Abstract
Aminopeptidases in the endoplasmic reticulum (ER) can cleave antigenic peptides and in so doing either create or destroy MHC class I-presented epitopes. However the specificity of this trimming process overall and of the major ER aminopeptidase ERAP1 in particular is not well understood. This issue is important because peptide trimming influences the magnitude and specificity of CD8 T cell responses. By systematically varying the N-terminal flanking sequences of peptides in a cell free biochemical system and in intact cells, we elucidated the specificity of ERAP1 and of ER trimming overall. ERAP1 can cleave after many amino acids on the N-terminus of epitope precursors but does so at markedly different rates. The specificity seen with purified ERAP1 is similar to that observed for trimming and presentation of epitopes in the ER of intact cells. We define N-terminal sequences that are favorable or unfavorable for antigen presentation in ways that are independent from the epitopes core sequence. When databases of known presented peptides were analyzed, the residues that were preferred for the trimming of model peptide precursors were found to be overrepresented in N-terminal flanking sequences of epitopes generally. These data define key determinants in the specificity of antigen processing.
Keywords: Antigen Presentation/Processing, Antigens/Peptides/Epitopes, MHC
Introduction
Peptides capable of binding major histocompatibility complex (MHC) class I molecules are generated during intracellular protein degradation. These can then be presented on the cell surface and non-native peptides (e.g., from viral proteins) recognized by circulating cytotoxic T lymphocytes (1–5).
A single type of MHC class I molecule can bind to a large repertoire of different peptides in the ER. This is due to the fact that the MHC binding groove interacts with main chain atoms plus a few amino acid side chains of the peptide (6). However, the groove also interacts with the α-amino (N) and carboxy (C) terminal groups of the epitope, thus limiting antigenic peptides to a length of eight, nine or ten residues, depending on the particular MHC class I molecule (6). In spite of the promiscuity of peptide binding, MHC class I molecules typically present only a small fraction of potential peptides encoded by viral genomes, and even fewer peptides trigger a potent T cell response; for example, of the five potential H-2Kb-binding peptides in ovalbumin, only one (SIINFEKL (S-L)) stimulates a strong immune response (7). The reasons for this phenomenon (“immunodominance”) are unclear and probably complex (reviewed in (8)), but antigen processing and ultimately epitope abundance at the cell surface can be a major factor (9–11).
It is now firmly established that the proteasome pathway is responsible for the initial degradation of most proteins within the cytoplasm and through this process generates peptides which feed into the antigen presentation pathway (2, 3, 12). The peptides generated by the proteasomes can either be mature epitopes which are capable of binding MHC class I molecules without further trimming or precursors that are extended on the N-terminus (13–15). These longer peptides could be further trimmed to the length required for antigen presentation by aminopeptidases residing in the cytoplasm or ER. In the presence of interferon γ this tendency for generating precursors with N-terminal extensions may be enhanced due to proteasomes containing different catalytic subunits (“immunoproteasomes”) (16).
The majority of peptides generated by the proteasome are destroyed before they are transported by the transporter associated with antigen presentation (TAP) into the ER (17). Although TAP efficiently translocates peptides between 8 and 16 residues long (18), it is often more efficient in transporting N-extended precursors than mature epitopes (19–21). This is especially important for certain MHC binding peptides. For example, although many epitopes have a proline at position 2, TAP transports such peptides inefficiently (22, 23). This means that these epitopes must be generated as N-terminal extended precursors in order to be translocated efficiently into the ER. Taken together, these findings suggest that for many epitopes processing is a multistep event involving proteasomal release of precursors followed by downstream N-terminal trimming either before or after transport into the ER by TAP (5, 24). Each step in the pathway adds a layer of specificity which is potentially capable of limiting/increasing peptide display on the cell surface. While the roles some components play have been extensively analyzed the contribution of peptidases is not as well understood.
Endoplasmic reticulum (ER) aminopeptidase 1 (ERAP1; ERAAP), an IFN-γ inducible metallopeptidase, is responsible for the trimming of many N-extended precursors in the ER (25–27). Unlike other aminopeptidases, ERAP1 trims precursors to 8 or 9 residues, the optimal length for binding to MHC class I, before cleavage slows or stops completely (28). In cells lacking ERAP1 there is a marked reduction of peptides, such as S-L, supplied to the MHC class I presentation pathway from precursors extended on the N-terminus (11, 25, 26, 29, 30). In addition mice lacking ERAP1 had markedly different presentation of a variety of epitopes (11, 29–31), leading to differences in immunodominance hierarchies (11, 31). These results suggest that ability of ERAP1 to remove N-terminal amino acids from epitope precursors is a key determinant of the amount of epitope displayed on the cell surface and the specificity of the immune response to invading pathogens. This led us to initiate the present study to systematically examine the trimming of N-terminal sequences from antigenic precursors in the ER and the role ERAP1 plays in this process.
Materials and Methods
Peptide trimming assays
Peptides were synthesized by Sigma-Aldrich (St. Louis, MO) and were >80% by MS analysis. Peptides (100 μM) were incubated with purified recombinant human ERAP1 (3.5 μg/ml) (A kind gift from T. Nguyen and L.J. Stern, University of Massachusetts Medical School, Worcester, MA) at 37°C in 50 mM Tris HCl (pH 7.8) and 0.5 μg/ml protease-free BSA (Sigma). Reactions were terminated by adding 0.6% trifluoroacetic acid. The peptide-containing supernatant was analyzed by RP-HPLC on a 4.6 mm × 250 mm C18 column (Vydac) and eluted with a 7–50% acetonitrile gradient in 10 mM sodium phosphate buffer (pH 6.8) or a gradient of 7% acetonitrile in 0.06% trifluoroacetic acid (TFA) to 41% acetonitrile in 0.06% TFA. The amount of each peptide was calculated by integration of the area under the peak.
Plasmids
N-terminal S-L (OVA254-264) precursors were targeted to the ER using a signal sequence (ss) derived from the adenoviral E3/19K protein (32). To ensure efficient cleavage by the signal peptidase an alanine reside was encoded between the signal sequence and the C-terminally fused peptide. Synthetic complementary oligonucleotides encoding N-terminal extended epitope precursors were annealed and inserted into pBS-ss-L-OVA digested with Pst I and Xba I. The oligonucleotides used were: for pBS-ss-XXS-L, 5′-CTGCAGCGCT GCT XXX XXX AGT ATA ATC AAC TTT GAA AAA CTG TAGTTCTAGA-3′; for pBS-ss-X S-L, 5′-CTGCAGCGCT GCT XXX AGT ATA ATC AAC TTT GAA AAA CTG TAGTTCTAGA-3′; for pBS-ss-XXYY S-L, 5′-CTGCAGCGCT GCT XXX XXX TAC TAT AGT ATA ATC AAC TTT GAA AAA CTG TAGTTCTAGA-3′; for pBS-ss-XXF-L (Sendai NP321-332), 5′-CTGCAGCGCT GCT XXX XXX TTC GCC CCC GGC AAC TAC CCC GCC CTG TAGTTCTAGA-3′; and for pBS-ss-XXR-Y (HCV NS5b2588-2596), 5′-CTGCAGCGCT GCT ATG ATG AGA GTG TGC GAG AAG ATG GCC CTG TAC TAGTTCTAGA-3′. These were subcloned into pTracer-CMV2 (Invitrogen Life Technologies) a plasmid containing a GFP/zeocin resistance fusion protein, using Kpn I and Xba I to generate ss XXS-L, ss XS-L, ss-XXYYS-L, ss XXF-L and ss XXR-Y respectively. For ss- 4N XS-L (ss LEQLXS-L) constructs, synthetic complementary oligonucleotides (5′-CTGCAG C GCC GCT CTAGA-3′) where annealed and inserted into pBS-ss-L-OVA digested with Pst I and Xba I to generate pTracer-ss-L. Complementary synthetic oligonucleotides encoding EQLXS-L (5′-TCTAGAG CAG CTG XXX AGT ATA ATC AAC TTT GAA AAA CTG TAGTTCTAGA-3′) were annealed and inserted into pTracer-ss-AL digested with Xba I to generate ss- 4N XS-L. The most common codons for each amino acid based on frequency of usage of each codon (per thousand) in human coding regions were used; ala (A), GCT; arg (R), CGC; asn (N), AAC; asp (D), GAC; cys (C), TGT; gln (Q), CAG; glu (E), GAG; gly (G), GGC; his (H), CAC; ile (I), ATC; leu (L), CTG; lys (K), AAG; met (M), ATG; phe (F), TTC; pro (P), CCC; ser (S), TCC; thr (T), ACC. All plasmids were sequenced to confirm correct sequences and reading frames.
