Abstract
Single-molecule force spectroscopy constitutes a powerful method for probing RNA folding: it allows the kinetic, energetic, and structural properties of intermediate and transition states to be determined quantitatively, yielding new insights into folding pathways and energy landscapes. Recent advances in experimental and theoretical methods, including fluctuation theorems, kinetic theories, novel force clamps, and ultrastable instruments, have opened new avenues for study. These tools have been used to probe folding in simple model systems, for example, RNA and DNA hairpins. Knowledge gained from such systems is helping to build our understanding of more complex RNA structures composed of multiple elements, as well as how nucleic acids interact with proteins involved in key cellular activities, such as transcription and translation.
Introduction
RNA plays diverse roles in the cell, ranging from the transmission of genetic information to the regulation of genes, ligand binding, and catalysis. Driven by the desire to understand the myriad functions of RNA, there has been an intense effort to unravel details of structure formation and dynamics in RNA molecules. RNA folding is simplified by the independent stability of the (generally predictable) secondary structures, leading to a picture of “hierarchical” folding [1] wherein secondary structure forms prior to tertiary structure. Folding nevertheless remains a formidable problem, characterized by multiple conformations arrayed across rugged energy landscapes, important chain entropy effects, and a sensitive dependence on electrostatic interactions between RNA and various metal ions [2,3].
One of the newest techniques for studying RNA folding is single-molecule force spectroscopy (SMFS), where the extension of an individual molecule is measured under an applied tension [4]. By varying the load, a single RNA molecule can be unfolded and refolded repeatedly. Force thus functions as a mechanical denaturant acting selectively on a given molecule, in contrast to traditional denaturants, such as temperature or urea. This property allows individual folding trajectories to be observed, subpopulations and rare/transient states (including partially-folded intermediates) to be distinguished, and the behavior of molecules with widely different stabilities to be compared under identical buffer conditions. Because unfolded states are fully stretched under load, the unfolded state is simplified from an ensemble of energetically-similar, high-entropy configurations to a single low-entropy configuration: both the initial and final states of the folding reaction are thus well-defined. The vectorial nature of force also imposes a preferential direction upon the folding reaction, biasing particular pathways which can be isolated for study. The molecular extension measured in SMFS supplies a natural coordinate for describing the course of the reaction and can be interpreted in terms of specific structural elements. All these features make SMFS particularly well-suited for probing folding reactions. Spatial resolution reaching the ångström level [5] permits sub-nucleotide extension changes to be measured. The broad temporal range currently achievable, ~10−4–103 s [6•], is well-matched to the time scales of RNA folding.
This review will focus on work with RNA using optical traps, as this has been the most commonly employed SMFS technique. We organize the discussion around four themes characterizing recent advances: (1) work on model systems for RNA folding, (2) advances in experimental and theoretical methods, (3) folding of complex functional RNAs, and (4) interactions between RNA folding and nucleic acid enzymes.
Model systems for RNA folding
The first SMFS study of RNA folding [7] probed the properties of the P5abc domain from the T. thermophila ribozyme. A number of subsequent studies have examined simple secondary structures, such as RNA and DNA hairpin loops [6•,8–11,12•,13,14••], and simple tertiary structures, such as kissing loops [15•] and pseudoknots [16,17]. A focus on model systems has not only allowed specific structural elements and interactions to be examined in isolation, but also established the utility and general validity of SMFS [18].
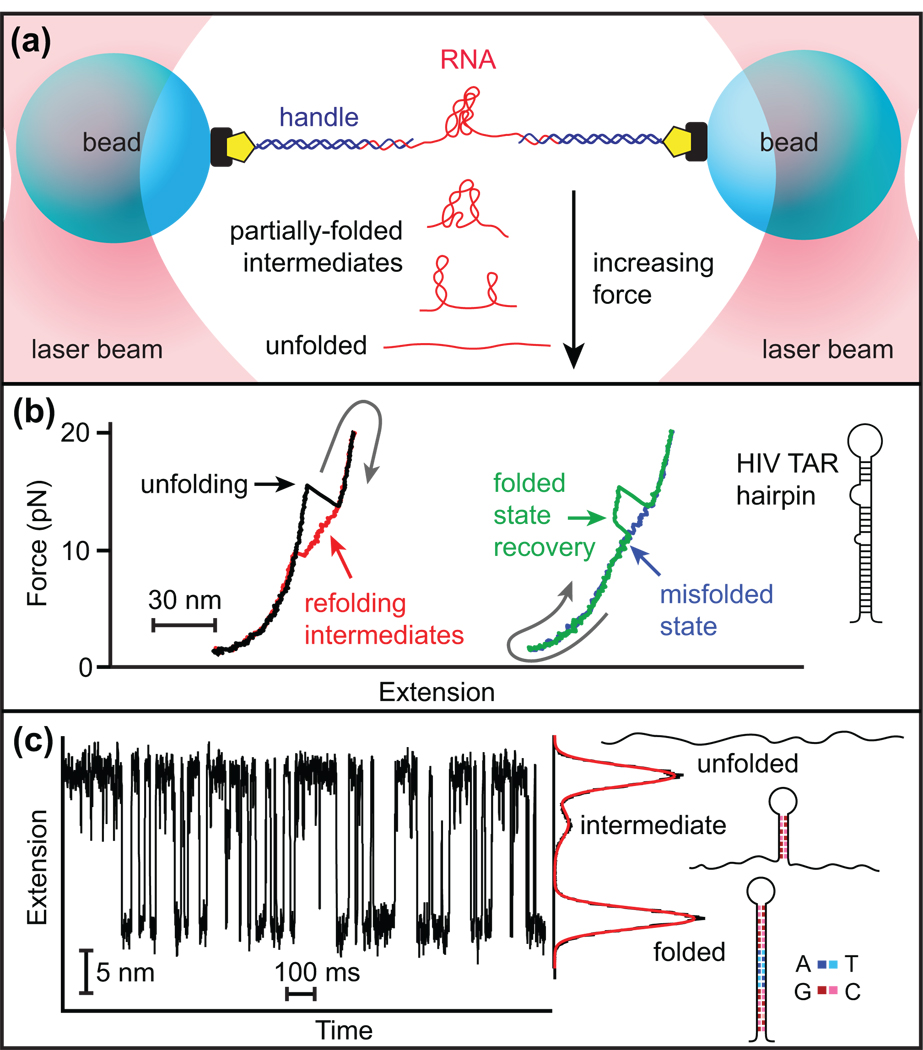
Force is generally applied to the RNA using duplex “handles” attached to microspheres held either in two optical traps (Figure 1a) or between one trap and a fixed mechanical support [19]. Measurements typically take one of two forms: force-extension curves (FECs), where the extension is monitored as the force is ramped up and down (Figure 1b), or extension trajectories, during which the load is held constant by means of a force clamp (Figure 1c). Common variants of the latter include “force-jumps,” in which the constant-force extension is measured after the load is changed abruptly [11], and “unclamped” measurements, in which the load varies due to the finite trap stiffness in the absence of a force clamp [6•,11].
