Abstract
Mouse bladder mesenchyme differentiates into smooth muscle under the influence of urothelium at gestational day 13.5 (E13.5). The genes downstream of the Sonic hedgehog (Shh) that signal the mesenchyme and regulate smooth muscle cell differentiation have not been clarified. We hypothesize that gene expression across the full thickness of bladder mesenchyme is a function of proximity to the inducing bladder urothelium. Accordingly, we predict differences in gene expression in the future submucosa area adjacent to the urothelium, which lacks smooth muscle, versus the peripheral bladder mesenchyme, where smooth muscle differentiation normally occurs. Embryonic bladders from FVB mice were collected at E12.5, 13.5, 15, and 16, cryosectioned followed by microdissection. and mRNA expression profiles were measured. Smooth muscle α-actin (SMAA) and smooth muscle myosin heavy chain (SM-MHC) were expressed in the E13.5, E15 and E16 bladders in the peripheral layer of mesenchyme but not in the prospective submucosa. Patched 1 (Ptc1), Gli1 and Bone morphogenetic protein (Bmp) 4 were consistently elevated in the mesenchymal layer immediately adjacent to the urothelium compared to the peripheral location at E12.5. After E12.5 Ptc1 gene decreased to an undetectable level throughout the bladder mesenchyme. The level of TGF-β1 was highest in the mesenchymal layer adjacent to the serosa at E13.5. The level of Serum response factor (SRF) was also highest at E15 in the peripheral mesenchyme. The present study discovered genes downstream from Shh are differentially expressed in the prospective submucosa versus the peripheral bladder mesenchyme as a function gestation age and smooth muscle differentiation.
Keywords: Bladder, Laser capture microdissection, Mesenchyme, Signaling, Smooth muscle
Introduction
Reciprocal epithelial–mesenchymal interactions are essential for growth, differentiation, and patterning of many vertebrate organs, including the bladder (Baskin et al., 1996; Liu et al., 2000). Differentiation of smooth muscle in the embryo is temporally and spatially coordinated within the bladder mesenchyme by signals arising from the bladder epithelium (Baskin et al., 1996; Li et al., 2006; Shiroyanagi et al., 2007).
Previous work has shown that an epithelial signal is necessary for the induction of bladder smooth muscle (Baskin et al., 1996; Shiroyanagi et al., 2007), and that the smooth muscle-inducing signal is a diffusible (Liu et al., 2000). The paradox is that smooth muscle develops in the peripheral bladder mesenchyme adjacent to the serosa surface and not in the mesenchyme immediately adjacent to the smooth muscle inducing urothelium. The presumption is that the epithelial signal is transmitted through the submucosal zone (which does not undergo smooth muscle differentiation) to the peripheral bladder mesenchyme. Perhaps the submucosal connective tissue and/or basement membrane of the bladder act as inductive or repressive signaling channels or mediators of smooth muscle differentiation in their own rights (Liu et al., 2000; Chuang et al., 2000).
Insight into bladder smooth muscle differentiation comes from both cellular and genetic studies. Smooth muscle differentiation in the mouse urinary bladder occurs at gestational day 13.5 (E13.5) (Li et al., 2006; Shiroyanagi et al., 2007). cDNA microarray gene expression profiles of intact embryonic murine bladders have identified regulators of vascular smooth muscle differentiation in bladder mesenchyme, including serum response factor (SRF) and its cofactors, ELK1 and SRF accessory protein (SAP) 1, as well as two SRF-associated pathways, angiotension receptor II and transforming growth factor-beta (TGF-β) 2 (Li et al., 2006). These results suggest that bladder smooth muscle differentiation may share a similar gene expression program as occurs during vascular smooth muscle differentiation (Wijgerde et al., 2002; Yu et al., 2002; Kumar and Owens, 2003; Weaver et al., 2003; Owens et al., 2004; Li et al., 2006; White et al., 2006; Haraguchi et al., 2007; Jenkins et al., 2007; Morrow et al., 2007; Shiroyanagi et al., 2007).
To analyze differences in gene expression among different cellular populations in the bladder, gene expression before and after smooth muscle differentiation was evaluated using laser capture microdissection (Bonner et al., 1997; Green et al., 2003; Hong et al., 2004; Jacquet et al., 2005; Kerman et al., 2006).