Cells
HeLa-Kb and HeLa-Kb-ICP47 (HeLa-Kb-A3-47 cells (HeLa cells stably transfected with H-2Kb, HLA-A3 and ICP47)) cells have been described previously (26, 33). COS-Kb cells (COS7 cells stably transfected with H-2Kb) have been previously described (33).
siRNA
Cells were transfected with siRNA for both ERAP1 and control mTOP (directed against murine TOP in a region that differs from human TOP) using oligofectamine (Invitrogen, CA) as previously described (26).
Antibodies and flow cytometry
Cells were transiently transfected with plasmid (1 μg) using TransIT HeLa Monster (Mirus, Madison, WI) according to the manufacturer’s protocol and incubated for 24–48hrs. The mAb 25.D1.16 (anti-H-2Kb+S-L) (34), B8-24.3 (anti-H-2-Kb) (35) and GAP-A3 (anti-HLA-A3) (36) were used as primary antibodies. When HeLa-Kb-ICP47 cells were transfected with ss XXF-L constructs, cells were heated to 40°C for one hour (to denature any empty MHC class I molecules) before primary antibody binding. The cells were analyzed by flow cytometry, gating on GFP-expressing cells. Unless otherwise stated, data are representative of three independent experiments.
On-line databases and epitope analysis
4934 naturally-presented MHC class I epitopes from SYFPEITHI (http://www.syfpeithi.de) and The Internet Epitope Database (IEDB) (http://www.immuneepitope.org/) were selected in April 2009 (MHC class II epitopes in these databases were excluded from analysis) and the sequences of the proteins from which they originated were obtained from Genbank. The 15 amino acids immediately N-terminal to each epitope sequence (or, if the epitope was less than 15 amino acids from the origin of the protein, as many as possible) were identified and stored in a SQLite database. The epitopes, the MHC class I alleles to which they bind, and the upstream sequences are listed in Supplemental Table 1. Python scripts were used to calculate the frequency of each amino acid in each position N-terminal of the epitope. As a control, the protein precursors were pooled, and 4934 15-amino-acid-long peptides were randomly selected from this pool and analyzed in the same way; random selection and analysis was repeated 500 times and the average frequency and standard deviation of each amino acid was calculated.
Results
In vitro ERAP1 specificity
The observation that the loss of ERAP1 reduces the presentation of many peptides indicates that these epitopes must be initially produced as longer precursors that are subsequently trimmed in the ER. Moreover, the finding that ERAP1 deficiency affects the presentation of different peptides in different ways suggests that either some peptides are made as precursors while others are not (and hence manifest a differential requirement for trimming), or that ERAP1 has specificity and trims some precursors better than others (or both). Microsome preparations have previously been shown to be capable of trimming some precursor peptides (37, 38). However, since the specificity of ERAP1 for trimming polypeptide substrates has not yet been well defined, we sought to initially test the second possibility in vitro using recombinant human ERAP1.
In order to systematically examine the specificity of trimming of N-extended precursors of MHC class I-presented peptides we synthesized a series of peptides containing the model epitope S-L (the immunodominant H-2Kb-restricted epitope SIINFEKL from chicken ovalbumin) and compared the rates of degradation. Each peptide was generated with a single amino acid extension on the N-terminus of SIINFEKL. In total the rates of removal of 16 amino acids were analyzed (four of the possible twenty were excluded due to impurities or insufficient resolution). Time courses of peptide degradation were evaluated (Figure 1A). As shown previously the mature epitope SIINFEKL is a very poor substrate for ERAP1 and is not degraded further (27). On the other hand LSIINFEKL is trimmed very rapidly to SIINFEKL (Figure 1B). The amount of mature 8-residue peptide steadily increased over time and the trimming process stopped when the 9-residue peptide was converted to SIINFEKL. The rates of degradation of all 16 N-terminal extended peptides are shown in Figure 1B arranged from highest to lowest, left to right. This analysis revealed that the N-terminal flanking residue had a marked and highly reproducible influence on the rate of mature epitope generation; e.g. leucine and methionine were both efficiently removed from the N-terminus, in comparison aspartic acid and glutamic acid were poorly removed.
Figure 1. Trimming of NH2-extended precursors by ERAP1.
Time course of peptide digestion (A). XS-L peptides, differing only in their N-terminal residue (X), were incubated with purified ERAP1. At various intervals samples were processed and RP-HPLC was used to determine XS-L remaining. Rate of peptide trimming by ERAP1, from highest to lowest (B). In the inset, the elution profile of products following LS-L trimming by ERAP1 (t= 0, 15 and 30 minutes).
N-terminal flanking amino acids affect presentation from ER targeted precursors
ERAP1 is localized in the ER. We next wanted to determine whether the N-terminus of epitope precursors, specifically targeted to the compartment where ERAP1 resides, would affect the amount of peptide seen on the cell surface in vivo.
