigen-specific T cell function during tumor progression. J. Exp. Med. 2004;200:1581–1592. doi: 10.1084/jem.20041240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 17.Mullen AC, High FA, Hutchins AS, Lee HW, Villarino AV, Livingston DM, Kung AL, Cereb N, Yao TP, Yang SY, Reiner SL. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science. 2001;292:1907–1910. doi: 10.1126/science.1059835. [DOI] [PubMed] [Google Scholar]
- 18.Ramsdell F, Fowlkes BJ. Maintenance of in vivo tolerance by persistence of antigen. Science. 1992;257:1130–1134. doi: 10.1126/science.257.5073.1130. [DOI] [PubMed] [Google Scholar]
- 19.Pape KA, Merica R, Mondino A, Khoruts A, Jenkins MK. Direct evidence that functionally impaired CD4+ T cells persist in vivo following induction of peripheral tolerance. J. Immunol. 1998;160:4719–4729. [PubMed] [Google Scholar]
- 20.Drake CG, Doody AD, Mihalyo MA, Huang CT, Kelleher E, Ravi S, Hipkiss EL, Flies DB, Kennedy EP, Long M, et al. Androgen ablation mitigates tolerance to a prostate/prostate cancer-restricted antigen. Cancer Cell. 2005;7:239–249. doi: 10.1016/j.ccr.2005.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adler AJ. Mechanisms of T cell tolerance and suppression in cancer mediated by tumor-associated antigens and hormones. Curr. Cancer Drug Targets. 2007;7:3–14. doi: 10.2174/156800907780006931. [DOI] [PubMed] [Google Scholar]
- 22.Kirberg J, Baron A, Jakob S, Rolink A, Karjalainen K, von Boehmer H. Thymic selection of CD8+ single positive cells with a class II major histocompatibility complex-restricted receptor. J. Exp. Med. 1994;180:25–34. doi: 10.1084/jem.180.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adler AJ, Marsh DW, Yochum GS, Guzzo JL, Nigam A, Nelson WG, Pardoll DM. CD4+ T cell tolerance to parenchymal self-antigens requires presentation by bone marrow-derived antigen presenting cells. J. Exp. Med. 1998;187:1555–1564. doi: 10.1084/jem.187.10.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grubin CE, Kovats S, deRoos P, Rudensky AY. Deficient positive selection of CD4 T cells in mice displaying altered repertoires of MHC class II-bound self-peptides. Immunity. 1997;7:197–208. doi: 10.1016/s1074-7613(00)80523-3. [DOI] [PubMed] [Google Scholar]
- 25.Higgins AD, Mihalyo MA, Adler AJ. Effector CD4 cells are tolerized upon exposure to parenchymal self-antigen. J. Immunol. 2002;169:3622–3629. doi: 10.4049/jimmunol.169.7.3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long M, Higgins AD, Mihalyo MA, Adler AJ. Effector CD4 cell tolerization is mediated through functional inactivation and involves preferential impairment of TNF-α and IFN-γ expression potentials. Cell Immunol. 2003;224:114–121. doi: 10.1016/j.cellimm.2003.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long M, Slaiby AM, Hagymasi AT, Mihalyo MA, Lichtler AC, Reiner SL, Adler AJ. T-bet down-modulation in tolerized Th1 effector CD4 cells confers a TCR-distal signaling defect that selectively impairs IFN-γ expression. J. Immunol. 2006;176:1036–1045. doi: 10.4049/jimmunol.176.2.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fields PE, Kim ST, Flavell RA. Cutting edge: changes in histone acetylation at the IL-4 and IFN-γ loci accompany Th1/Th2 differentiation. J. Immunol. 2002;169:647–650. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- 29.Lyman MA, Aung S, Biggs JA, Sherman LA. A spontaneously arising pancreatic tumor does not promote the differentiation of naive CD8+ T lymphocytes into effector CTL. J. Immunol. 2004;172:6558–6567. doi: 10.4049/jimmunol.172.11.6558. [DOI] [PubMed] [Google Scholar]
- 30.Marzo AL, Klonowski KD, Le Bon A, Borrow P, Tough DF, Lefrancois L. Initial T cell frequency dictates memory CD8+ T cell lineage commitment. Nat. Immunol. 2005;6:793–799. doi: 10.1038/ni1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hataye J, Moon JJ, Khoruts A, Reilly C, Jenkins MK. Naive and memory CD4+ T cell survival controlled by clonal abundance. Science. 2006;312:114–116. doi: 10.1126/science.1124228. [DOI] [PubMed] [Google Scholar]
- 32.Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat. Immunol. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- 33.Ohashi PS. T-cell signalling and autoimmunity: molecular mechanisms of disease. Nat. Rev. Immunol. 2002;2:427–438. doi: 10.1038/nri822. [DOI] [PubMed] [Google Scholar]
- 34.Adler AJ. Peripheral tolerization of effector and memory T cells: implications for autoimmunity and tumor-immunity. Curr Immunol. Rev. 2005;1:21–28. doi: 10.2174/1573395052952879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doody AD, Kovalchin JT, Mihalyo MA, Hagymasi AT, Drake CG, Adler AJ. Glycoprotein 96 can chaperone both MHC class I- and class II-restricted epitopes for in vivo presentation, but selectively primes CD8+ T cell effector function. J. Immunol. 2004;172:6087–6092. doi: 10.4049/jimmunol.172.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz RH. T cell anergy. Annu. Rev. Immunol. 2003;21:305–334. doi: 10.1146/annurev.immunol.21.120601.141110. [DOI] [PubMed] [Google Scholar]
- 37.Mueller DL. E3 ubiquitin ligases as T cell anergy factors. Nat. Immunol. 2004;5:883–890. doi: 10.1038/ni1106. [DOI] [PubMed] [Google Scholar]
- 38.Stavely-O’Carroll K, Sotomayor E, Montgomery J, Borrello I, Hwang L, Fein S, Pardoll D, Levitsky H. Induction of antigen-specific T cell anergy: an early event in the course of tumor progression. Proc. Natl. Acad. Sci. USA. 1998;95:1178–1183. doi: 10.1073/pnas.95.3.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hung K, Hayashi R, Lafond-Walker A, Lowenstein C, Pardoll D, Levitsky H. The central role of CD4+ T cells in the antitumor immune response. J. Exp. Med. 1998;188:2357–2368. doi: 10.1084/jem.188.12.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin Z, Blankenstein T. CD4+ T cell–mediated tumor rejection involves inhibition of angiogenesis that is dependent on IFN gamma receptor expression by nonhematopoietic cells. Immunity. 2000;12:677–686. doi: 10.1016/s1074-7613(00)80218-6. [DOI] [PubMed] [Google Scholar]
- 41.Ikeda H, Old LJ, Schreiber RD. The roles of IFN γ in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev. 2002;13:95–109. doi: 10.1016/s1359-6101(01)00038-7. [DOI] [PubMed] [Google Scholar]
- 42.Winter H, Hu HM, McClain K, Urba WJ, Fox BA. Immunotherapy of melanoma: a dichotomy in the requirement for IFN-gamma in vaccine-induced antitumor immunity versus adoptive immunotherapy. J. Immunol. 2001;166:7370–7380. doi: 10.4049/jimmunol.166.12.7370. [DOI] [PubMed] [Google Scholar]