Results
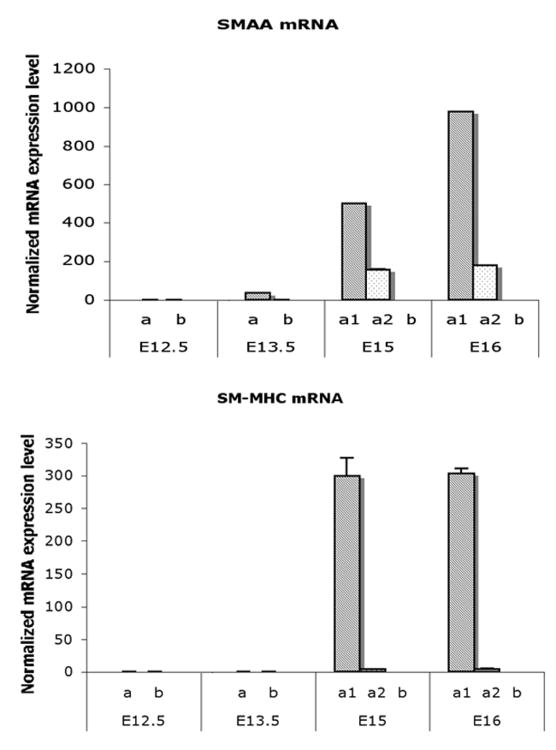
At E12.5, mRNA of smooth muscle markers, α-actin (SMAA) and smooth muscle myosin heavy chain (SM-MHC), were undetectable in bladder mesenchyme. Messenger RNA of SMAA was initially detected in the E13.5 bladders in the peripheral mesenchyme and remained expressed in the peripheral mesenchyme in both the E15 and E16 bladders (Mann-Whitney test, p<0.001) (Fig. 3). In contrast, SMAA and SM-MHC mRNAs were undetectable in the submucosal. SM-MHC was detectable slightly later in gestation in the peripheral mesenchyme than SMAA. SMAA was also detectable in the loosely organized intermediate mesenchymal layer (a2) between the smooth muscle and subepithelial layer of E15 – E16 (Fig. 3).
Fig. 3. Real time RT-PCR analysis on mRNA expressions of SMAA and SM-MHC in laser captured mesenchymal components of each gestational stage.
Expression of SMAA mRNA was detectable in the peripheral cellular population at E13.5 and increased to significantly high levels in smooth muscle cells at E15 and E16. SM-MHC was detectable temporal slightly later than SMAA and increasing in expression at E15 and E16 (Mann-Whitney test, p<0.001). a, Serosal zone. a1, Smooth muscle layer. a2, Intermediate zone. b, Submucosal zone.
Downstream genes of sonic hedgehog (Shh) signaling pathway
At E12.5, Patched1 (Ptc1), Gli1, and bone morphogenetic protein (Bmp4) mRNAs were expressed many folds higher in the submucosa mesenchymal cells immediately adjacent to the epithelium relative to that in the peripheral serosal mesenchymal layer (Fig. 4). Ptc1 and Bmp4 genes decreased significantly in all mesenchymal layers in the E13.5 bladder (Mann-Whitney test, p<0.001). Ptc1 remained low in the serosal and intermediate zones (a1, a2) in the E15 and E16 bladder mesenchyme (Mann-Whitney test, p<0.001). In contrast, the level of Bmp4 mRNA increased in the E15 and E16 bladders. Gli1 gene was expressed at high levels in the submucosa at E12.5 and E13.5 and decreased to low levels thereafter. At E15 the expression of Gli1 increased to a higher level in the smooth muscle layer (Mann-Whitney test, p<0.001) (Fig. 4). Gli1 increased steadily in the peripheral mesenchyme and the smooth muscle layer from E12.5 to E15 and decreased thereafter.
Fig. 4. Real time RT-PCR analysis on mRNA expressions of Ptc1, Gli1, and Bmp4 in laser captured mesenchymal components of each gestational stage.
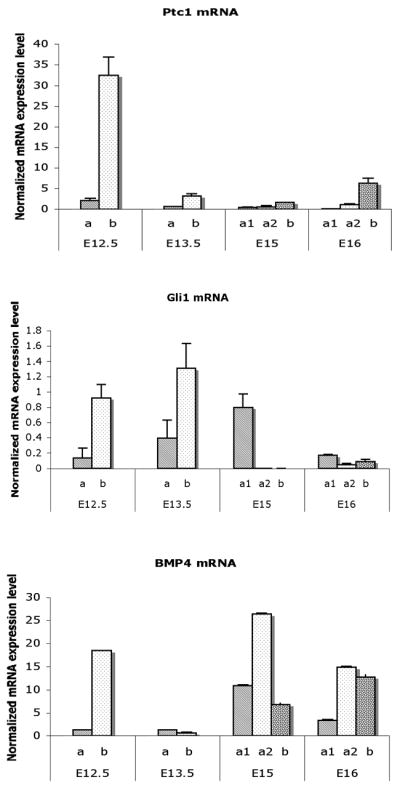
Ptc1, Gli1, and Bmp4. expression were restricted to a thin layer of mesenchymal cells in the submucosa immediately adjacent to the epithelium. Both Ptc1 and Bmp4 genes were significantly decreased in all mesenchymal cells in the E13.5 bladder. Ptc1 remained at a low level in contrast to Bmp4 in which was increased at E15 and E16. Gli1 gene expression was up regulated in the submucosa at E13.5. At E15 the expression changed to the smooth muscle layer (Mann-Whitney test, p<0.001). a, Serosal zone. a1, Smooth muscle layer. a2, Intermediate zone. b, Submucosal zone.
