Abstract
Genomic and proteomic studies of brain regions of specialized function provide evidence that communication among neurons is mediated by systems of diverse chemical messengers. These analyses are largely tissue- or population-based, whereas the actual communication is from cell-to-cell. To understand the complement of intercellular signals produced by individual neurons, new methods are required. We have developed a novel neuron-to-neuron, serum-free coculture approach that was used to determine the higher-level cellular peptidome of individual primary mammalian neurons. We isolated magnocellular neurons from the supraoptic nucleus of early postnatal rat and maintained them in serum-free low-density cultures without glial support layers; under these conditions, they required low-density cocultured neurons. Coculturing magnocellular neurons with hippocampal neurons permitted local access to individual neurons within the culture for mass spectrometry. Using direct sampling, we obtained peptide profiles for spatially distinct, identifiable neurons within the coculture. We repeatedly detected 10 peaks that we assign to previously characterized peptides and 17 peaks that remain unassigned. Peptides from the vasopressin prohormone and secretogranin-2 are attributed to magnocellular neurons, whereas neurokinin A, peptide J, and neurokinin B are attributed to cultured hippocampal neurons. This approach enables the elucidation of cell-specific prohormone processing and the discovery of cell−cell signaling peptides.
Keywords: Magnocellular neuron, supraoptic nucleus, hippocampus, cell culture, mass spectrometry, neuropeptide, neuron coculture
Cell-to-cell communication in the brain is achieved via a complex interplay of diffusive molecules, including peptides, neurotransmitters, neurotrophins, and cytokines. Signaling molecules of the mammalian nervous system have been isolated and identified from the complex chemical milieu of brain tissue extracts (1−5) and neuron cultures (6). The goal of this work is to resolve neuron-specific profiles of signaling peptides from cultured mammalian neurons using mass spectrometry (MS).
A recent advance in peptide measurement is the ability to perform MS to determine the peptides found in individual cells. A number of earlier studies demonstrated the ability to characterize unique neuropeptides and prohormone processing in larger invertebrate neurons (7−9). Recently, single cell protocols and instrument performance have improved to the point where individual mammalian cells can be profiled for their peptides (10,11).
To expand on this prior work and to profile neuron-specific peptide cues, we selected mammalian neurons from the central nervous system that produce known peptides in abundance. Magnocellular neurons from the rat supraoptic nucleus are large neuroendocrine cells that form long axons that transport oxytocin and vasopressin to the pituitary where they are released into the blood via the portal system. The neuroendocrine functions of these neuropeptides are well-characterized and include regulation of parturition and lactation and osmoregulation (12−14). Magnocellular neurons also are capable of region-specific, somatic, dendritic, and axonal peptide release (15−18), although the total complement and functional context of these peptides is not established. While these neuroendocrine cells are conventionally studied in the intact brain and acute brain slice, cultures of dissociated magnocellular neurons can be achieved (19−24) but are rarely used due to the level of culture difficulty and low yields (22).
To analyze the peptide complement of individual magnocellular neurons, defined culture conditions that remove serum and glial support layers are necessary. We hypothesized that by developing low-density primary cultures and then directly applying MS to individual cells, we could obtain peptide profiles of magnocellular neurons and their networks. To this end, we optimized conditions for establishing magnocellular neurons in primary culture with serum-free, defined media conditions compatible with MS. However, through systematic investigation, we found that these neurons required additional cellular support. Because hippocampal neurons are easy to culture in defined media, we incorporated them in cocultures to support magnocellular neurons for direct matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS.
The hippocampus, a key brain structure in memory acquisition and recall whose function is altered in Alzheimer’s disease, schizophrenia, depression, and epilepsy, is well-defined anatomically. Hippocampal neuronal morphology, connectivity, and cell culture protocols are very well established (25−27). Because we found that hippocampal neurons are beneficial for sustaining magnocellular neurons in serum-free cultures without glial support layers, we extended our MS analysis to include hippocampal neurons. By profiling the neuropeptide content of individual hippocampal neurons in coculture using MS, we expand the peptidome profiles that have been produced for homogenized hippocampi (3,28).
To identify candidate signaling compounds involved in hippocampal neuron-assisted magnocellular neuron development, cocultures were directly analyzed for their peptide content following the application of a one-step sample extraction method utilizing 2,5-dihydroxybenzoic acid (DHB) (29). This approach preserves, extracts, and stabilizes signaling peptides for MS. As is typical with single-cell or low-cell number MS, instrument parameters are optimized for sensitivity and not resolution; in addition, the low amount of material often precludes tandem MS. Nonetheless, we recently completed a peptidomic analysis of the supraoptic nucleus (1), and similar lists are available for the hippocampus (3). Using this information along with common prohormone processing information (30,31) and our accurate mass data, we reliably detect 27 masses, 17 of which are currently unassigned and 10 assigned as being from provasopressin, protachykinins A and B, secretogranin-2, and proopiomelanocortin.
Our work demonstrates that magnocellular neurons from the supraoptic nucleus of early postnatal rat require contact with other cells to develop long, peptide-rich processes in defined media and that mass-to-charge-based neuropeptide profiles of cultured mammalian neurons using MALDI-TOF MS permit the classification of mammalian cells by the profiles observed.
Results and Discussion
Considerations for Direct Cellular Peptidomics of Neurons
A variety of MS-based peptidomic studies have characterized a number of known and novel peptides from whole brains to brain regions. Most prior MS studies have used microliters of tissue containing hundreds of thousands of cells. These samples include neurons, glia, immune cells, and vasculature, so it becomes difficult to assign peaks or peptides to specific cell types. When we move from the tissue to single-cell level, the signal-to-noise ratio of the peptide peaks in the mass spectra is reduced but not to the extent that the cell population is reduced. This is because some peptides are present at locally high concentrations in specific types of cells, such as the magnocellular neurons.
While neuron-specific peptide studies using single-cell MS have been performed using a variety of preparations (8,11), investigations profiling cultured mammalian neurons have been limited. One reason has been that the complexities of heterogeneous cell populations in the brain do not permit precise analysis of only the neurons but include multiple cell types. Consequently, some peaks have yet to be assigned to a specific cell type. Given the importance of neuron-to-neuron communication for brain information processing, compared with other types of cell-to-cell signaling, such data are of considerable interest.
A second issue is that mammalian neurons are small with long, delicate neurites; thus isolation and manipulation of individual mammalian neurons directly from tissue for MS analysis is challenging, and cellular yields are low. In acute isolation of single dissociated neurons from brain tissue, longer neurites often are lost. Because axons and dendrites transport, store, and release neuropeptides, the desired analytes may be reduced or even eliminated in acute neuron isolation protocols.
Third, neurons of the mammalian central nervous system are sensitive to their chemical environment and thus require precise culture conditions for optimal growth (22,26,27,32); often the ideal media formulations contain sugars, salts, and other components, which are problematic for MS. Nevertheless, cell culture approaches offer great advantages by eliminating competitive signals and confounding variables of tissue preparations, thereby allowing single mammalian cell detection (10,11). For successfully profiling the peptides of magnocellular neuron, serum-free culturing conditions are optimal. The serum used in media preparation contains a surprising range of peptides that make spectra from serum-containing media problematic to interpret with direct proteomic analysis. To this end, we developed neuroendocrine magnocellular neuron cultures in serum-free conditions compatible with MALDI-TOF MS.