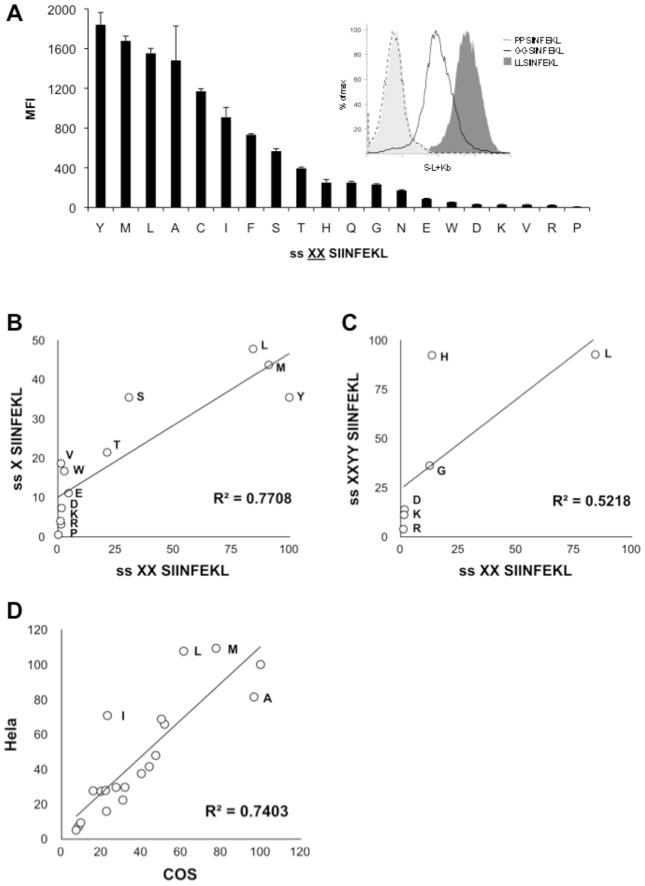
In order to do this systematically we generated a series of minigene constructs containing S-L. Each minigene was constructed with an N-terminus containing one of the possible twenty amino acids. In order to amplify any differences between constructs in the rate of removal of their flanking residues, they were expressed with two identical amino acids upstream of the epitope (XXS-L, where “X” represents any amino acid). These sequences were then fused to an N-terminal signal sequence derived from the adenoviral E3/19K protein (MRYMILGLLALAAVCSAAXXSIINFEKL) so that they would be cotranslationally transported by Sec61 into the ER and the signal sequence removed during this process (these constructs will be referred to as ss XXS-L). To ensure equal trimming by signal peptidase an alanine reside was encoded between the signal sequence and the C-terminally fused peptide. The ss XXS-L constructs were transiently transfected into HeLa-Kb cells stably transfected with the TAP blocker ICP47 (39), to specifically analyze ER processing. The presence of S-L-Kb complexes on the cell surface of transfected (GFP+) cells was quantified by staining with the mAb 25.D1.16. This assay gave highly reproducible results between replicate groups and independent experiments. This analysis revealed that N-terminal flanking amino acids had a marked influence on the levels of presented peptide detected on the cell surface (Figure 2A). N-terminal amino acids such as methionine, leucine and tyrosine appear to be efficiently removed from precursors to generate mature S-L epitope. On the other hand residues such as arginine and proline were processed inefficiently by ER resident aminopeptidases resulting in presentation that was <1% of that observed with ss YYS-L (the construct leading to the greatest presentation). The effects of other amino acids on presentation fell between these two extremes resulting in a hierarchy of presentation among the 20 constructs tested that was highly reproducible (Figure 2A). In fact 14 of the 20 amino acids were associated with presentation that was ≤50% of that observed with ss YYS-L demonstrating that many amino acids are removed rather inefficiently in the ER. Although amino acids with acidic, amide and basic side chains appear to be processed poorly there is a broad range of efficiencies of presentation within each group when amino acids are grouped by the chemical nature of their side chain.
Figure 2. Antigen presentation in HeLa-Kb-ICP47 cells transfected with ER targeted S-L precursors.
(A) Cells were transfected with ER targeted XXS-L (ss XXS-L, where X represents one of the twenty amino acids). After 24 hrs, cells were stained for S-L + H-2Kb complexes with 25.D1.16. Graphs represent the average mean fluorescent intensity (MFI) of two independent transfections. Error bars represent the difference within each group. In the inset, FACS showing cells stained with 25.D1.16 following transfection with ss GGS-L, ss PPS-L and ss LLS-L; background, dark shaded histogram. (B) Correlation between presentation of S-L in cells transfected with ss XS-L and ss XXS-L. (C) Correlation between presentation of S-L in cells transfected with ss XXYYS-L and xx XXS-L. (D) Correlation between presentation of S-L in Hela-Kb and COS-Kb transfected with ss XXS-L.
We next examined whether a single amino acid showed the same effect as the corresponding doublet when targeted to the ER. To this end we generated a subset of ss XS-L constructs which encoded amino acids which were processed efficiently (L, M and Y) and inefficiently in the ER (D, E, K, P, R, V and W) as well as amino acids which lie in between these two extremes (S and T). The data obtained with these constructs are largely consistent with those obtained with the corresponding doublet (Figure 2B). To test whether the efficiency of amino acid removal was length-dependent, we measured processing of constructs in which Leu (efficiently processed), Arg, Asp or Lys (poorly processed), or His or Gly (moderately processed) amino acids were present at the N-terminus of a 12-mer (ss XXYYS-L, since YYS-L is efficiently processed) rather than a 10mer (ss XXS-L). Again, removal of XX from XXYYS-L was consistent with the efficiency of removal of XX from XXS-L (Figure 2C).
It was important to determine whether the results we obtained with HeLa cells were generalizable to another cell type. Moreover, the HeLa cells used in our analyses express ERAP1 but not the homologous peptidase ERAP2, which may also trim peptides in the ER. Of importance, ERAP2 has a different specificity than ERAP1 and is more active in removing basic residues, at least in cell free systems (40). Therefore, to test the generality of the results obtained with HeLa, we performed a similar analysis using a second cell line (COS 7) that expresses both ERAP1 and ERAP 2 (supplemental data Figure 1). All 20 of the ss-XXS-L constructs were transfected into COS-Kb cells and the generation of SIINFEKL-Kb complexes quantified with 25D1. The results of this analysis correlated extremely well (correlation coefficient of 0.74) with those obtained with HeLa cells, even for charged residues that were potential substrates for ERAP2 (correlation between Hela-Kb and COS-Kb for charged residues = 0.995) (Figure 2D).
Analysis of the trimming of ss XXS-L in the ER by ERAP1
For presentation of S-L precursors with the natural N-terminal flanking sequence from ovalbumin (LEQLE), ERAP1 appears to be the only important peptide-trimming enzymatic activity in the ER (25, 26). We therefore examined whether this enzyme was vital for epitope liberation from the various ss XXS-L constructs. In these experiments, the ER-targeted precursors were transfected into HeLa-Kb ICP47 cells that were treated with control or ERAP1-specific siRNA under conditions where ERAP1 protein expression is reduced by at least 90% (26).
The loss of ERAP1 reduced the presentation of all 20 constructs, most of them to little more than background levels (Figure 3). The presentation of a few of the best presented constructs was inhibited by 70–80 percent, leaving significant presentation of H-2Kb-S-L (e.g. ss YYS-L and ss MMS-L); whether the remaining presentation is due to residual amounts of ERAP1, or whether another peptidase can contribute to processing of these peptides, remains to be determined. The construct that was least dependent on ERAP1 was ss AAS-L; this may be because the signal peptidase cleaves preferentially after small, uncharged residues such as alanine and therefore may be able to generate the mature epitope from this construct. In any case, this analysis clearly shows that ERAP1 is required for the bulk of presentation of all 20 constructs. Therefore the differences in trimming and presentation of the various constructs must be due to the specificity of ERAP1, allowing us to define the specificity of ERAP1 as it functions in intact cells.
Figure 3. Antigen presentation in HeLa-Kb-ICP47 cells transfected with ss XXS-L precursors, with and without treatment with ERAP1 siRNA.
HeLa-Kb-ICP47 cells were treated with siRNA targeting ERAP1 (white bars) or control (ctrl) siRNA (black bars) then transfected with ss XXS-L. On day three S-L presentation was determined by staining with with 25.D1.16. Graphs represent the average percentage presentation of two independent experiments using the MFI of cells treated with ctrl siRNA and transfected with ss YYS-L as 100% presentation. Error bars represent the difference within each group. In the inset, only amino acids with presentation under 10% are displayed for a more detailed look
N-terminal flanking residues affect epitope liberation from a large OVA-precursor
We next investigated how incorporating different residues directly upstream of S-L in a subset of longer precursor peptides, LEQLXS-L, influenced liberation of the mature epitope, compared to the natural context in chicken ovalbumin of LEQLES-L (Figure 4A). The ER targeted constructs LEQLLS-L, LEQLMS-L and LEQLYS-L were presented very efficiently, while epitope generation from LEQLKS-L and LEQLRS-L was lower than that from LEQLES-L. The data obtained with these constructs are highly consistent with those obtained with simpler constructs where the residue was expressed as a doublet (Figure 4B) or a single residue (Figure 4C) upstream of S-L.