Expression of TGF-β1 gene
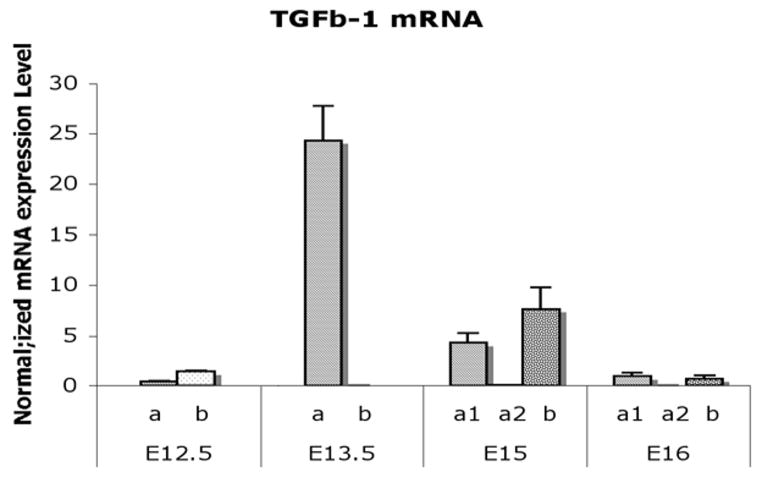
TGF-β1 was minimally expressed in E12.5 bladder mesenchyme. At E13.5 TGF-β1 increased dramatically in the serosal mesenchymal layer and then decreased in all mesenchymal compartments in the E15 and E16 bladders (Mann-Whitney test, p<0.001) (Fig. 5).
Fig. 5. Real time RT-PCR analysis on mRNA expression of TGF-β1 in laser captured mesenchymal components of each gestational stage.
TGF-β1 expression was minimal in the E12.5 bladder mesenchyme. TGF-β1 increased to the highest level in the smooth muscle differentiation layer adjacent to the serosa at E13.5 and then decreased in all mesenchymal compartments in the E15 and E16 (Mann-Whitney test, p<0.001). a, Serosal zone. a1, Smooth muscle layer. a2, Intermediate zone. b, Submucosal zone.
Expression of serum response factor (SRF)
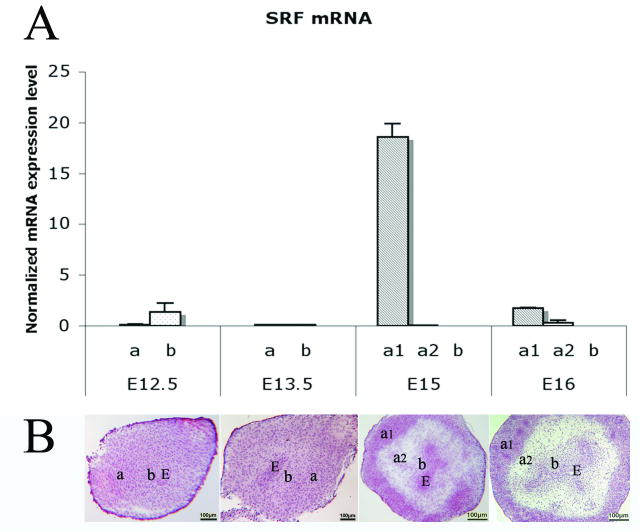
Messenger RNA expression of SRF was significantly upregulated in smooth muscle cells at E15 by real time RT-PCR analysis compared to other mesenchymal components in E12.5, E13.5, and E16 (Mann-Whitney test, p<0.001) (Fig. 6).
Fig. 6. Real time RT-PCR analysis on mRNA expression of SRF in laser captured mesenchymal components of each gestational stage and mouse bladder developmental illustrations.
(A) SRF was significantly up regulated in smooth muscle cells at E15 compared to other mesenchymal components at E12.5, E13.5, and E16 (Mann-Whitney test, p<0.001). (B) Mouse bladder developmental illustrations with HE staining show locations of a1, a2 and b nomenclatures. a, Serosal zone. a1, Smooth muscle layer. a2, Intermediate zone. b, Submucosal zone. E, Bladder epithelium. Scale bar, 100μm.
Meanwhile, bladder developmental illustrations with HE staining were shown with locations of a1, a2 and b nomenclatures in Fig. 6.
A summary of the expression patterns of SMAA, SM-MHC, Ptc1, Gli1, Bmp4, TGF-β1, and SRF is shown in Table 2.