Characteristics of Magnocellular Neuron Development in Dissociated Cultures
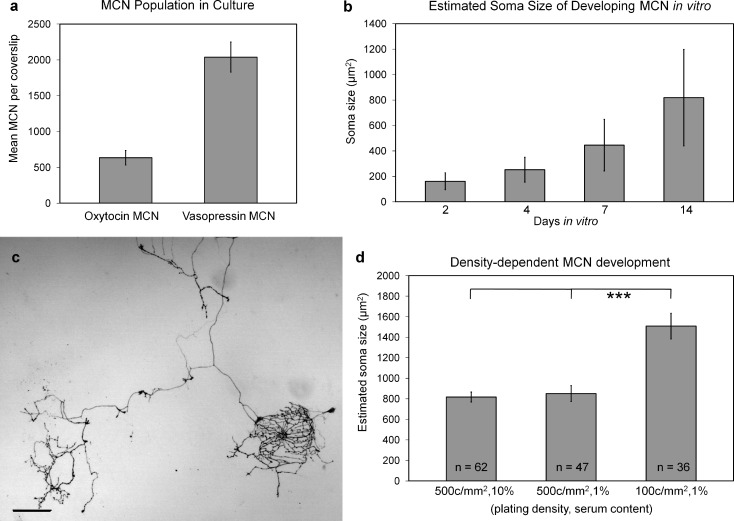
To establish baseline characteristics, initial cultures of magnocellular neurons were plated at 500 cells/mm2 in 10% serum (see Methods) and analyzed at 7 days in vitro (DIV) (Figure 1a). Under these conditions, glia rapidly proliferate to confluence and support magnocellular neuron development in culture. Magnocellular neurons were evaluated via immunochemistry. Coverslips from parallel cultures were labeled separately using PS-38 and PS-41 antibodies against oxytocin−neurophysin and vasopressin−neurophysin, respectively. The numbers of individual oxytocin−neurophysin and vasopressin−neurophysin magnocellular neurons per coverslip were counted from coverslips of five independent cultures (Figure 1a). The viability of vasopressin-producing, but not oxytocin-producing, magnocellular neurons in culture is influenced by media osmolarity, with lower osmolarity (255 mOsm) increasing survival over higher media osmolarities (300−330 mOsm) (22). Neurobasal culture media has low osmolarity (205−245 mOsm); therefore, vasopressin−neurophysin survival is favored under these conditions.
Figure 1.
Magnocellular neuron soma size in dissociated cultures depends on DIV and plating density. (a) The average number of vasopressin- and oxytocin-containing magnocellular neurons in baseline neuron cultures as identified through immunochemistry with PS-41 and PS-38 NP antibodies, respectively. Baseline cultures were seeded at 500 cells/mm2, cultured with 10% fetal bovine serum, and immunolabeled at day 7 DIV. Data (mean ± SEM) represents five separate cultures. (b) To assess magnocellular neuron development in high-density cultures (plated at 500 cells/mm2), soma size was measured at 2, 4, 7, and 14 DIV. (c) Axon terminals can develop distinct morphologies, presumably due to the recognition of different substrate cell types. Scale bar = 100 μm. (d) Magnocellular neuron size in culture is dependent on plating density rather than serum level. For comparison, the 14 DIV data of panel b is included (500 cells/mm2, 10%). In panels b and c, soma size was estimated as the mean product of the length and width. Mean ± SEM, unpaired t-test, p = 0.0001.
Our baseline data are in general agreement with a previously published trend demonstrating a preponderance of vasopressin−neurophysin over oxytocin−neurophysin in dissociated cultures (22). The morphological development of magnocellular neurons in culture was measured at 2, 4, 7, and 14 DIV. The product of perikaryon length and width demonstrates scatter of magnocellular neuron sizes from a single postnatal dissociated culture (Figure 1b) and is representative of the general trend of increasing soma size across three separate cultures. At ≥4 DIV, magnocellular neurons in culture develop characteristically long axons and short broad dendrites; Figure 1c displays a single axon bifurcation with distinctly different peptide-rich terminals.
To evaluate serum- and density-dependent growth of perikaryon size, we measured the number of neurons obtained from 15 high-resolution images per sample acquired at random intervals across the coverslips from a single magnocellular neuron culture at 14 DIV (Figure 1d; 500 cells/mm2, 1%, and 100 cells/mm2, 1%). By comparing these data with those of Figure 1b (500 cells/mm2, 10%, 14 DIV), we found that magnocellular neuron soma size did not change with serum reduction, but as cell density decreased, soma size increased substantially.
While we show that baseline cultures of postnatal magnocellular neurons express oxytocin- and vasopressin-producing neurons (Figure 1a) and soma size expands throughout 14 DIV (Figure 1b), cultures of embryonic magnocellular neurons also have been shown to increase cell size in culture depending on the age of tissue harvest (embryonic day 15−19) (33). In culture, magnocellular neurons develop long axonal processes with many varicosities and elaborated axon terminals for peptide transport and release. These terminals appear to recognize cell-to-cell contacts and develop into specialized structures (Figure 1c). The data indicate that magnocellular neurons undergo density-dependent development in vitro that influences perikaryon size (Figure 1d). While the serum reduction does not have a pronounced influence on magnocellular neuron soma size, growth factors in serum may provide beneficial signals to sustain extensive growth. Such benefits may include other changes than those we measured, for example, in rates of neuropeptide production. By using serum-free culture conditions and neuron-permissive media, we have almost completely eliminated non-neural support cells (i.e., glia) (27), further reducing confounds for peptide analysis.
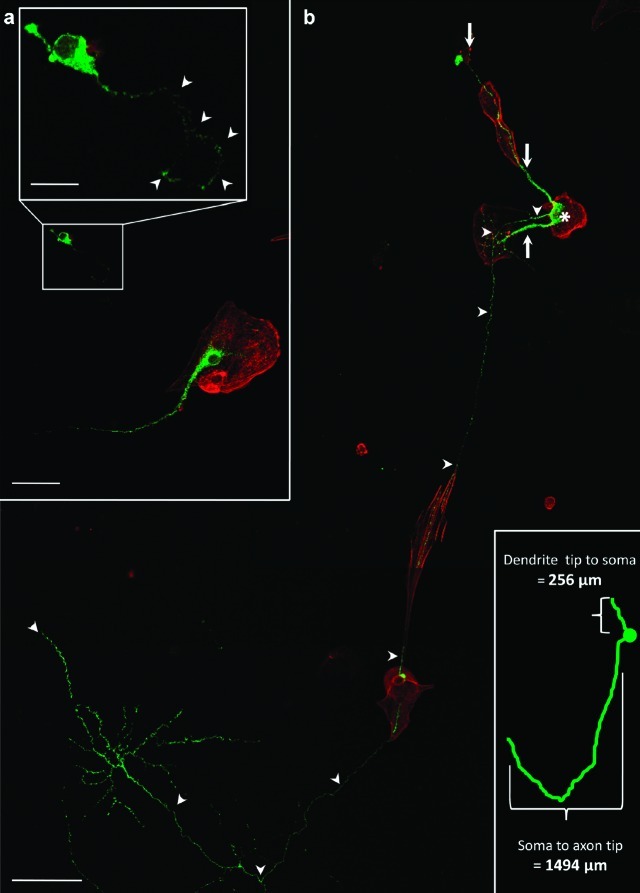
To establish culture conditions for neuropeptide analysis through direct MS, we analyzed magnocellular neuron development under serum-free conditions. To generate low-density cultures devoid of serum and glia, we plated cells at 100 cells/mm2 in serum-free media. Under these conditions, we observed two different fates for magnocellular neurons. In one case, when they attached only to laminin- or poly-d-lysine-coated coverslips, neuron development appeared stunted (Figure 2a); in the other case, when magnocellular neurons were in contact with another cell, development improved substantially yielding highly differentiated neurons with ramified axons (Figure 2a,b). Both outcomes were repeatable between cultures, and they were internally consistent within the same culture dish. Under these conditions, the numbers of magnocellular neurons with extensive neurite development are few.
Figure 2.