Figure 4. The amino acid in the P1 position influences presentation from an OVA-derived precursor.
HeLa-Kb-ICP47 cells were transfected with LEQLXS-L targeted to the ER (A). After 24 hrs antigen presentation was analyzed by staining for S-L + Kb complexes with 25.D1.16 and flow cytometry. Graphs represent the average MFI of two independent transfections. Error bars represent the difference within each group. Correlation between presentation of S-L in cells transfected with ss LEQLXS-L and ss XXS-L (B) or ss XS-L (C).
Poorly cleaved residues are dominant in a doublet
We next investigated the effect of juxtaposing, in different orientations, residues that were efficiently removed from precursors in the ER and those that were not. We generated a series of constructs expressing XZS-L and ZXS-L in the ER (where X and Z are two different amino acids which were efficiently (Leu, Met and Tyr) or poorly (Lys, Arg and Val) trimmed from S-L and compared antigen presentation to that observed with the single amino acid counterparts (XS-L and ZS-L). Juxtaposing lysine and valine, both of which are poorly removed, leads to low epitope generation. On the other hand when the N-terminal flanking doublet is composed of leucine and methionine, which are both efficiently removed, S-L presentation on the cell surface is high (Figure 5).
Figure 5. The effect of juxtaposing different amino acids in a doublet on S-L presentation from N-extended precursors.
Cells were transfected with ss XS-L, ss ZS-L, ss XZS-L or ss ZX S-L (where Z represents a different amino acid than X). H-2Kb-S-L presentation was determined by staining cells with 25.D1.16. Graphs represent the average MFI of two independent transfections. Error bars represent the difference within each group.
We next tested constructs that had an efficiently trimmed residue juxtaposed with a poorly trimmed one. When methionine and lysine or tyrosine and arginine were juxtaposed in either orientation, presentation is significantly lower than that of the single methionine or tyrosine residue precursors. However, epitope generation from these constructs was not reduced to the level of the corresponding single poor residue (Lys or Arg). The difference in presentation between these constructs and the corresponding single efficiently trimmed residue was statistically significant (P < 0.05, Student t test). This suggests that poorly removed amino acids have a large impact on the efficiency of presentation when located at either the P1 or P2 position. Interestingly, we found one exception to this rule. Although the level of presentation from the LVS-L construct was consistent with the above observations in that it was significantly lower than that of the single leucine precursor, presentation from the VLS-L construct was almost as high as that of the LS-L construct (Figure 5). These results suggest that not only the identity of the amino acid, but also adjacent residues may sometimes be important in determining the efficiency of removal.
Comparison of in vitro and in vivo ERAP1 specificity
Given that the specificity of N-terminal amino acid removal from epitope precursors observed in vivo appears to dependent on ERAP1 (Figure 3) we next compared these observations to that seen in vitro with recombinant ERAP1 (Figure 1). There is a good correlation between presentations from ER targeted XXS-L and the specificity of recombinant ERAP1 (Figure 6A) again suggesting that ERAP1 is the major aminopeptidase within the ER responsible for epitope liberation and that its specificity determines which epitopes are generated from precursors with N-terminal extensions. This finding is also mirrored when comparing ERAP1s specificity to presentation from ER targeted XS-L (Figure 6B).
Figure 6. Comparison of in vitro and in vivo ERAP1 specificity.
Correlation between generation of S-L from XS-L in vitro using purified ERAP1 and presentation of S-L in vivo following HeLa-Kb-ICP47 transfection with ss XS-L (A) or ss XXS-L (B).
The influence of upstream amino acids is the same on multiple epitopes
When precursors are expressed in the ER the residues upstream of the model epitope S-L have a marked impact on presentation by MHC class I molecules. Given this result we next wanted to broaden the observation and determine whether the same amino acids upstream of other epitopes had a similar effect on presentation. The two epitopes chosen for this analysis were FAPGNYPAL (F-L), a H-2Kb restricted epitope derived from Sendai virus nucleoprotein, and RVCEKMALY (R-Y), a HLA-A3 restricted epitope derived from the Hepatitis C virus NS5b. We generated signal sequence fusion constructs which allowed expression of XXF-L and XXR-Y in the ER, where the N-terminal doublet (XX) consisted of amino acids which were efficiently removed from S-L in the ER (Leu, Met and Tyr) and those which were not (Lys, Arg and Val).
We were unable to measure the presentation of these Sendai and Hepatitis virus epitopes using the same approach used for S-L because there are no antibodies available that are specific for F-L/H2-Kb or R-Y/HLA-A3 complexes. We therefore developed an alternate quantitative assay. In HeLa Kb cells stably transfected with ICP47 endogenous peptides are prevented from gaining access into the ER (39). In the absence of peptides most H-2Kb and HLA-A3 molecules are retained in the ER and very few are transported to the cell surface. However, if binding peptides are delivered into the ER via Sec61, the MHC class I molecules are then transported to the cell surface. Quantitation of cell-surface MHC class I levels, by staining with anti-H-2Kb- or anti-HLA-A3-specific antibodies, is therefore a measure of peptide supply to the molecules in the ER and of precursor N-terminal trimming. Transfecting the HeLa-Kb-A3-47 cells with ER-targeted MMS-L, SSS-L and KKS-L increased H-2Kb expression on the cell surface, and the amount of this increase paralleled the level of H-2Kb/S-L complexes (measured with 25.D1.16) (Figure 7A). Similarly, the ER-targeted R-Y peptide increased the surface expression of HLA-A3 in HeLa-Kb-A3-47 cells (Figure 7B). In contrast, S-L (which does not bind HLA-A3), when targeted into the ER, did not restore surface expression.
Figure 7. Effect of N-terminal flanking residues on presentation of alternate epitopes.
HeLa-Kb-ICP47 cells were transfected with ER targeted MMS-L, SSS-L or KKS-L and peptide/MHC complexes quantified using flow cytometry and either antibodies for overall H-2Kb (B8.24.3) or specific for H-2Kb/S-L complexes (25.D1.16) (A). (B) Hela-Kb-ICP47 cells were transfected with ER targeted R-Y (a HLA-A3 binding peptide), or with control peptide (S-L). Surface expression of HLA-A3 was measured with the monoclonal antibody GAP-A3. Cells were transfected with (C) ER targeted XXF-L or (D) ER targeted XXR-Y. Cells were stained for (C) H-2Kb with B8.24.3 or (D) HLA-A3 with GAP-A3. In the inset (C), correlation between presentation of F-L from ss XXF-L and S-L from ss XXS-L. Graphs represent the average MFI of two independent transfections. Error bars represent the difference within each group.