TABLE 2. Primer Sequences for Target Genes.
| Genes | Primers Sequences (5′-3′) | Production (bp) |
|---|---|---|
| SMAA | F: ATTGTGCTGGACTCTGGAGATGGT | 187 |
| R: TGATGTCACGGACAATCTCACGCT | ||
| SM-MHC | F: TGA GCT CAG TGA CAA GGT CCA CAA | 109 |
| R: GGA AGC CAC ATC TTT GGC CAG TTT | ||
| Ptc1 | F: TGGCCCATGCATTCAGTGAAACAG | 147 |
| R: TAGGGATCAATGCGGCCATGAAGA | ||
| Gli1 | F: ACAAGTGCACGTTTGAAGGCTGTC | 116 |
| R: GCTGCAACCTTCTTGCTCACACAT | ||
| Bmp4 | F: CGTTACCTCAAGGGAGTGGA | 116 |
| R: ATGCTTGGGACTACGTTTGG | ||
| TGF-β1 | F: GTG CGG CAG CTG TAC ATT GAC TTT | 127 |
| R: TGT ACT GTG TGT CCA GGC TCC AAA | ||
| SRF | F: TCAATGCCTTCTCTCAGGCACCAT | 151 |
| R: ATCACAGCCATCTGGTGAAGCTGA | ||
| GapDH | F: TGT GAT GGG TGT GAA CCA CGA GAA | 130 |
| R: GAG CCC TTC CAC AAT GCC AAA GTT |
F, forward; R, reverse.
Discussion
Smooth muscle differentiation in the murine bladder is initiated at 13.5 days gestation under the influence of bladder urothelium (Li et al., 2006; Shiroyanagi et al., 2007). It has been proposed that the urothelial factor responsible for smooth muscle induction is sonic hedgehog (Shh) (Yu et al., 2002; Shiroyanagi et al., 2007). Shh in turns acts through its receptor Ptc1 and a number of down stream mesenchymal genes such as Gli1, Bmp4, TGF-β1, and SRF to both inhibit and induce smooth muscle as judged by SMAA and SM-MHC expression (Liu et al., 2000; Yu et al., 2002; Li et al., 2006; Shiroyanagi et al., 2007). Herein, we have mapped the temporal and spatial location of these genes during bladder development.
The Shh signaling pathway functions throughout development. Shh is involved in the determination of cell fate and embryonic patterning during early vertebrate development. Later during organogenesis, Shh is involved in the formation and differentiation of a variety of tissues and organs (Chuang and Kornberg, 2000). In bladder development, Ptc1, the receptor for Shh, was expressed in the mesenchyme adjacent to the bladder epithelium before the initiation of smooth muscle differentiation at E12.5, and to a far lesser extent in the prospective and definitive SMAA-producing cells later in development. The expression patterns of Ptc1 and Gli1 in the bladder are similar to that in ureteral development, since both of these genes were expressed at high levels in the mesenchymal cells immediately adjacent to the epithelium and to a lesser extent in the peripheral mesenchyme about to undergo smooth muscle differentiation. Subsequently both Ptc1 and Gli1 decreased in all mesenchymal cells after smooth muscle differentiation (Yu et al., 2002).
Bone morphogenetic protein 4 (Bmp4) has been shown to promote smooth muscle differentiation in the ureter (Raatikainen-Ahokas et al., 2000). Bmp4, a member of the transforming growth factor-beta (TGF-β) family, is a downstream gene of Shh pathway, and regulates several developmental processes during animal development (Raatikainen-Ahokas et al., 2000; Bragg et al., 2001; Sadlon et al., 2004; Shiroyanagi et al., 2007). We found Bmp4 gene expresses initially at high level in submucosa layer in E12.5 bladder (as Ptc1 gene), it is rather strong in intermediate connective tissue in E15, and later both in connective tissue and smooth muscle cell. Our results are consistent with Bmp4 inhibiting smooth muscle formation in the submucosa adjacent to the urothelium and initiating smooth muscle formation in the periphery of the bladder. It also further suggests that a similar Shh- and Bmp4-dependent pathway involves in bladder mesenchyme for smooth muscle differentiation as in ureter (Mendelsohn, 2006).
In the mouse 10T1/2 multipotent mesenchymal cell line, TGF-β1 stimulates cell growth with the up-regulation of several smooth muscle cell differentiation markers, such as SMAA, SM-MHC, smooth muscle protein 22-α (SM22α), and calponin (Lien et al., 2006). Our data are consistent with these studies in that TGF-β1 was dramatically up-regulated in the smooth muscle forming cells in the E13.5 bladder suggesting that TGF-β1 in outer layer cells of bladder mesenchyme promotes bladder smooth muscle differentiation. Previous studies discovered that TGF-β's are mediated by SMAD2 and SMAD3, while BMPs are mediated by SMAD1, SMAD5 and SMAD9 (Moustakas, 2002). BMP and Activin membrane bound inhibitor (BAMBI) has a similar extracellular domain as type I receptors. It serves as a negative regulator of TGF-β signaling and may limit TGF-β expression during embryogenesis. The present research shows TGF-β1 expression as a peak (especially in outer layer bladder mesenchyme at E13.5) rather for short period compared with Bmp4. This finding suggests that TGF/Bmp signaling plays corresponding role during process of bladder smooth muscle differentiation and development.