Cell-to-cell contact improves magnocellular neuron development. Neurons cultured on laminin show improved development when in contact with support cells rather than laminin alone. (a) Magnocellular neuron (green) on laminin alone and adjacent to magnocellular neuron on support cell (red). Magnified inset shows stunted magnocellular neuron growth; arrowheads identify the axon. (b) Extensive development of magnocellular neurons in low-density, serum-free culture for 8 d. The neurophysin-labeled magnocellular neurons (green) with soma (∗), axon (arrow heads), and dendrites (arrows), are situated on support cells (red), which are easily identified with rhodamine phalloidin. Distal dendrite tip to distal axon tip measurement = 1750 μm. Distance for distal dendrite to soma (∗) is 256 μm and for soma to axon tip is 1494 μm. Scale bars: top = 15 μm; center = 50 μm; bottom = 100 μm.
Under low-density, serum-free culture conditions, magnocellular neurons in contact with other cells demonstrate elaborate process maturation compared with those growing in isolation (Figure 2a,b). Relatively few cell contacts produced a pronounced difference in magnocellular neuron growth that enabled the formation of elaborate axon terminals that extended well over 1 mm (Figure 2b). This example was selected because the axon terminal demonstrates a bilaterally symmetric morphology on laminin and occurs in the absence of any direct cellular contacts. We conclude that cell-to-cell contacts provide some key, undefined support for optimal magnocellular neuron growth.
Cocultures Support Magnocellular Neuron Development in Serum-Free, Low-Density Cultures
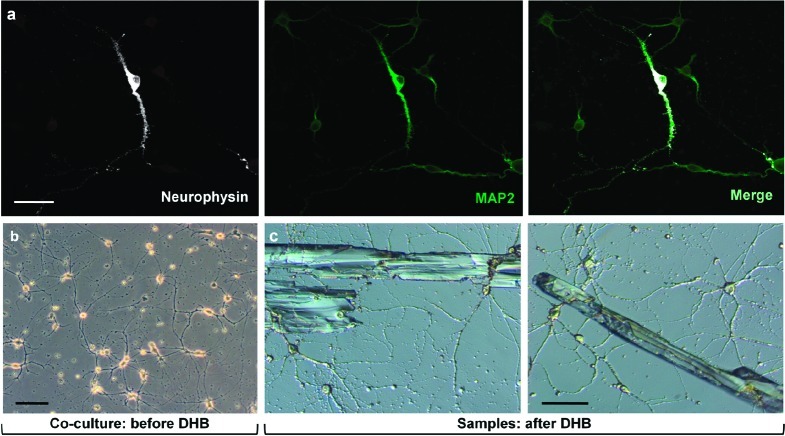
If cell-to-cell contact of non-neural, substrate cells support and improve magnocellular neuron development, then heterogeneous neurons may as well. To test this, we generated serum-free, hippocampal neuron/magnocellular neuron cocultures and evaluated magnocellular neuron development. Hippocampal neurons were tested due to their identifiable neuronal characteristics, easy dissection, and well-defined culture conditions. First, we established hippocampal neuron cultures for 4−9 days to allow the neurons to undergo axo-dendritic specification and axon elongation prior to seeding magnocellular neurons. To assess magnocellular neuron presence and development, cultures were double-labeled with antibodies against NP and MAP2, which recognize magnocellular neurons and all neurons (Figure 3), respectively. By plating magnocellular neurons onto established cultures of hippocampal neurons, development of the magnocellular neurons was improved to yield nearly pure neuronal populations consisting of arborized magnocellular and hippocampal neurons at low densities (Figure 3). In these cocultures, hippocampal neurons were the predominant neuronal type.
Figure 3.
Magnocellular neuron development is improved by hippocampal neurons in coculture. (a) Magnocellular neurons were plated onto a 5-day-old culture of hippocampal neurons and maintained for 7 days. Magnocellular neurons show abundant NP (white), while hippocampal neurons lack NP. MAP2 content (green) of the neurosecretory magnocellular neuron is abundant, compared with hippocampal neurons. (b) Low magnification, phase-contrast image of primary magnocellular neuron−hippocampal neuron coculture demonstrates neuronal connectivity. (c) After cultures were rapidly rinsed with DHB, crystals form to embed neurons and locally extract peptides (differential interference contrast microscopy). For this type of MS analysis, neuronal cells must be embedded in matrix crystals. Samples were dried for 1−2 days in a desiccator prior to performance of MALDI analysis. All scale bars = 50 μm.
As demonstrated in Figure 3a, hippocampal neurons effectively support magnocellular neuron development in serum-free conditions. Previous studies determined that hippocampal neuron density influences neuron viability, possibly due to achieving threshold concentrations of diffusive cues or neuron connectivity (27). We improved the microenvironment for magnocellular neurons by preplating hippocampal neurons to provide the close associations produced by coculturing. In this way, hippocampal neurons can provide both trophic and synaptic support to magnocellular neurons. We recognize the possibility that other neuronal populations also may be able to support magnocellular neuron development, by directly innervating magnocellular neurons or otherwise.
In contrast to our methods, prior protocols for dissociated magnocellular neuron cultures require serum or high-density feeder layers or both to achieve survival and magnocellular neuron differentiation (19−24,33). While the magnocellular neuron−hippocampal neuron coculture protocol is more labor intensive than the culture process of an individual brain area, this coculture process produces a chemical environment that is free of the observed serum-derived mass spectral peaks. Not only does this heterogeneous coculture have promise for profiling candidate neuropeptides from cultured magnocellular and hippocampal neurons, it reveals a novel developmental dependence relating to the cell biology of magnocellular neurons.
Direct MALDI-TOF MS and Peptide Profiles
By incorporating magnocellular neurons in coculture, we retain a positive control for validating our approach using the aforementioned single-step sample preparation. Due to the presence of magnocellular neurons, levels of expected peptides, such as arginine vasopressin, provide a meaningful baseline against which unidentified neuropeptides can be compared.
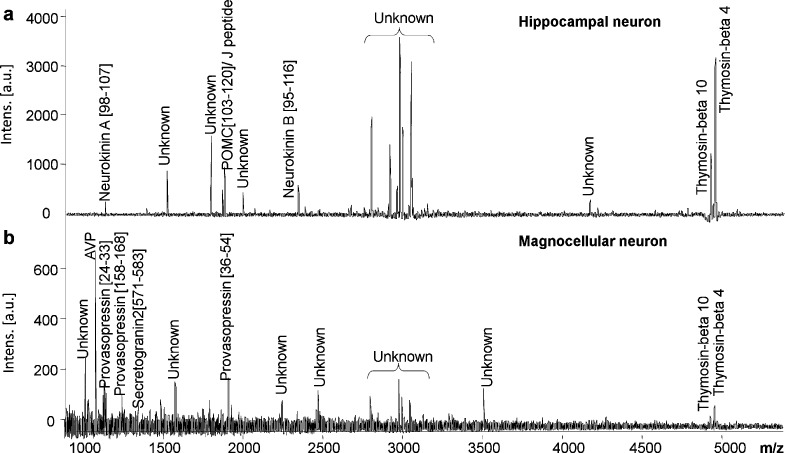
The supraoptic nucleus and hippocampus are regions of the brain that are known to exhibit neuroplasticity throughout the lifespan; therefore, our direct peptidomic approach provides a potential means for identifying bidirectional signaling molecules involved in neuron development and repair. To generate neuropeptide profiles of individual magnocellular and hippocampal neurons in cocultures, a total of six coverslips from three separate cocultures were used. DHB matrix rinse applications removed media from the cells, leaving a trace amount of matrix solution barely wetting the coverslip. As the crystals form on and around the neurons, endogenous peptides are locally extracted from the neurons and incorporated into the crystal matrix. This extraction protocol retains the structural integrity of the cultured neurons (Figure 3c) while enabling peptide detection (Figure 4 and Table 1). Coverslips with DHB crystals were directly analyzed using MALDI-TOF MS, and neuropeptide content was confirmed (Figure 4 and Table 1). The formation of DHB crystals can, but do not always, encapsulate neuronal soma or neurites (Figure 3). From the MS results, two characteristic profiles emerged repeatedly (Figure 4) and with relative consistency. Small variations were observed and are to be expected with mass spectrometry; for this reason, we replicated the results using six coverslips from three independent cocultures. Table 1 reports the peaks associated with the characteristic profiles. One contained predominantly vasopressin and other fragments from the same prohormone, while the other profile showed few vasopressin prohormone identifications.