Using this system we then tested the effect of different N-terminal amino acids on presentation of F-L (Figure 7C) and R-Y (Figure 7D). For 9 of the 10 constructs the effects of the various N-terminal residues was the same as those observed with S-L. This led to a highly significant correlation between presentation from ss XXS-L and ss XXF-L (Figure 7C inset). Methionine and tyrosine (which are efficiently removed from S-L) led to high presentation of both F-L and R-Y, while lysine and arginine led to poor presentation of the two alternate epitopes. Valine in the flanking doublet also led to poor presentation of F-L as it had done for S-L. However, in contrast with S-L, valine was efficiently removed from the N-terminus of R-Y, leading to good presentation. Valine was also efficiently processed in the context of a long peptide, ss-VVYYS-L (data not shown), suggesting that amino acid or length context may be particularly important for trimming of this amino acid. Interestingly the natural flanking residue of R-Y is valine, suggesting that the context of the amino acids to be removed may occasionally also play a role in determining efficient cleavage.
Efficiently removed amino acids are overrepresented upstream of natural epitopes
We reasoned that if the findings with model epitopes are broadly applicable, then residues that permit high-level expression of epitopes should be over-represented upstream of naturally-presented epitopes, and those that were associated with poor presentation should be underrepresented. This issue has previously been investigated by Schatz et al (38), but it was worth revisiting using a larger number of epitopes (4,394 vs 1543) from the IEDB as well as SYFPEITHI databases allowing an analysis with substantially more statistical power. Therefore, we selected 4394 naturally-processed epitopes from the SYFPEITHI and IEDB on-line databases, and identified the 15 amino acids N-terminal to the epitopes in their precursor proteins. At each position (where “1” represents the amino acid adjacent to the epitope, and “15” represents the most distal amino acid) we calculated the frequency of each amino acid. As controls, we also selected 4394 random non-epitope peptides of the same size from the pool of precursor proteins, and analyzed their N-terminal flanking residues. This random selection was repeated 500 times, and the average and standard deviations of these “background” frequencies were calculated. Comparison of sample to background frequencies shows that amino acids in positions 1 and (to a lesser extent) 2 and 3 diverge furthest from background frequencies, consistent with previous observations that MHC class I epitopes presented on MHC class I are often imported into the ER with 1, 2, or 3-amino acid extensions (23); the probability of this variation being due to chance (Chi-squared test) is <10−17 (Figure 8A). The amino acids most responsible for this skewed distribution are indicated in Figure 8B. Ala, Cys, Leu, Met, Ser, and Tyr are all more than 2 standard deviations greater than “background” frequency, and Val and (especially) Pro are greater than 2 standard deviations lower than background. Charged residues (Asp, Glu, His, Lys, and Arg) all showed a strong trend towards being underrepresented, although this trend was not statistically significant for any of these residues when analyzed individually; However, when analyzed as a group, charged residues were more than 2 standard deviations lower than background frequency. Other groups of amino acids (aromatic, hydrophobic, nucleophilic) did not differ significantly from their respective background frequencies. These results, particularly for charged residues, differ somewhat from an earlier study (38) presumably because the present analysis analyzed a much larger set of peptides and was therefore more highly powered.
Figure 8. Aminopeptidases influence amino acid frequency N-terminal of naturally-presented MHC class I epitopes.
Regions N-terminal to naturally-processed MHC class I epitopes, or selected randomly from protein precursors, were identified as described in the Methods. (A) Probability of divergence occuring randomly (Chi-squared test) vs. position relative to epitope start site. (B) Observed amino acid frequencies at position 1 (P1) relative to epitope start, vs. no divergence from background (45-degree line). The amino acids that diverge more than 2 standard deviations from background frequency are indicated.
Thus the residues that we find empirically to lead to high-level antigen presentation (Tyr, Met, Leu, and Cys) are all over-represented residues N-terminal to natural epitopes in our analysis and that of Shatz et al (38) (although in the latter study Tyr and Met are more abundant than background frequency but do not reach a significant diffrence). Ala is also over represented and better presented, however as discussed some of its trimming in our in vivo system is not due to ERAP1. In our experiments, charged residues were processed poorly, correlating to some extent with the overall under-representation of charged residues immediately adjacent to natural epitopes.
The bond upstream of the most under-represented residue, proline, is poorly trimmed by ERAP1 (Figure 1 and 2A) and cannot be trimmed by many other aminopeptidases (41); therefore we expect that constraints of aminopeptidase trimming would select against proline being in the P1 and P2 position. In our experiments, Val showed evidence of context-dependent trimming: when immediately adjacent to the epitope Val was generally processed poorly, but when separated from the epitope by one or more residues, Val was processed relatively well. Similarly, in natural epitopes Val is under-represented immediately adjacent to epitopes (P1; more than 2 SD less than background) but is present at background frequency at P2 and more distant positions (not shown).
Similar results were obtained when we performed an analysis only of epitopes presented on human MHC class I molecules (supplemental data table II). This is not surprising because they account for 85% of the sequences in the databases we have analyzed (4231 of 4934 total). It should be noted that the epitopes presented on human MHC class I molecules are primarily (~90%) longer then 8 amino acids, meaning that in some cases they could be further trimmed by ERAP1.
Discussion
MHC class I epitopes are generated through a complex pathway that is influenced by proteasomes, peptidases, TAP, and tapasin as well as physical binding of mature epitopes to MHC class I alleles. The efficiencies by which potential epitopes are generated from precursor proteins influences epitope abundance and recognition, which is important in vaccine design and the development of immunodominance hierarchies. While some steps in the pathway are relatively well understood (e.g. TAP translocation, proteasome processing) the importance and direction of some other steps (e.g. peptidase processing) have not been as well studied.
N-terminally-extended epitopes, generated by the proteasome (16), can be trimmed by aminopeptidases to generate peptides of the correct length for MHC class-I binding (24). However, only a small fraction of the peptide pool bind MHC class-I molecules and even fewer trigger a significant T-cell response. This immunodominance can be a result of antigen processing and ultimately epitope abundance at the cell (9–11). In cells lacking the ER-resident aminopeptidase ERAP1, ER-targeted precursors are presented poorly if at all and ERAP1-deficient mice have markedly different presentation of many MHC class I epitopes (11, 29–31). Previous studies have also shown that this aminopeptidase can contribute to the pattern of immunodominance (11, 42). ERAP1 is a unique aminopeptidase in that its substrate preferences are guided in part by the C-terminus of the recognized peptide (28). However, its specificity is not otherwise well characterized.
Most previous analyses of the N-terminal sequence specificity of ERAP1 have been done with non-physiological substrates (such as dipeptides) and it has been clear that in some cases such substrates don’t model ERAP1’s behavior with longer “physiological” epitope precursors (43) Moreover, these analyses have been primarily performed in cell free systems with purified enzymes or microsomes and it is not clear whether these assays replicate what occurs in living cells. Since ERAP1 is such a key player in the antigen presentation pathway we set out to understand its specificity for N-terminal amino acids on N-extended epitopes and how this specificity could influence peptide delivery to MHC class-I molecules.
Using purified ERAP1 we showed that this aminopeptidase was capable of removing many different amino acids from the N-terminus of an epitope precursor. This broad activity is of obvious biological importance in allowing ERAP1 to trim the panoply of MHC class I-presented epitopes. However, the rates at which N-terminal residues were cleaved varied considerably and reproducibly between different amino acids. This suggests that ERAP1s specificity could limit peptide supply from ER precursors if epitopes are extended on the N-terminus by unfavorable amino acids. Conversely, more efficient trimming by ERAP1 of other amino acids could lead to greater epitope production for competition and presentation to the immune system. Previous studies characterizing ERAP1 or microsomal extracts, using a limited set of simple single amino acid fluorogenic substrates (X-AMC) (38, 44) have shown activity primarily for cleaving Leu and Met residues. This is consistent with our results that show that these amino acids are the most rapidly removed from the XS-L peptides and presumably reflects the higher affinity of Leu and Met for the ERAP1 active site. However, it is clear from our work that ERAP1 can remove many other amino acids, although more slowly. Our results are largely consistent with an analysis of the trimming of a limited number of peptide precursors by microsomes in vitro (38, 44); the few variations between these studies, particularly for charged amino acids, may reflect differences between our using a pure enzyme versus the earlier study using crude microsome preparations (potentially contaminated with cytosolic peptidases).