Extensive studies have established the central role of serum response factor (SRF) in controlling smooth muscle cell specific gene expression. The SMAA gene is known be regulated at the transcriptional level through the binding of SRF in vascular smooth muscle (Lien et al., 2006). We have found that the SRF expression pattern directly precedes SMAA expression and that SRF localizes to the peripheral bladder mesenchyme 24 hours prior to smooth muscle differentiation (Li et al., 2006). A similar finding was also reported in gizzard smooth muscle development in which increased expression of SRF immediately preceded SMGA expression (Browning et al., 1998). SRF activates genes involved in smooth muscle differentiation and proliferation by recruiting muscle-restricted cofactors, such as the transcriptional coactivator myocardin, GATA-6, and complex factors of the ETS-domain family, respectively (Wang et al., 2002; Kanematsu et al., 2007). In the present study, expression of SRF was significantly up regulated in smooth muscle cells in the E15 bladder after smooth muscle formation at E 13.5 implying that SRF plays its regulatory role as secondary factor as opposed to an initiator of smooth muscle differentiation.
Interestingly, SMAA gene was also detectable in the loosely organized mesenchymal cells between smooth muscle layer and subepithelial layer both in the E15 and E16 bladders. One possibility is that these cells may be smooth muscle progenitor cells at early stages of bladder smooth muscle differentiation.
In summary, the current study has generated a “snapshot view” of gene expression during bladder smooth muscle differentiation. The expression profiles of these genes provide a necessary framework for interpreting the signaling pathway that drives bladder smooth muscle formation.
Experimental Procedures
Animal
FVB mice were purchased from Charles River Laboratories (Wilmington, MA) and housed in separate animal cages (20 × 25 × 47 cm) with laboratory-grade pine shavings (heat-treated to remove resins) as bedding. They were acclimated to 68 ∼ 74°F and 40% ∼ 50% relative humidity on a reversed light schedule (14 hrs light and 10 hrs dark) and received ad libitum mouse chow and water. Timed mating was set up according experiment schedule. Noon on the day of the vaginal plug was designated as embryonic day 0.5. All animal-related procedures described here were approved by the laboratory animal resource center at University of California, San Francisco.
Laser capture microdissection (LCM)
Embryonic mouse bladders were collected at E12.5, 13.5, 15, and E16, respectively. The embryonic bladders were cut into 7μm sections and stained using Histogene™ (HS) solution (Arcturus Engineering Inc., Mountain View, CA).
Laser capture microdissection was used to localize specific compartments of the bladder mesenchyme in relationship to the urothelium. Each sample was cryosectioned consecutively. Mesenchymal locations were chosen either next to the serosal layer or next to the urothelium in the E12.5 and E 13.5 bladders before smooth muscle differentiation (Fig. 1). In the E15 and E16 bladders, in which smooth muscle differentiation has already occurred, three locations were captured: the smooth muscle layer, the subepithelial layer, and the loosely organized mesenchymal cell layer between (Fig. 2). 50∼100 cells were microdissected from each compartment of bladder mesenchyme to assure sufficient RNA. Each sample from each mesenchymal location at the same gestational stage was pooled from three fetuses from different dams.
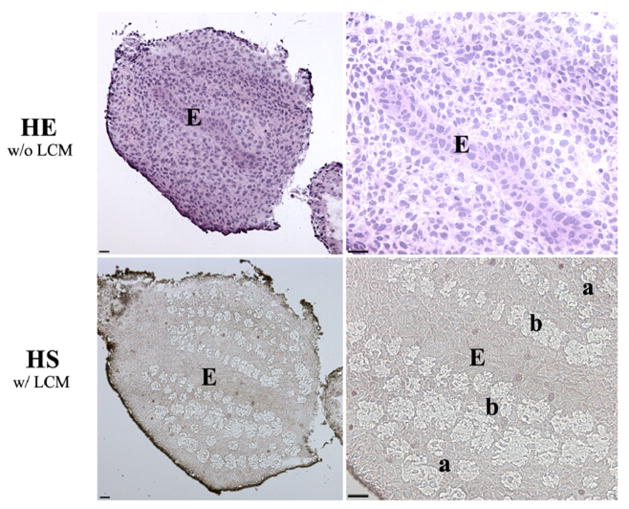
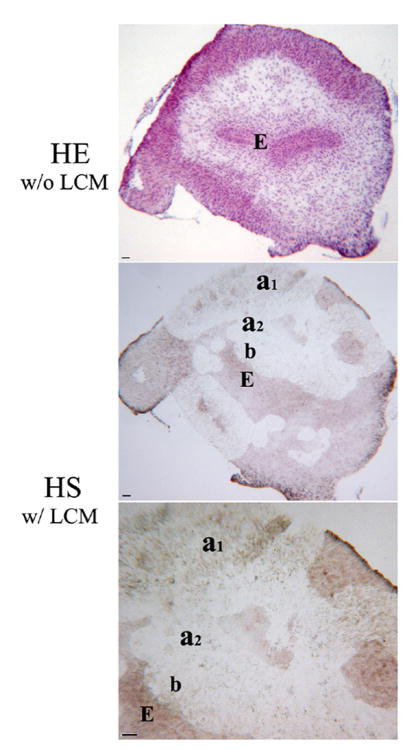
Fig. 1. Laser-assisted microdissection of mesenchyme in the E13.5 mouse bladder. (HE w/o LCM).