Figure 4.
Peptide profiles of primary magnocellular and hippocampal neurons in coculture. Analysis of cocultures repeatedly produced distinct peptide profiles for each neuronal type; putative peptides are identified, and unknowns are labeled as such. Seventeen “unknown” peptides peaks were observed repeatedly. Cultures were analyzed by a Bruker Ultraflex II TOF/TOF instrument; mass spectra were accumulated over two to three individual acquisitions of 300 laser shots each with external calibration covering the 700−3000 Da mass range.
Table 1. Peptides Detected Directly from Supraoptic Magnocellular and Hippocampal Neurons in Coculturea.
| prohormone | obsd (m/z) | theor. (m/z) | error (Da) | putative magnocellular neuron | putative hippocampal neuron | presence in supraoptic nucleus (MSMS)b |
|---|---|---|---|---|---|---|
| neurokinin A [98−107] | 1132.87 | 1132.55 | −0.32 | x | ||
| POMC[103−120]/J peptide | 1882.93 | 1882.83 | −0.10 | x | x | x |
| neurokinin B[95−116] | 2345.58 | 2344.51 | −1.48 | x | ||
| provasopressin[24−32] | 1084.49 | 1084.44 | −0.05 | x | x | |
| provasopressin[24−33] | 1142.49 | 1142.46 | −0.03 | x | neuropred-rat model | |
| provasopressin[158−168] | 1250.49 | 1250.37 | −0.86 | x | x | |
| provasopressin[36−54] | 1920.36 | 1920.20 | −0.16 | x | neuropred-rat model | |
| secretogranin2[571−583] | 1354.49 | 1354.67 | 0.18 | x | x | |
| thymosin beta-10 | 4932.10 | 4932.51 | 0.41 | x | x | x |
| thymosin beta-4 | 4959.30 | 4960.48 | 1.18 | x | x | x |
| unknown | 863.74 | x | x | |||
| unknown | 1021.50 | x | ||||
| unknown | 1044.75 | x | x | |||
| unknown | 1521.62 | x | ||||
| unknown | 1586.61 | x | x | |||
| unknown | 1799.60 | x | ||||
| unknown | 1870.50 | x | ||||
| unknown | 1997.85 | x | ||||
| unknown | 2256.34 | x | ||||
| unknown | 2481.59 | x | ||||
| unknown | 2808.30 | x | x | |||
| unknown | 2923.80 | x | ||||
| unknown | 2985.70 | x | x | |||
| unknown | 3004.60 | x | ||||
| unknown | 3056.38 | x | x | |||
| unknown | 3516.44 | x | x | |||
| unknown | 4174.70 | x |
Summary of direct MALDI-TOF MS results for magnocellular neurons and hippocampal neurons in serum-free coculture after a single-step application of DHB matrix.
Reference (1).
On average, 17 peptides were observed per coverslip. This includes the combined total of unknown and putative signaling peptides for both magnocellular and hippocampal neurons; Table 1 lists the masses of the 27 peptide peaks observed. From direct MALDI-TOF MS, two reproducible, characteristic peptide profiles were observed (Figure 4). Molecular mass profiles show 10 peptides having mass matches to provasopressin, protachykinins A and B, proopiomelanocortin (POMC), and secretogranin-2 (Table 1). Vasopressin was used to confirm the identification of putative magnocellular neurons. In addition, provasopressin[24−32], [24−33], [36−54], and [158−168], POMC[103−120], and secretogranin2[571−583] of our current study (Table 1) were mass matched to values of our previous supraoptic nucleus peptidomics study, wherein the peaks were characterized by tandem MS (1) and by NeuroPred software. The cleavage tool NeuroPred predicted four putative provasopressin-related peptides, including provasopressin[24−32], [24−33], and [36−54], corresponding to 1084.46 m/z, 1142.46 m/z, and 1920.20 m/z, respectively. Based on the mass matches from these two sources, we assigned the above peaks to magnocellular neurons.
Mass profiles characteristic of putative hippocampal neurons are expected to contain a different set of peptides. Neurokinin A[98−107] was detected previously in two peptidomic studies of the hippocampus (3,28). Based on these studies, predicted peptides from neurokinin A and B are attributed to putative hippocampal neurons (Figure 4). Under these conditions, six unknown masses (m/z = 2808.30, 2923.80, 2985.70, 3004.60, 3056.38, and 3516.44) present a stronger signal in the mass profile of putative hippocampal neurons. A mass peak corresponding to the J peptide from proopiomelanocortin (POMC) is observed in profiles characteristic of hippocampal neurons. Previous studies have identified POMC in the human hippocampus (34), but it was not detected in the mouse hippocampus (3). However, POMC fragments were detected in the rat supraoptic nucleus (1). Either POMC is produced in the rat hippocampus and detected here, or axons from magnocellular neurons intermingled with hippocampal neurons to contribute POMC to these spectra; therefore, J peptide could be attributed to either magnocellular neurons, hippocampal neurons, or both. A thorough comparative peptidomic analysis of tissue from the rat hippocampus is warranted for future studies, work that would clarify this issue. In addition, thymosin β-4 and thymosin β-10 were dominant features of both type of neurons. These results demonstrate that a high number of peptides are detectable from mammalian neurons in serum-free cultures.
As part of a large neuropeptide family, tachykinins are active peptides that are involved in a range of physiological processes (35) and have been detected in the hippocampus of rats and humans (36−38), but not the mouse (37). Furthermore, the hippocampal formation contains abundant tachykinin binding sites (NK1, NK2, and NK3) (39), thus indicating a potential role of tachykinins in hippocampal neuron function. The specific localization and functional significance of neurokinin A and B, J peptide of POMC, and secretogranin-2[571−583] in the supraoptic nucleus and hippocampus are worthy of further investigation. Several unassigned peaks are observed in the mass range of m/z 2000−3600 (Table 1). Identifying these and placing them within a functional context will be important to more fully understand communication within the highly complex, interconnected structure of the brain.
In this direct neuronal proteomic study, we characterized 10 known peptides. The presence of several putative provasopressin fragments in the magnocellular neuron profile may indicate unique prohormone processing or cleavages, because these are often observed in direct MALDI MS profiling. Alternatively, these results may be attributed to peptide degradation during sample preparation or drying. An important finding is the ability to obtain peptide profiles from mammalian neurons in dissociated cell culture while retaining some spatial information relative to the neurons. We do observe occasional magnocellular neuron peptides from provasopressin in hippocampal neuron profiles. This can be explained by the fact that neurons develop long, branching processes that intermingle with other neurons in culture. As demonstrated here, axons can exceed 1.4 mm (Figure 2b); in the intact animal, axons of magnocellular neurons navigate several millimeters from the supraoptic nucleus to their pituitary target. Thus, it is possible that axons of magnocellular neurons can contribute a relatively small amount of neuropeptides to hippocampal neuron profiles.