Another finding of interest was that regardless of the N-terminal flanking residues, purified ERAP1 virtually stopped trimming when the mature S-L epitope was generated. This is consistent with our previous findings, that ERAP1 trims with a molecular ruler down to a peptide of 8 or 9 residues in length (27), and indicates that this ruler is not influenced by the identity of the P1 residue. This unique function may contribute to the dominant role ERAP1 plays in antigen presentation and epitope generation due to the fact that trimming ceases once peptides capable of MHC class-I binding are generated. Kanaseki et al recently suggested that ERAP1 does not have a molecular ruler because it hydrolyzed and destroyed a mature antigenic epitope (45); however this epitope was a 9mer and we have previously established that ERAP hydrolyzes about 50% of 9 mers to 8mers (27). In other words, the concept of a molecular ruler is that ERAP1 trims down to a final core size (which can be 8 or 9 residues) and this core may or may not result in a mature epitope. This explains why ERAP1 can help to both create and destroy epitopes (11, 26). Importantly, the specificity of ERAP1 for N-terminal residues was not just seen at the 9mer to 8mer conversion but also at sequences further upstream of the epitope (e.g. the conversion of 12mers to 11mers to 10mers). To determine the relevance of these findings to the in vivo situation we transfected cells with minigenes encoding ER-targeted (signal sequence) epitope precursors. The amino acid in the P1 or P2 positions strongly influenced the amount of MHC class I presentation from precursors in the ER. The same trends were seen whether the residue upstream of the epitope was present as a single amino acid or as a doublet, when the residue was preceded by a longer sequence (e.g. LEQLXS-L), when the precursor peptide was a 10mer or a 12mer, and when different unrelated epitopes were used including 8mers and 9mers (S-L, F-L, or R-Y). When residues that were efficiently or poorly removed were paired in either orientation, the poor residue usually dominated and led to low presentation, as expected since trimming of the N-terminal extension would be affected whether it is the first or second residue that is removed slowly. Taken together these results demonstrate that in vivo the amino acids in the P1 or P2 positions of both long and short precursors are critical determinants of the amount of epitope presented on MHC class I molecules.
The specificity of trimming of the ER-targeted precursors was similar in both HeLa and COS7 cells. This suggests that our findings are not cell type specific but more general, although additional cell types need to be examined to fully evaluate this issue. Another point is that since COS7 cells express ERAP1 and ERAP2, our findings suggest that ERAP1 is the dominant ER-trimming enzyme; of note this is true even for charged residues that ERAP2 could potentially trim. Nevertheless, it is possible that the few differences in presentation that were observed between HeLa and COS7 cells were due to the presence or absence of ERAP2. Moreover, it remains possible that ERAP2 plays a more important role for other sequences and/or in different cells.
We were not able to identify any simple chemical feature that determined whether an amino acid is a good substrate or not. In general, charged residues are processed slowly; but the large hydrophobic Trp, the polar non-charged Asn, and the small non-polar Gly are each also processed slowly. Thus while there is clear specificity in the trimming process it is not simply defined by the chemical class of amino acid side chains.
In almost all cases when two amino acids from either end of the presentation hierarchy were combined the inefficient residue was dominant and led to poor presentation. One exception suggests that context (i.e. adjacent amino acids, or peptide length) may also influence processing of some residues. When valine was immediately adjacent to S-L or F-L, the precursors were slowly trimmed, whereas when Val was separated from S-L by one residue (e.g. VLS-L) or in the peptide VR-Y, processing was efficient. It is also interesting that even when the presentation of epitopes preceded by an efficient/inefficient or inefficient/efficient pair was low, it was better than was observed with the inefficient residue alone, suggesting that the rate of trimming in vivo is also influenced to some extent by adjacent amino acids or peptide length. Similarly, presentation of the long XXYYS-L peptides was generally higher than for the same residue as an XXS-L peptide. Nevertheless, it is important to emphasize that while context may influence trimming (particularly of valine), there clearly are definable rules of efficiency and these rules apply with different epitopes.
Knocking down ERAP1 strongly inhibited presentation from the vast majority of ER targeted epitope precursors, suggesting a major role in trimming. We interpret the difference in presentation between the different precursors as reflecting the specificity of ERAP1 trimming. This conclusion makes the assumption that the various XX residues do not influence cleavage by the signal peptidase (which liberates the epitope precursor from the signal peptide). Two pieces of data support this assumption. The first is that we have observed essentially the same results for sequences that are adjacent (ss X-S-L) or 6 residues away (ss LEQLXS-L) from the signal sequence cleavage site. The second is that the results with the presentation from minigenes and trimming of the same sequences by purified ERAP1 correlates well with one another. Nevertheless, it should be noted that this in vitro to in vivo correlation is not 100%. Where concordance is not perfect there must be other factors that influence peptide trimming in vivo. In these situations we can’t exclude a minor contribution from the signal peptidase although the difference could equally well reflect the participation of other ER molecules (chaperones, other peptidases) or that the conditions in the ER and cell free buffer system are not identical and this somehow influences ERAP1’s properties. A complete understanding of ERAP1’s specificity will require further study and may be aided when its crystal structure is solved.
We believe our results are potentially contributory to helping to define some of the specificity of antigen processing. The power of current algorithms to predict presented peptides from the sequence of an antigen is limited presumably because they do not take into account all of the events, such as peptide trimming, that are needed to generate (or destroy) epitopes. Consistent with these idea that peptide trimming is an important factor in determining the repertoire of MHC class I-presented peptides (the presented “peptidome”) we find that the residues that empirically lead to high-level antigen presentation in our system (Tyr, Met, Leu, Ala, and Cys) are all over-represented N-terminal to natural epitopes, by at least 2 standard deviations above background frequency; while residues that are underrepresented adjacent to natural epitopes (Pro and Val, and charged residues as a group) are poorly processed in our system. Overall these results are mostly similar qualitatively to the earlier analysis of a smaller set of sequences by Schatz et al (38). It should be noted that one limitation of the databases used for these analyses is that the majority of the epitopes were defined based of their ability to optimally stimulate specific CD8 T cells and because of this some may not be identical to the naturally processed peptides. However, it is thought that only a small minority of epitopes are incorrectly assigned and therefore this should have a minimal effect on such analyses. Therefore, nonrandom frequencies of various amino acids are almost certainly due to differences in antigen processing.
While it is possible that the upstream residues also affect proteasome cleavage or TAP transport, the residues that are consistently associated with high-level presentation of our model epitopes are not particularly preferred for proteasomal cleavage (46, 47) or TAP translocation (23, 48). This implies that aminopeptidase specificities, specifically ERAP1, play an important role in vivo in determining the generation of presented peptides, and that our system accurately defines at least some of these specificities. It is in fact remarkable that correlations are seen between ERAP1’s specificity and the presented peptidome because there are other peptidases (e.g. cytosolic ones) that can contribute to trimming N-terminal residues.