Morphology of HE staining on serial section for laser capture microdissection. (HS w/ LCM) Morphology of HS staining after laser capture microdissection. Mesenchymal locations were chosen either immediately adjacent to the urothelium in the prospective submucosa or next to the serosal layer in the peripheral bladder mesenchyme, an area of future smooth muscle differentiation in E12.5 and E13.5 bladders. a, Serosal zone. b, Submucosal zone. E, Epithelium. Scale bar, 50μm.
Fig. 2. Laser-assisted microdissection of mesenchyme in the E15 mouse bladder. (HE w/o LCM).
Morphology of HE staining on serial section for laser capture microdissection. (HS w/ LCM) Morphology of HS staining after laser capture microdissection. Bladder mesenchymes were captured in the submucosa and in the area of peripheral smooth muscle in E15 and E16. a1, Smooth muscle layer. a2, Intermediate zone. b, Submucosal zone. E, Epithelium. Scale bar, 50μm.
For mesenchyme microdissection, slides were immediately transferred to a PixCell® II laser capture microscope (Arcturus). This technique employs low-power infrared laser to melt a thermoplastic film over the cells of interest, to which the cells become attached. Pictures of samples were taken using HITACHI KP-D580 high sensitivity autogain thermoelectrically cooled digital signal processor color CCD camera.
Cell clusters were transferred to caps with the thermoplastic polymer film by laser hits. Each cap was placed in an ExtracSure™ device (Arcturus), and RNA extraction was performed with the PicoPure™ RNA Isolation Kit (Arcturus). Two rounds of linear RNA amplifications were performed with RiboAmp™ RNA Amplification Kit (Arcturus) according to the manufacturer's protocol.
Real-time RT-PCR
To compare the expression levels of mRNAs within the bladder mesenchyme in respect to location relative to the urothelium, SYBR Green real-time RT-PCR technique and data analysis were performed using 96-well microwell plates and an ABI PRISM 7300 sequence detector (Applied BioSystems, Foster City, CA). Nanodrop spectrophotometer (NanoDrop Technologies, Wilmington, DE) quantification was performed as an accurate measure of the amount of intact starting RNA.
All primers for real-time PCR were designed according target gene sequences published on PubMed and synthesized by Integrated Device Technology, Inc. (San Jose, CA) (Table 1). The reaction was run online at 50°C for 2 min and 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 55°C for 30 s and an extension phase of 1 cycle at 95°C for 60 s, 60°C for 60 s, and 95°C for 60 s (ramp time, 19.59 min). All PCRs were performed in triplicate.
TABLE 1. Smooth Muscle Development Related Gene Expression Profiles in the Mouse Embryonic Bladder Mesenchyme.
| Bladder mesenchyme | E12.5 | E13.5 | > smooth muscle differentiation< | E15 | E16 |
|---|---|---|---|---|---|
| Peripheral (Layer a) |
- | SMAA TGF-β1 Gli1 |
Smooth muscle layer (layer a1) |
SMAA | SMAA |
| SM-MHC Gli1 Bmp4 SRF TGF-β1 |
Bmp4 | ||||
|
| |||||
| Loosely organized layer (Layer a2) |
SMAA | SMAA | |||
| Bmp4 | Bmp4 | ||||
|
| |||||
| Submucosa (layer b) |
Ptc1 | Gli1 | TGF-β1 | Ptc1 | |
| Gli1 | Bmp4 | Bmp4 | |||
| Bmp4 | |||||
The housekeeping gene GapDH was amplified as a reference for the amount loaded and the quality of the cDNA. The specificity of the reaction is given by the detection of the Tms of the amplification products immediately after the last reaction cycle. Results were analyzed with the melting curve analysis software (Dissociation Curve 1.0) provided with the ABI PRISM 7300 sequence detector.
Statistical analysis
To determine the significance of differential expression in the laser captured cells, a two-sided Mann-Whitney U nonparametric analysis was performed, for which a p-value of <0.05 was considered significant.
Acknowledgments
We gratefully acknowledge many discussions with Joseph Pham and the cooperation of Lia Banie. This study was supported by the NIH grant DK073449 (to Dr. Baskin LS) and California Urology Foundation Award P0009249 (to Dr. Liu B).