Although we obtained cell-specific peptide profiles on primary cultured neurons, direct peptide sequencing through this approach was not possible due to low amounts of analytes available from the cultured neuronal samples. Therefore, not every peptide identified by mass was confirmed by tandem MS sequencing. For example, oxytocin (theoretical mass of m/z 1006.44) was not detected even though it is a known magnocellular neuron product and is in the ideal mass range for detection. This could be due to low amounts of oxytocin, the low number of magnocellular neurons in the culture, or a yet unidentified molecular interaction or post-translational modification that may alter the ability to detect oxytocin at its expected mass. While any of these explanations may be valid, it should be noted that oxytocin was not detected in peptidome studies of rat supraoptic nucleus extracts (1) nor was it reported in extracts of the entire rat hypothalamus (40). Nevertheless, it was detected in the mouse hypothalamus via MS by utilizing different reagents for sample preparations (3) and after N-terminal peptide modification (4).
Although extracting brain tissue will provide a greater number of peptides, the present approach enables a direct measure of mammalian neuropeptide content within the context of specific cell types. In either case, complete profiles are not attained (1). Rather, different preparation methods provide different peptide signatures, and by combining data from the range of sample types, extraction methods, and peptide characterization approaches, we arrive at a more complete understanding of peptides in the context of cell−cell signaling. In our previous work, tissue was collected from 6−12 week old rats (1), whereas cultured neurons used here are comparable to 4−13 day old, age-adjusted rats. Allowing neurons to age longer in culture or culturing from older animals may enable more, or at least different, peptides to be evident in the mass profiles. Only a fraction of those peptides observed in our former work are observed here, possibly due to differences in age-dependent development and peptide production, the amount of peptide in the sample, or the fewer number of cells. While our study demonstrates the ability to directly sample primary neurons from the mammalian brain in culture, we expect that technological advances will provide improved opportunities for sample preparation and peptide detection toward advancing our understanding of the complete peptide complement in individual mammalian neurons.
Conclusion and Future Directions
In conclusion, we demonstrate that low-density coculturing of magnocellular and hippocampal neurons improves magnocellular neuron development over other methods; our results highlight the importance of intercellular chemotrophic support. To better understand the chemotrophic neuropeptides of these cocultures, we confirmed the neuropeptide content of established magnocellular neuron marker peptides and profiled cultured hippocampal neurons using MALDI-TOF MS. We demonstrate the ability to perform direct mass profiling of cultured and cocultured mammalian neurons and to distinguish spectra attributable to each type of neuron. This is accomplished in serum-free media, a chemical environment free of interfering mass spectral peaks. Manipulations of tiny samples as small as individual rat neurons directly makes it possible to access intact mammalian neurons within cocultures using MALDI-TOF techniques for spatially sensitive characterization of intracellular neuropeptides. This approach enables unprecedented access to individual, developed, intact mammalian neurons for cell-specific mass spectrometry. Now, reliable detection of neuropeptides from dissociated cultures of mammalian neurons can be achieved by using a one-step sampling extraction application.
Future directions will focus on improving peptide yield for sequence identification, understanding the roles these peptides play in magnocellular neuron development and clarifying the nature of previously established peptides, specifically oxytocin. We will explore the chemical environment provided by cell-to-cell contacts, whether neuron-to-neuron or glia-to-neuron, and how they provide support for optimal magnocellular neuron growth. A thorough comparative peptidomic analysis of tissue from the rat hippocampus is warranted to clarify our observation of a mass peak corresponding to the J peptide from pro-opiomelanocortin (POMC) in profiles characteristic of hippocampal neurons. To more fully address context-dependent and temporal facets of neuropeptide production, neurons can be sustained in low-density culture by controlling the local microenvironment of the neuron (41), which can be manipulated to retain secreted factors in the immediate proximity for temporal analysis of released peptides (42). Local environments can be accessed to stimulate altered cellular functions, and local changes in peptides can be measured. Our approaches enable unprecedented access to individual neurons and their neurite extensions, raising new prospects for subcellular peptidomic analyses.
Methods
Chemicals
All reagents were obtained from Sigma-Aldrich Chemical Co. (St. Louis, MO), unless otherwise noted.
Cell Culture
To generate cultures of magnocellular neurons and hippocampal neurons, postnatal (P1−P2) Long−Evans BluGill rats (University of Illinois at Urbana−Champaign) were used in accordance with protocols established by the University of Illinois Institutional Animal Care and Use Committee and in accordance with all state and federal regulations. Procedures for culturing each neuronal type are defined below. Neurons were grown on glass coverslips for microscopic imaging and MS. Prior to cell culture, coverslips were cleaned by immersion in undiluted sulfuric acid (overnight minimum) and rinsed with copious amounts of sterile-filtered deionized water (dH2O, Milli-Q, Millipore, Bedford, MA). Dry coverslips were coated for at least 2 h with laminin (100 μg/mL) for initial magnocellular neuron cultures at high density and with poly-d-lysine (PDL, 1−2 h, 100 μg/mL; MW = 30 000−70 000) for hippocampal neuron cocultures and magnocellular neuron low-density cultures.
Magnocellular Neurons in Primary Culture
Supraoptic nuclei were isolated by dissecting the optic tracts and immediately adjacent tissue using a modified brain dissection (Figure 5) from Bilinski and colleagues (19). Dissection, enzymatic digestion, trituration, and centrifugation steps were carried out in Hibernate-A without phenol red (Brain Bits, Springfield, IL) and supplemented with 0.5 mM l-glutamine (Invitrogen, Eugene, OR), B-27 (Invitrogen), 100 U/mL penicillin and 0.1 mg/mL streptomycin. For each experiment, eight or fewer supraoptic nucleus regions were pooled and digested with papain (25.5 U/mL, Worthington, Lakewood, NJ) for 45 min at 37 °C; the number of supraoptic nucleus regions used were ∼1.0−1.5 per high-density coverslip (500 cells/mm2) or 1.0 supraoptic nucleus for 5−6 coverslips at 100−125 cells/mm2. After papain incubation, the enzyme was removed, and the tissue was rinsed and mechanically dissociated through trituration using a fire-polished Pasteur pipet. After the undissociated optic tracts settled, the supernatant was transferred to a new 15 mL vial and centrifuged at 1400 rpm for 5 min. Hibernate supernatant was aspirated, and the cellular pellet was resuspended in culture media.
Figure 5.
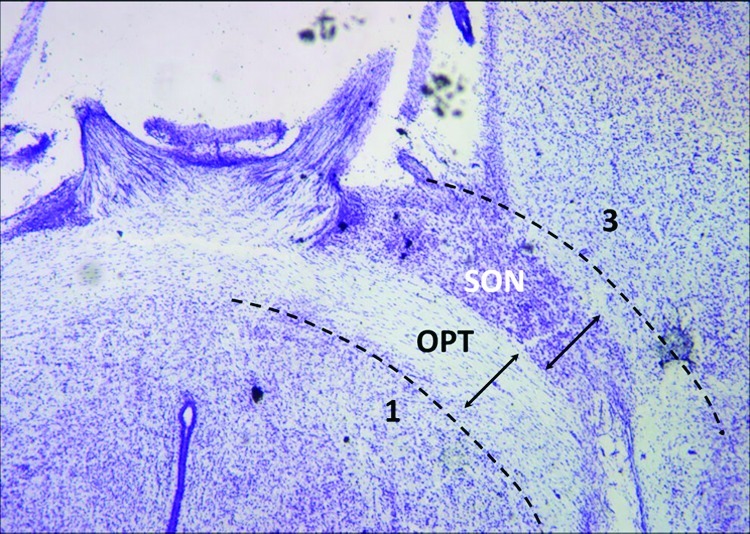
Horizontal hypothalamic brain slice of postnatal rat. Using the optic tracts (OPT) as landmarks, four dissection steps were used to remove the tissue containing the supraoptic nucleus (SON). (1) Posterior cuts (shown): bilateral incisions were made along the immediate posterior surface of the optic tract to a point where the tracts enter into the brain and are no longer visible from the ventral surface. (2) Lateral cuts (not shown): one cut per side of the brain to sever the optic tract where it penetrates into the brain near the rostral cerebral artery. (3) Anterior cuts (shown): these incisions were performed away from the surface of the optic tract, by about the same width of the OPT, to ensure that the supraoptic nucleus would be retained and not damaged in the cutting. (4) Dorsal cut (not shown): this last cut is made as close to the dorsal border of the optic tract as possible. Upon completion of these four steps, the excised tissue containing the supraoptic nucleus and a portion of the optic tract is removed. The images are paraformaldehyde-fixed sections from a single postnatal rat brain, sectioned horizontally by a cryostat and histochemically stained with cresyl violet to identify the Nissl substance. The supraoptic nucleus appears as a darkly stained cluster of cells adjacent to the optic tracts.