Presumably other peptidases do contribute to the trimming of the sequences that are poorly trimmed by ERAP1; this may especially be the case for epitopes flanked by charged residues. Such flanking residues show only a modest reduction in frequency in the databases of presented peptides yet are very poorly trimmed by ERAP1 in vitro and in vivo. This might be because cytosolic aminopeptidases such as leucine aminopeptidase can remove charged residues (15). Alternatively, the proteasome’s tryptic active site, which cleaves preferentially on the carboxlylic side of basic residues and its caspase-like site, which cleaves after acid residues, might remove these charged residues. Consistent with this later idea, the presentation from ovalbumin of SIINFEKL (which is flanked on its N-terminus with a charged E residue) is not affected in ERAP1 deficient cells. Interestingly, although ERAP2 can hydrolyze basic residues (40), we find that precursors with charged flanking residues are still poorly presented in ERAP2-expressing cells.
In summary our findings reveal that the amount of MHC-peptide complex presented to the immune system is determined in part by the amino acids upstream of epitope precursors and the ability of ERAP1 to remove these. The specificity of this trimming process for ERAP1 in vivo and in vitro for multiple epitopes is defined and reveals that that ERAP1 influences the extent of presentation in predictable ways.
Supplementary Material
Acknowledgments
We thank T. Nguyen and L.J. Stern for recombinant ERAP1 and S. Zendzian for technical assistance.
Abbreviations
- ER
endoplasmic reticulum
- ERAP1
Endoplasmic reticulum aminopeptidase 1
- MFI
mean fluorescence intensity
- MHC
major histocompatibility complex
- TAP
transporter associated with Ag processing
Footnotes
This work was supported by grants from the National Institutes of Health (to K.L.R.) and core resources supported by the Diabetes Endocrinology Research Center Grant.
References
- 1.Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg AL, Cascio P, Saric T, Rock KL. The importance of the proteasome and subsequent proteolytic steps in the generation of antigenic peptides. Mol Immunol. 2002;39:147–164. doi: 10.1016/s0161-5890(02)00098-6. [DOI] [PubMed] [Google Scholar]
- 3.Rock KL, I, York A, Saric T, Goldberg AL. Protein degradation and the generation of MHC class I-presented peptides. Adv Immunol. 2002;80:1–70. doi: 10.1016/s0065-2776(02)80012-8. [DOI] [PubMed] [Google Scholar]
- 4.Trombetta ES, Mellman I. Cell biology of antigen processing in vitro and in vivo. Annu Rev Immunol. 2005;23:975–1028. doi: 10.1146/annurev.immunol.22.012703.104538. [DOI] [PubMed] [Google Scholar]
- 5.Shastri N, Cardinaud S, Schwab SR, Serwold T, Kunisawa J. All the peptides that fit: the beginning, the middle, and the end of the MHC class I antigen-processing pathway. Immunol Rev. 2005;207:31–41. doi: 10.1111/j.0105-2896.2005.00321.x. [DOI] [PubMed] [Google Scholar]
- 6.Rammensee HG. Chemistry of peptides associated with MHC class I and class II molecules. Curr Opin Immunol. 1995;7:85–96. doi: 10.1016/0952-7915(95)80033-6. [DOI] [PubMed] [Google Scholar]
- 7.Lipford GB, Hoffman M, Wagner H, Heeg K. Primary in vivo responses to ovalbumin. Probing the predictive value of the Kb binding motif. J Immunol. 1993;150:1212–1222. [PubMed] [Google Scholar]
- 8.Yewdell JW, Bennink JR. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu Rev Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]
- 9.Restifo NP, Bacik I, Irvine KR, Yewdell JW, McCabe BJ, Anderson RW, Eisenlohr LC, Rosenberg SA, Bennink JR. Antigen processing in vivo and the elicitation of primary CTL responses. J Immunol. 1995;154:4414–4422. [PMC free article] [PubMed] [Google Scholar]
- 10.Gallimore A, Dumrese T, Hengartner H, Zinkernagel RM, Rammensee HG. Protective immunity does not correlate with the hierarchy of virus-specific cytotoxic T cell responses to naturally processed peptides. J Exp Med. 1998;187:1647–1657. doi: 10.1084/jem.187.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.York IA, Brehm MA, Zendzian S, Towne CF, Rock KL. Endoplasmic reticulum aminopeptidase 1 (ERAP1) trims MHC class I-presented peptides in vivo and plays an important role in immunodominance. Proc Natl Acad Sci U S A. 2006;103:9202–9207. doi: 10.1073/pnas.0603095103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 13.Craiu A, Akopian T, Goldberg A, Rock KL. Two distinct proteolytic processes in the generation of a major histocompatibility complex class I-presented peptide. Proc Natl Acad Sci U S A. 1997;94:10850–10855. doi: 10.1073/pnas.94.20.10850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mo XY, Cascio P, Lemerise K, Goldberg AL, Rock K. Distinct proteolytic processes generate the C and N termini of MHC class I-binding peptides. J Immunol. 1999;163:5851–5859. [PubMed] [Google Scholar]
- 15.Beninga J, Rock KL, Goldberg AL. Interferon-gamma can stimulate post-proteasomal trimming of the N terminus of an antigenic peptide by inducing leucine aminopeptidase. J Biol Chem. 1998;273:18734–18742. doi: 10.1074/jbc.273.30.18734. [DOI] [PubMed] [Google Scholar]
- 16.Cascio P, Hilton C, Kisselev AF, Rock KL, Goldberg AL. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. EMBO J. 2001;20:2357–2366. doi: 10.1093/emboj/20.10.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reits E, Griekspoor A, Neijssen J, Groothuis T, Jalink K, van Veelen P, Janssen H, Calafat J, Drijfhout JW, Neefjes J. Peptide diffusion, protection, and degradation in nuclear and cytoplasmic compartments before antigen presentation by MHC class I. Immunity. 2003;18:97–108. doi: 10.1016/s1074-7613(02)00511-3. [DOI] [PubMed] [Google Scholar]
- 18.Momburg F, Roelse J, Hammerling GJ, Neefjes JJ. Peptide size selection by the major histocompatibility complex-encoded peptide transporter. J Exp Med. 1994;179:1613–1623. doi: 10.1084/jem.179.5.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauvau G, Kakimi K, Niedermann G, Ostankovitch M, Yotnda P, Firat H, Chisari FV, van Endert PM. Human transporters associated with antigen processing (TAPs) select epitope precursor peptides for processing in the endoplasmic reticulum and presentation to T cells. J Exp Med. 1999;190:1227–1240. doi: 10.1084/jem.190.9.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knuehl C, Spee P, Ruppert T, Kuckelkorn U, Henklein P, Neefjes J, Kloetzel PM. The murine cytomegalovirus pp89 immunodominant H-2Ld epitope is generated and translocated into the endoplasmic reticulum as an 11-mer precursor peptide. J Immunol. 2001;167:1515–1521. doi: 10.4049/jimmunol.167.3.1515. [DOI] [PubMed] [Google Scholar]
- 21.Neisig A, Roelse J, Sijts AJ, Ossendorp F, Feltkamp MC, Kast WM, Melief CJ, Neefjes JJ. Major differences in transporter associated with antigen presentation (TAP)-dependent translocation of MHC class I-presentable peptides and the effect of flanking sequences. J Immunol. 1995;154:1273–1279. [PubMed] [Google Scholar]