Abbreviations in this paper
- Bmp
bone morphogenetic protein
- E
gestational day
- LCM
laser capture microdissection
- Shh
sonic hedgehog
- SMAA
smooth muscle α-actin
- SM-MHC
smooth muscle myosin heavy chain
- SRF
serum response factor
- TGF-β
transforming growth factor-beta
Contributor Information
Benchun Liu, Email: benchunliu@yahoo.com.
Dongxiao Feng, Email: fengdx@hotmail.com.
Mei Cao, Email: mcao@urology.ucsf.edu.
Yuet Wai Kan, Email: yw.kan@ucsf.edu.
Gerald R. Cunha, Email: gcunha@urology.ucsf.edu.
Laurence S. Baskin, Email: lbaskin@urology.ucsf.edu.
References
- Airik R, Bussen M, Singh MK, Petry M, Kispert A. Tbx18 regulates the development of the ureteral mesenchyme. J Clin Invest. 2006;116:663–74. doi: 10.1172/JCI26027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin LS, Hayward SW, Young P, Cunha GR. Role of mesenchymal–epithelial interactions in normal bladder development. J Urol. 1996;156:1820–7. [PubMed] [Google Scholar]
- Bonner RF, Emmert-Buck M, Cole K, Pohida T, Chuagui R, Goldstein S, Littoa LA. Laser capture microdissection: molecular analysis of tissue. Science. 1997;278:1481–1483. doi: 10.1126/science.278.5342.1481. [DOI] [PubMed] [Google Scholar]
- Bragg AD, Moses HL, Serra R. Signaling to the epithelium is not sufficient to mediate all of the effects of transforming growth factor beta and bone morphogenetic protein 4 on murine embryonic lung development. Mech Dev. 2001;109:13–26. doi: 10.1016/s0925-4773(01)00508-1. [DOI] [PubMed] [Google Scholar]
- Browning CL, Culberson DE, Aragon IV, Fillmore RA, Croissant JD, Schwartz RJ, ZiImmer WE. The developmentally regulated expression of serum response factor plays a key role in the control of smooth muscle-specific genes. Dev Biol. 1998;194:18–37. doi: 10.1006/dbio.1997.8808. [DOI] [PubMed] [Google Scholar]
- Chuang PT, Kornberg TB. On the range of Hedgehog signaling. Curr Opin Genet Dev. 2000;10:515–522. doi: 10.1016/s0959-437x(00)00121-0. [DOI] [PubMed] [Google Scholar]
- Green AR, Edwards RE, Greaves P, White IN. Comparison of the effect of oestradiol, tamoxifen and raloxifene on nerve growth factor-alpha expression in specific neonatal mouse uterine cell types using laser capture microdissection. J Mol Endocrinol. 2003;30:1–11. doi: 10.1677/jme.0.0300001. [DOI] [PubMed] [Google Scholar]
- Haraguchi R, Motoyama J, Sasaki H, Satoh Y, Miyagawa S, Nakagata N, Moon A, Yamada G. Molecular analysis of coordinated bladder and urogenital organ formation by Hedgehog signaling. Development. 2007;134:525–33. doi: 10.1242/dev.02736. [DOI] [PubMed] [Google Scholar]
- Hong SH, Nah HY, Lee JY, Gye MC, Kim CH, Kim MK. Analysis of estrogen-regulated genes in mouse uterus using cDNA microarray and laser capture microdissection. J Endocrinol. 2004;181:157–67. doi: 10.1677/joe.0.1810157. [DOI] [PubMed] [Google Scholar]
- Jacquet R, Hillyer J, Landis WJ. Analysis of connective tissues by laser capture microdissection and reverse transcriptase-polymerase chain reaction. Anal Biochem. 2005;337:22–34. doi: 10.1016/j.ab.2004.09.033. [DOI] [PubMed] [Google Scholar]
- Jenkins D, Winyard PJ, Woolf AS. Immunohistochemical analysis of Sonic hedgehog signalling in normal human urinary tract development. J Ana. 2007;211:620–9. doi: 10.1111/j.1469-7580.2007.00808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanematsu A, Ramachandran A, Adam RM. GATA-6 mediates human bladder smooth muscle differentiation: involvement of a novel enhancer element in regulating alpha-smooth muscle actin gene expression. Am J Physiol Cell Physiol. 2007;293:C1093–102. doi: 10.1152/ajpcell.00225.2007. [DOI] [PubMed] [Google Scholar]
- Kerman IA, Buck BJ, Evans SJ, Akil H, Watson SJ. Combining laser capture microdissection with quantitative real-time PCR: effects of tissue manipulation on RNA quality and gene expression. J Neurosci Methods. 2006;153:71–85. doi: 10.1016/j.jneumeth.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Ownes GK. Combinatorial control of smooth muscle-specific gene expression. Arterioscler Thromb Vasc Biol. 2003;23:737–47. doi: 10.1161/01.ATV.0000065197.07635.BA. [DOI] [PubMed] [Google Scholar]