Viable cells, identified by Trypan-blue exclusion, were counted with a hemacytometer, diluted in culture media, and plated at 500 cells/mm2 for high-density cultures and 100−125 cells/mm2 for low-density cultures. By dissecting neurons in this manner from early postnatal development (P1−P2) animals, we achieved a nearly 6-fold increase of total magnocellular neurons in culture over a previous report by Kusano et al. (22). Based on their work, we used the media formulation of Neurobasal-A media without phenol red (Invitrogen) supplemented with 2% B-27 (Invitrogen), 100 U/mL penicillin, 0.1 mg/mL streptomycin, and 50 ng/mL recombinant human brain-derived neurotrophic factor (BDNF, PeproTech, Rocky Hill, NJ). Fetal bovine serum (Hyclone, Logan, UT) was initially at 10% and systematically reduced through separate experiments, then eliminated over the course of developing the serum-free culture protocol. Finally, magnocellular neurons in serum-free culture were plated onto cultures of differentiated hippocampal neurons, established previously for at least 4 d, and examined with MS.
Hippocampal Neurons
For cocultures with magnocellular neurons, hippocampal neuron cultures were prepared with a similar tissue digestion and cell isolation process (41). Postnatal tissue was used to reduce glial proliferation evident with embryonic tissue (43). Briefly, the hippocampi of postnatal (P1−P2) Long−Evans BluGill rats were dissected, enzymatically digested (30 min), rinsed, dissociated, centrifuged, counted, and then plated (100−125 cells/mm2) on PDL-coated coverslips. Due to our (1) use of postnatal brains as the cell source (44), (2) method of cell isolation, and (3) choice of Neurobasal-A media with B-27, which substantially eliminates glial cells in serum-free cultures (27), we observed the viable cell yield to be between 30% and 40% of initial plating density. We estimated final hippocampal neuron yield to be 30−50 cells/mm2 when plated at 100−125 cells/mm2. For MALDI-TOF MS and microscopy, Hibernate-A and Neurobasal-A were both phenol red-free solutions, and both were supplemented as defined above to include 0.5 mM l-glutamine, B-27, 100 U/mL penicillin, and 0.1 mg/mL streptomycin. BDNF was omitted during the establishment of the hippocampal neurons prior to the addition of the dissociated magnocellular neurons; fetal bovine serum was not used in hippocampal neuron cultures or cocultures because serum-free media is required for MS, but is not needed for magnocellular neuron cocultures. Phase contrast and differential interference contrast images of cocultures and crystallized DHB on cultures were acquired using a Zeiss Axiovert 40 microscope.
Immunocytochemistry
The following protocol is based on previously reported methods from Banker and Goslin (25). Cell cultures were immediately rinsed and incubated with 4% paraformaldehyde (37 °C) for 30 min, then with PBS (Fisher Scientific, Pittsburgh, PA), followed by 0.25% Triton X-100 (Biorad, Hercules, CA) application for 5−10 min to permeabilize the membrane. For 3,3′-diaminobenzidine (DAB) immunochemistry, samples were treated with hydrogen peroxide (<15%) in PBS to inactivate endogenous peroxidases prior to blocking (10% BSA for 30 min). The primary antibody was incubated for 1−2 h at room temperature or overnight at 4 °C in 2.5% BSA in PBS. Primary mouse monoclonal antibodies against oxytocin−neurophysin (PS-38) and vasopressin−neurophysin (PS-41) were kindly supplied by Dr. Harold Gainer, NIH, Bethesda, MD. A rabbit polyclonal antineurophysin (NP) antibody (product number V2045, Biomeda, Foster City, CA) that recognizes both forms of NP was used in low-density cultures and cocultures to identify all magnocellular neurons of the supraoptic nucleus that were present. The neuron-specific marker, microtubule associated protein 2 (MAP2), was immunolabeled with rabbit polyclonal antibodies (AB5622, Chemicon, Temecula, CA).
After primary antibody incubation, cultures were thoroughly rinsed with PBS, and then secondary antibodies were incubated for 1 h at room temperature in 2.5% BSA in PBS, followed by PBS washes. For MAP2 and NP immunofluorescence double labeling, the magnocellular neurons were first labeled with the polyclonal NP immunoreagent, then Alexa 633 secondary antibody, followed by the MAP2 immunoreagent and Alexa 488 secondary antibody (Alexa antibodies, Molecular Probes, Eugene, OR). When rhodamine phalloidin (Invitrogen) was used to label F-actin, it was applied to fixed cells in PBS for 20 min followed by PBS rinsing for at least another 20 min. For DAB labeling, only monoclonal antibodies were used; the Vector ABC Biotin Kit (Burlingame, CA) was used in conjunction with biotin-conjugated secondary antibodies. Prior to mounting samples, coverslips were briefly rinsed with dH2O to prevent crystallization of salts. The samples were dried and mounted with Prolong Gold Antifade reagent (Molecular Probes) for fluorescent samples and Permount (Fisher Scientific) for DAB-labeled samples. Images were acquired using a laser-equipped, confocal microscope (Zeiss LSM-510 Meta NLO).
Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS)
Peptide standards used for external calibration were applied to corners of the coverslips, regions where no cellular profiles were analyzed, but in close proximity to the cells of interest. Standards were obtained from Bruker Daltonics, Inc. (Billerica, MA). Standards used included angiotensin II, angiotensin I, substance P, bombesin, ACTH clip 1−17, bradykinin, ACTH clip 18−39, and somatostatin 28 covering a mass range of ∼700−3200 Da. Coverslips containing magnocellular neuron−hippocampal neuron cocultures were prepared for MALDI-TOF MS following an optimized modification of a previously published protocol of a one-step sample extraction method (29). In addition to this protocol, we attempted to extract peptides from neurons cultured on coverslips using standard protocols and using solid-phase extraction beads. We were unable to obtain any reliable signal with these standard methods being applied to cultured neurons from the postnatal rat brain.
For the one-step extraction protocol, cocultures were quickly and carefully rinsed three to four times with approximately 1 mL of DHB (20 mg/mL, 70% acetonitrile, 0.1% trifluoroacetic acid); small crystals of various morphologies formed over the cell surface of the coverslip. Samples were dried for 1−2 days under desiccation at room temperature. Crystals of various sizes and morphology were surveyed across the coverslip. Experimental results were acquired from neurons embedded in sharp DHB crystals found on the glass (Figure 3c). Slender crystals (50−100 μm in width) for MS measurement form on the coverslips that are barely wetted by the matrix, rather than in higher volumes of salts, media, and cellular debris. Due to the coverslip size and shape (22 mm square) and the rinsing process, the corners and coverslip periphery are rapidly rinsed clean. The central portion (∼25−50%) is difficult to clean completely and is often rendered unsuitable for reliable MS analysis due to residual media salt retention that accumulates in matrix crystals. To control crystal formation, each culture application should be optimized with the following considerations: the precise method of application (rinsing or dipping) of DHB matrix solution and removal of excess matrix (wicking or aspirating), coverslip size and shape, and cell density on the glass. Even though MALDI-TOF MS is salt tolerant, maintaining a clean, dry sample with good crystal formation is essential (Figure 3c).