- 22.van Endert PM, Riganelli D, Greco G, Fleischhauer K, Sidney J, Sette A, Bach JF. The peptide-binding motif for the human transporter associated with antigen processing. J Exp Med. 1995;182:1883–1895. doi: 10.1084/jem.182.6.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doytchinova I, Hemsley S, Flower DR. Transporter associated with antigen processing preselection of peptides binding to the MHC: a bioinformatic evaluation. J Immunol. 2004;173:6813–6819. doi: 10.4049/jimmunol.173.11.6813. [DOI] [PubMed] [Google Scholar]
- 24.Rock KL, I, York A, Goldberg AL. Post-proteasomal antigen processing for major histocompatibility complex class I presentation. Nat Immunol. 2004;5:670–677. doi: 10.1038/ni1089. [DOI] [PubMed] [Google Scholar]
- 25.Serwold T, Gonzalez F, Kim J, Jacob R, Shastri N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature. 2002;419:480–483. doi: 10.1038/nature01074. [DOI] [PubMed] [Google Scholar]
- 26.York IA, Chang SC, Saric T, Keys JA, Favreau JM, Goldberg AL, Rock KL. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8–9 residues. Nat Immunol. 2002;3:1177–1184. doi: 10.1038/ni860. [DOI] [PubMed] [Google Scholar]
- 27.Saric T, Chang SC, Hattori A, York IA, Markant S, Rock KL, Tsujimoto M, Goldberg AL. An IFN-gamma-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat Immunol. 2002;3:1169–1176. doi: 10.1038/ni859. [DOI] [PubMed] [Google Scholar]
- 28.Chang SC, Momburg F, Bhutani N, Goldberg AL. The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a “molecular ruler” mechanism. Proc Natl Acad Sci U S A. 2005;102:17107–17112. doi: 10.1073/pnas.0500721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammer GE, Gonzalez F, Champsaur M, Cado D, Shastri N. The aminopeptidase ERAAP shapes the peptide repertoire displayed by major histocompatibility complex class I molecules. Nat Immunol. 2006;7:103–112. doi: 10.1038/ni1286. [DOI] [PubMed] [Google Scholar]
- 30.Yan J, V, Parekh V, Mendez-Fernandez Y, Olivares-Villagomez D, Dragovic S, Hill T, Roopenian DC, Joyce S, Van Kaer L. In vivo role of ER-associated peptidase activity in tailoring peptides for presentation by MHC class Ia and class Ib molecules. J Exp Med. 2006;203:647–659. doi: 10.1084/jem.20052271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammer GE, Gonzalez F, James E, Nolla H, Shastri N. In the absence of aminopeptidase ERAAP, MHC class I molecules present many unstable and highly immunogenic peptides. Nat Immunol. 2006;8:101–108. doi: 10.1038/ni1409. [DOI] [PubMed] [Google Scholar]
- 32.Wold WS, Cladaras C, Deutscher SL, Kapoor QS. The 19-kDa glycoprotein coded by region E3 of adenovirus. Purification, characterization, and structural analysis. J Biol Chem. 1985;260:2424–2431. [PubMed] [Google Scholar]
- 33.York IA, Grant EP, Dahl AM, Rock KL. A mutant cell with a novel defect in MHC class I quality control. J Immunol. 2005;174:6839–6846. doi: 10.4049/jimmunol.174.11.6839. [DOI] [PubMed] [Google Scholar]
- 34.Porgador A, Yewdell JW, Deng Y, Bennink JR, Germain RN. Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity. 1997;6:715–726. doi: 10.1016/s1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
- 35.Ozato K, Hansen TH, Sachs DH. Monoclonal antibodies to mouse MHC antigens. II. Antibodies to the H-2Ld antigen, the products of a third polymorphic locus of the mouse major histocompatibility complex. J Immunol. 1980;125:2473–2477. [PubMed] [Google Scholar]
- 36.Berger AE, Davis JE, Cresswell P. Monoclonal antibody to HLA-A3. Hybridoma. 1982;1:87–90. doi: 10.1089/hyb.1.1982.1.87. [DOI] [PubMed] [Google Scholar]
- 37.Saveanu L, Carroll O, Lindo V, Del Val M, Lopez D, Lepelletier Y, Greer F, Schomburg L, Fruci D, Niedermann G, van Endert PM. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat Immunol. 2005;6:689–697. doi: 10.1038/ni1208. [DOI] [PubMed] [Google Scholar]
- 38.Schatz MM, Peters B, Akkad N, Ullrich N, Martinez AN, Carroll O, Bulik S, Rammensee HG, van Endert P, Holzhutter HG, Tenzer S, Schild H. Characterizing the N-terminal processing motif of MHC class I ligands. J Immunol. 2008;180:3210–3217. doi: 10.4049/jimmunol.180.5.3210. [DOI] [PubMed] [Google Scholar]
- 39.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D. Herpes simplex virus turns off the TAP to evade host immunity. Nature. 1995;375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 40.Tanioka T, Hattori A, Masuda S, Nomura Y, Nakayama H, Mizutani S, Tsujimoto M. Human leukocyte-derived arginine aminopeptidase. The third member of the oxytocinase subfamily of aminopeptidases. J Biol Chem. 2003;278:32275–32283. doi: 10.1074/jbc.M305076200. [DOI] [PubMed] [Google Scholar]
- 41.Vanhoof G, Goossens F, De Meester I, Hendriks D, Scharpe S. Proline motifs in peptides and their biological processing. FASEB J. 1995;9:736–744. [PubMed] [Google Scholar]
- 42.Blanchard N, Gonzalez F, Schaeffer M, Joncker NT, Cheng T, Shastri AJ, Robey EA, Shastri N. Immunodominant, protective response to the parasite Toxoplasma gondii requires antigen processing in the endoplasmic reticulum. Nat Immunol. 2008;9:937–944. doi: 10.1038/ni.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evnouchidou I, Momburg F, Papakyriakou A, Chroni A, Leondiadis L, Chang SC, Goldberg AL, Stratikos E. The internal sequence of the peptide-substrate determines its N-terminus trimming by ERAP1. PLoS One. 2008;3:e3658. doi: 10.1371/journal.pone.0003658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsujimoto M, Hattori A. The oxytocinase subfamily of M1 aminopeptidases. Biochim Biophys Acta. 2005;1751:9–18. doi: 10.1016/j.bbapap.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 45.Kanaseki T, Blanchard N, Hammer GE, Gonzalez F, Shastri N. ERAAP synergizes with MHC class I molecules to make the final cut in the antigenic peptide precursors in the endoplasmic reticulum. Immunity. 2006;25:795–806. doi: 10.1016/j.immuni.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tenzer S, Peters B, Bulik S, Schoor O, Lemmel C, Schatz MM, Kloetzel PM, Rammensee HG, Schild H, Holzhutter HG. Modeling the MHC class I pathway by combining predictions of proteasomal cleavage, TAP transport and MHC class I binding. Cell Mol Life Sci. 2005;62:1025–1037. doi: 10.1007/s00018-005-4528-2. [DOI] [PubMed] [Google Scholar]
- 47.Bhasin M, Raghava GP. Pcleavage: an SVM based method for prediction of constitutive proteasome and immunoproteasome cleavage sites in antigenic sequences. Nucleic Acids Res. 2005;33:W202–207. doi: 10.1093/nar/gki587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peters B, Bulik S, Tampe R, Van Endert PM, Holzhutter HG. Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors. J Immunol. 2003;171:1741–1749. doi: 10.4049/jimmunol.171.4.1741. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.