- Li J, Shiroyanagi Y, Lin G, Haqq C, Lin CS, Lue TF, Willingham E, Baskin LS. Serum response factor, its cofactors, and epithelial-mesenchymal signaling in urinary bladder smooth muscle formation. Differentiation. 2006;74:30–9. doi: 10.1111/j.1432-0436.2006.00057.x. [DOI] [PubMed] [Google Scholar]
- Lien SC, Usami S, Chien S, Chiu JJ. Phosphatidylinositol 3-kinase/Akt pathway is involved in transforming growth factor-beta1-induced phenotypic modulation of 10T1/2 cells to smooth muscle cells. Cellular Signalling. 2006;18:1270–8. doi: 10.1016/j.cellsig.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Liu W, Li Y, Cunha S, Hayward G, Baskin L. Diffusable growth factors induce bladder smooth muscle differentiation. In Vitro Cell Dev Biol Anim. 2000;36:476–84. doi: 10.1290/1071-2690(2000)036<0476:dgfibs>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Mendelsohn C. Going in circles: conserved mechanisms control radial patterning in the urinary and digestive tracts. J Clin Invest. 2006;116:635–37. doi: 10.1172/JCI27985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow D, Sweeney C, Birney YA, Guha S, Collins N, Cummins PM, Murphy R, Walls D, Redmond EM, Cahill PA. Biomechanical regulation of hedgehog signaling in vascular smooth muscle cells in vitro and in vivo. Am J Physiol Cell Physiol. 2007;292:C488–96. doi: 10.1152/ajpcell.00337.2005. [DOI] [PubMed] [Google Scholar]
- Moustakas A. Smad signalling network. J CELL SCI. 2002;115:3355–6. doi: 10.1242/jcs.115.17.3355. [DOI] [PubMed] [Google Scholar]
- Ownes GK, Kumar MS, Wamhoff BR. Molecular Regulation of Vascular Smooth Muscle Cell Differentiation in Development and Disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- Raatikainen—Ahokas A, Hytonen M, Tenhunen A, Sainio K, Sariola H. BMP-4 affects the differentiation of metanephric mesenchyme and reveals an early anterior-posterior axis of the embryonic kidney. Dev Dyn. 2000;217:146–58. doi: 10.1002/(SICI)1097-0177(200002)217:2<146::AID-DVDY2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Sadlon TJ, Lewis I, D'Andrea RJ. BMP4: its role in development of the hematopoietic system and potential as a hematopoietic growth factor. Stem Cells. 2004;22:457–74. doi: 10.1634/stemcells.22-4-457. [DOI] [PubMed] [Google Scholar]
- Shiroyanagi Y, Liu B, Cao M, Agras K, Li J, Hsieh MH, Willingham E, Baskin LS. Urothelial sonic hedgehog signaling plays an important role in bladder smooth muscle formation. Differentiation. 2007;75:968–77. doi: 10.1111/j.1432-0436.2007.00187.x. [DOI] [PubMed] [Google Scholar]
- Wang DZ, Li S, Hockemeyer D, Sutherland L, Wang Z, Schratt G, Richardson JA, Nordheim A, Olson EN. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc Natl Acad Sci USA. 2002;99:14855–60. doi: 10.1073/pnas.222561499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver M, Batts L, Hogan BL. Tissue interactions pattern the mesenchyme of the embryonic mouse lung. Dev Biol. 2003;258:169–84. doi: 10.1016/s0012-1606(03)00117-9. [DOI] [PubMed] [Google Scholar]
- White AC, Xu J, Yin Y, Smith C, Schmid G, Ornitz DM. FGF9 and SHH signaling coordinate lung growth and development through regulation of distinct mesenchymal domains. Development. 2006;133:1507–17. doi: 10.1242/dev.02313. [DOI] [PubMed] [Google Scholar]
- Wijgerde M, Mcmahon JA, Rule M, McMahon AP. A direct requirement for Hedgehog signaling for normal specification of all ventral progenitor domains in the presumptive mammalian spinal cord. Genes Dev. 2002;16:2849–64. doi: 10.1101/gad.1025702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Carroll TJ, McMahon AP. Sonic hedgehog regulates proliferation and differentiation of mesenchymal cells in the mouse metanephric kidney. Development. 2002;129:5301–12. doi: 10.1242/dev.129.22.5301. [DOI] [PubMed] [Google Scholar]
- Yucel S, Liu W, Cordero D, Donjacour A, Cunha G, Baskin LS. Anatomical studies of the fibroblast growth factor-10 mutant, Sonic Hedge Hog mutant and androgen receptor mutant mouse genital tubercle. Adv Exp Med Biol. 2004;545:123–48. doi: 10.1007/978-1-4419-8995-6_8. [DOI] [PubMed] [Google Scholar]