Coverslips were mounted on a MALDI target for MS analysis. Samples were grounded using silver paint to prevent charging of the glass to achieve optimal peptide signals (Figure S1, Supporting Information). Each cell culture was analyzed by a Bruker Ultraflex II TOF/TOF instrument with a delayed ion extraction and a frequency-tripled Nd:YAG laser operating at 1064 nm and 50 Hz (Bruker Daltonics). Mass spectra were accumulated over two to three individual acquisitions of 300 laser shots each. The instrument settings were optimized for reflection mode and internal calibration peptides covered the mass range of 700−3000 Da.
Data Analysis
Magnocellular Neuron Development
To determine the neuron content of the baseline cultures, total neurons per coverslip were tallied by scanning the entire coverslip in a serpentine formation using a Plan 10×/0.3 N.A. dark low (DL) objective mounted on a Nikon Optiphot-2 microscope equipped with fluorescence. Smaller neuron identities were confirmed at higher objectives (Plan 20×/0.5 DL and Plan 40×/0.7 DL). To estimate soma size, the DAB-labeled magnocellular neurons were imaged using a Zeiss laser-scanning confocal microscope (LSM) with an argon laser and a transmitted light photomultiplier tube. Determination of soma size was performed by measuring perikaryon diameter at the long and short axis with Zeiss LSM software, as defined previously (33).
MALDI-TOF MS
Mass profile peaks were selected and exported to the FlexAnalysis 3.0 software program (Bruker Daltonics) for mass identification. While the amount of material from the crystals on the coverslip precluded tandem MS, a number of peptides were confirmed via tandem MS from our previous peptidomics analysis of the supraoptic nucleus tissue (1). The MS results presented here are assigned by matching our observed mass values to those of our previous study (1) and to databases. Using binary logistic models, we obtained the predicted mass of expected cleavage sites with the web-based tool NeuroPred (http://neuroproteomics.scs.uiuc.edu/neuropred.html) (31,45). For NeuroPred, the rat model was used for cleavage prediction of the provasopressin hormone with a confidence interval of 95%. All observed molecular weight masses were searched using the SwePep rat database (www.swepep.org) (46). For all mass matching, the mass tolerance used was 0.50 Da for masses lower than 2000 and 1 Da for masses higher than 2000 Da. For peptides forms not observed previously, the assignments should be considered tentatively.
Acknowledgments
We thank Harold Gainer, Laboratory of Neurochemistry, NINDS/NIH, for supplying PS-38 and PS-41 antibodies.
Abbreviations
DAB, 3,3′-diaminobenzidine; DHB, 2,5-dihydroxybenzoic acid; DIV, days in vitro; MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight; MAP2, microtubule-associated protein 2; MS, mass spectrometry; NP, neurophysin; PDL, poly-d-lysine; POMC, pro-opiomelanocortin.
Supporting Information Available
Figure showing mounted coverslip on MALDI target. This material is available free of charge via the Internet at http://pubs.acs.org.
Larry Millet contributed to experimental design, performed research, analyzed data, and contributed to manuscript preparation. Adriana Bora contributed to experimental design, performed research, analyzed data, and contributed to manuscript preparation. Jonathan Sweedler provided research oversight and technical direction and contributed to manuscript preparation. Martha Gillette provided research oversight and technical direction and contributed to manuscript preparation.
Larry Millet was supported by the National Institute of Child Health and Human Development (NICHD) Developmental Psychobiology and Neurobiology Training Grant (HD007333). The content is solely the responsibility of the authors and does not represent the official views of the NIH.
This study was supported by the National Heart, Lung and Blood Institute (NHLBI, Grant HL092571), the National Institute of Dental and Craniofacial Research and the Office of the Director of the National Institutes of Health (NIDCR, Grant DE018866), and the National Institute On Drug Abuse (NIDA, Grant DA018310). The W. M. Keck Foundation funded preliminary work.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Bora A.; Annangudi S. P.; Millet L. J.; Rubakhin S. S.; Forbes A. J.; Kelleher N. L.; Gillette M. U.; Sweedler J. V. (2008) Neuropeptidomics of the supraoptic rat nucleus. J. Proteome Res. 7, 4992–5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che F. Y.; Biswas R.; Fricker L. D. (2005) Relative quantitation of peptides in wild-type and Cpe(fat/fat) mouse pituitary using stable isotopic tags and mass spectrometry. J. Mass Spectrom. 40, 227–237. [DOI] [PubMed] [Google Scholar]
- Che F. Y.; Lim J.; Pan H.; Biswas R.; Fricker L. D. (2005) Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Mol. Cell. Proteomics 4, 1391–1405. [DOI] [PubMed] [Google Scholar]
- Che F. Y.; Zhang X.; Berezniuk I.; Callaway M.; Lim J.; Fricker L. D. (2007) Optimization of neuropeptide extraction from the mouse hypothalamus. J. Proteome Res. 6, 4667–4676. [DOI] [PubMed] [Google Scholar]
- Li L. J.; Sweedler J. V. (2008) Peptides in the brain: Mass spectrometry-based measurement approaches and challenges. Annu. Rev. Anal. Chem. 1, 451–483. [DOI] [PubMed] [Google Scholar]
- Spellman D. S.; Deinhardt K.; Darie C. C.; Chao M. V.; Neubert T. A. (2008) Stable isotopic labeling by amino acids in cultured primary neurons: application to brain-derived neurotrophic factor-dependent phosphotyrosine-associated signaling. Mol. Cell. Proteomics 7, 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garden R. W.; Shippy S. A.; Li L. J.; Moroz T. P.; Sweedler J. V. (1998) Proteolytic processing of the Aplysia egg-laying hormone prohormone. Proc. Natl. Acad. Sci. U.S.A. 95, 3972–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummon A. B.; Amare A.; Sweedler J. V. (2006) Discovering new invertebrate neuropeptides using mass spectrometry. Mass Spectrom. Rev. 25, 77–98. [DOI] [PubMed] [Google Scholar]
- Li L. J.; Garden R. W.; Sweedler J. V. (2000) Single-cell MALDI: A new tool for direct peptide profiling. Trends Biotechnol. 18, 151–160. [DOI] [PubMed] [Google Scholar]
- Rubakhin S. S.; Churchill J. D.; Greenough W. T.; Sweedler J. V. (2006) Profiling signaling peptides in single mammalian cells using mass spectrometry. Anal. Chem. 78, 7267–7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubakhin S. S.; Sweedler J. V. (2007) Characterizing peptides in individual mammalian cells using mass spectrometry. Nat. Protoc. 2, 1987–1997. [DOI] [PubMed] [Google Scholar]
- Burbach J. P.; Luckman S. M.; Murphy D.; Gainer H. (2001) Gene regulation in the magnocellular hypothalamo-neurohypophysial system. Physiol. Rev. 81, 1197–1267. [DOI] [PubMed] [Google Scholar]
- Hatton G. I. (2004) Dynamic neuronal-glial interactions: An overview 20 years later. Peptides 25, 403–411. [DOI] [PubMed] [Google Scholar]
- Russell J. A.; Leng G.; Douglas A. J. (2003) The magnocellular oxytocin system, the fount of maternity: Adaptations in pregnancy. Front. Neuroendocrinol. 24, 27–61. [DOI] [PubMed] [Google Scholar]
- Bergquist F.; Ludwig M. (2008) Dendritic transmitter release: A comparison of two model systems. J. Neuroendocrinol. 20, 677–686. [DOI] [PubMed] [Google Scholar]
- Pow D. V.; Morris J. F. (1989) Dendrites of hypothalamic magnocellular neurons release neurohypophysial peptides by exocytosis. Neuroscience 32, 435–439. [DOI] [PubMed] [Google Scholar]
- Sabatier N.; Caquineau C.; Douglas A. J.; Leng G. (2003) Oxytocin released from magnocellular dendrites: A potential modulator of alpha-melanocyte-stimulating hormone behavioral actions?. Ann. N.Y. Acad. Sci. 994, 218–224. [DOI] [PubMed] [Google Scholar]
- Sabatier N.; Rowe I.; Leng G. (2007) Central release of oxytocin and the ventromedial hypothalamus. Biochem. Soc. Trans. 35, 1247–1251. [DOI] [PubMed] [Google Scholar]
- Bilinski M.; Sanchez A.; Gonzalez Nicolini V.; Villar M. J.; Tramezzani J. H. (1996) Dispersion and culture of magnocellular neurons from the supraoptic nucleus of the adult rat. J. Neurosci. Methods 64, 13–18. [DOI] [PubMed] [Google Scholar]
- Jirikowski G.; Reisert I.; Pilgrim C. (1981) Neuropeptides in dissociated cultures of hypothalamus and septum: quantitation of immunoreactive neurons. Neuroscience 6, 1953–1960. [DOI] [PubMed] [Google Scholar]
- Jirikowski G.; Reisert I.; Pilgrim C. (1984) Angiotensin II promotes development of neurophysin neurons in dissociated culture. Brain Res. 316, 179–183. [DOI] [PubMed] [Google Scholar]
- Kusano K.; House S. B.; Gainer H. (1999) Effects of osmotic pressure and brain-derived neurotrophic factor on the survival of postnatal hypothalamic oxytocinergic and vasopressinergic neurons in dissociated cell culture. J. Neuroendocrinol. 11, 145–152. [DOI] [PubMed] [Google Scholar]
- Meeker R. B.; Curras M. C.; Stewart J.; Serje A.; Al-Ghoul W. (1999) Functional activation of punch-cultured magnocellular neuroendocrine cells by glutamate receptor subtypes. J. Neurosci. Methods 89, 57–67. [DOI] [PubMed] [Google Scholar]
- Schilling K.; Pilgrim C. (1987) Hypothalamo-neurohypophysial neurons in vitro: Developmental potentials depend on the donor rat stock. J. Neurosci. Res. 18, 432–438. [DOI] [PubMed] [Google Scholar]
- Banker G., Goslin K. (1991) Culturing Nerve Cells, MIT Press, Cambridge, MA. [Google Scholar]
- Brewer G. J. (1995) Serum-free B27/neurobasal medium supports differentiated growth of neurons from the striatum, substantia nigra, septum, cerebral cortex, cerebellum, and dentate gyrus. J. Neurosci. Res. 42, 674–683. [DOI] [PubMed] [Google Scholar]
- Brewer G. J.; Torricelli J. R.; Evege E. K.; Price P. J. (1993) Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J. Neurosci. Res. 35, 567–576. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Che F. Y.; Berezniuk I.; Sonmez K.; Toll L.; Fricker L. D. (2008) Peptidomics of Cpe(fat/fat) mouse brain regions: implications for neuropeptide processing. J. Neurochem. 107, 1596–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova E. V.; Rubakhin S. S.; Sweedler J. V. (2008) One-step sampling, extraction, and storage protocol for peptidomics using dihydroxybenzoic Acid. Anal. Chem. 80, 3379–3386. [DOI] [PubMed] [Google Scholar]
- Amare A.; Hummon A. B.; Southey B. R.; Zimmerman T. A.; Rodriguez-Zas S. L.; Sweedler J. V. (2006) Bridging neuropeptidomics and genomics with bioinformatics: Prediction of mammalian neuropeptide prohormone processing. J. Proteome Res. 5, 1162–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegge A. N.; Southey B. R.; Sweedler J. V.; Rodriguez-Zas S. L. (2008) Comparative analysis of neuropeptide cleavage sites in human, mouse, rat, and cattle. Mamm. Genome 19, 106–120. [DOI] [PubMed] [Google Scholar]
- Brewer G. J.; Cotman C. W. (1989) Survival and growth of hippocampal neurons in defined medium at low density: Advantages of a sandwich culture technique or low oxygen. Brain Res. 494, 65–74. [DOI] [PubMed] [Google Scholar]
- Di Scala-Guenot D.; Strosser M. T.; Sarlieve L. L.; Legros J. J.; Richard P. (1990) Development of neurophysin-containing neurons in primary cultures of rat hypothalami is related to the age of the embryo: morphological study and comparison of in vivo and in vitro neurophysins, oxytocin, and vasopressin content. J. Neurosci. Res. 25, 94–102. [DOI] [PubMed] [Google Scholar]
- Bai F.; Sozen M. A.; Lukiw W. J.; Argyropoulos G. (2005) Expression of AgRP, NPY, POMC and CART in human fetal and adult hippocampus. Neuropeptides 39, 439–443. [DOI] [PubMed] [Google Scholar]
- Strand F. L. (1999) Neuropeptides: Regulators of Physiological Processes, MIT Press, Cambridge, MA. [Google Scholar]
- Stenfors C.; Theodorsson E.; Mathe A. A. (1989) Effect of repeated electroconvulsive treatment on regional concentrations of tachykinins, neurotensin, vasoactive intestinal polypeptide, neuropeptide Y, and galanin in rat brain. J. Neurosci. Res. 24, 445–450. [DOI] [PubMed] [Google Scholar]
- Duarte C. R.; Schutz B.; Zimmer A. (2006) Incongruent pattern of neurokinin B expression in rat and mouse brains. Cell Tissue Res. 323, 43–51. [DOI] [PubMed] [Google Scholar]
- Warden M. K.; Young W. S. 3rd (1988) Distribution of cells containing mRNAs encoding substance P and neurokinin B in the rat central nervous system. J. Comp. Neurol. 272, 90–113. [DOI] [PubMed] [Google Scholar]
- Saffroy M.; Torrens Y.; Glowinski J.; Beaujouan J. C. (2003) Autoradiographic distribution of tachykinin NK2 binding sites in the rat brain: comparison with NK1 and NK3 binding sites. Neuroscience 116, 761–773. [DOI] [PubMed] [Google Scholar]
- Dowell J. A.; Heyden W. V.; Li L. (2006) Rat neuropeptidomics by LC-MS/MS and MALDI-FTMS: Enhanced dissection and extraction techniques coupled with 2D RP-RP HPLC. J. Proteome Res. 5, 3368–3375. [DOI] [PubMed] [Google Scholar]
- Millet L. J.; Stewart M. E.; Sweedler J. V.; Nuzzo R. G.; Gillette M. U. (2007) Microfluidic devices for culturing primary mammalian neurons at low densities. Lab Chip 7, 987–994. [DOI] [PubMed] [Google Scholar]
- Jo K.; Heien M. L.; Thompson L. B.; Zhong M.; Nuzzo R. G.; Sweedler J. V. (2007) Mass spectrometric imaging of peptide release from neuronal cells within microfluidic devices. Lab Chip 7, 1454–1460. [DOI] [PubMed] [Google Scholar]
- Nam Y.; Brewer G. J.; Wheeler B. C. (2007) Development of astroglial cells in patterned neuronal cultures. J. Biomater. Sci. 18, 1091–1100. [DOI] [PubMed] [Google Scholar]
- Brewer G. J. (1997) Isolation and culture of adult rat hippocampal neurons. J. Neurosci. Methods 71, 143–155. [DOI] [PubMed] [Google Scholar]
- Southey B. R.; Amare A.; Zimmerman T. A.; Rodriguez-Zas S. L.; Sweedler J. V. (2006) NeuroPred: A tool to predict cleavage sites in neuropeptide precursors and provide the masses of the resulting peptides. Nucleic Acids Res. 34, W267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falth M.; Skold K.; Norrman M.; Svensson M.; Fenyo D.; Andren P. E. (2006) SwePep, a database designed for endogenous peptides and mass spectrometry. Mol. Cell. Proteomics 5, 998–1005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.