Figure 1.
Representative SMFS measurements. (a) An RNA molecule is attached to two “handles” bound to beads and manipulated using two optical traps. Unfolded RNA is stretched by the force applied between the traps. As RNA unfolds or refolds, the resulting extension change is monitored. (b) FECs for the HIV TAR hairpin. After unfolding in a single step (black), the hairpin refolds through multiple intermediate states (red), producing hysteresis. Occasionally, an unfolded hairpin refolds partially to a metastable misfolded state (blue), as revealed by the return to the fully-folded state in a subsequent unfolding curve (green). Adapted from [12•]. (c) Extension of a model DNA hairpin at equilibrium under constant force, using a passive force clamp. The G:C content of the stem sequence was engineered to produce a landscape containing a metastable, partially-folded intermediate.
In both forms of measurement, unfolding increases the molecular extension, whereas folding decreases it. In FECs, extension changes are also accompanied by changes in force, producing characteristic “sawtooth” patterns (Figure 1b). Multiple states can be observed, including misfolded states (Figure 1b) and partially-folded intermediates (Figure 1c). Constant-force trajectories directly reveal properties such as the extension changes between states or the kinetics and energetics of the folding process. Moreover, the force dependence of the kinetics yields the locations of transition states along the reaction coordinate [4]. Similar information may be obtained from FECs by fitting results to kinetic models [20•,21] or by applying theorems relating the nonequilibrium work to the equilibrium free energy [9,10,22,23].
Several important results have been obtained using model systems. RNA and DNA hairpins with quasi-random sequences fold and unfold with apparently high cooperativity, behaving as two-state systems [6•,7], although additional (intermediate) states may occur in hairpins containing patterned (non-random) sequences (Figure 1c) or basepair mismatches [14••]. SMFS measurements of free energies have now been made on a sufficiently wide range of sequences to provide an independent verification of the nearest-neighbor energy parameters for nucleic acid stability obtained from ensemble measurements [24].The effects of monovalent salts on duplex stability have been measured [13], confirming and extending previous observations [2,25]. The stability provided by pseudoknot folds has also been determined [16,17]. These studies have firmly established SMFS as a viable method for measuring energetic stabilities, especially for very stable molecules that are difficult to probe by traditional methods.
Probably the most interesting results concern the location of the transition state, which is generally difficult to determine. For duplex formation in a hairpin, the transition state is often close to the unfolded state, involving the formation of just a few basepairs to close the loop [6•]. However, it can be located closer to the folded state—or, indeed, anywhere in between—if the GC content of the duplex is unevenly distributed [14••]. In contrast, tertiary interactions invariably appear to be “brittle:” their transition states are generally found within ~1–2 nm of the folded state, presumably reflecting the short-range nature of such interactions. Such brittle transition states have been now observed for kissing loops [15•,26••], pseudoknots [16,17], metal-ion cores [7], and aptamer binding sites [26••].
Advances in single-molecule force spectroscopy methods
Significant progress has been made in understanding the details of how SMFS measurements actually work, and thereby how they can best be implemented and interpreted. This knowledge has been especially useful in establishing confidence in SMFS and learning how to relate results to those found using more traditional techniques.
An improved understanding of instrument characteristics has been gained through studies of the effects of trap stiffness [27], duplex handle length [28•,29,30], and recording bandwidth or feedback loop-closure times [29] on measurements of kinetics or energy landscapes. Considerable experimental effort has also been devoted to increasing the spatiotemporal resolution and stability of optical traps. Differential extension measurements have been implemented [27,31], a helium atmosphere has been used to reduce beam-pointing fluctuations [5], and a passive force clamp that works without feedback loops has been developed [27]. This last development is particularly significant. The absence of feedback permits data acquisition at the maximum possible bandwidth, limited only by the physical properties of the molecule and beads in solution, and facilitates an ideal measurement in the Gibbs ensemble [32,33] where the force is truly constant throughout the folding reaction.
A variety of models for RNA folding under tension have been proposed and tested against experimental data, including kinetic models [34–37], quasi-equilibrium models [38], coarse-grained simulations [39], and self-organized polymer models [40]. More fundamentally, theoretical work has illuminated the question of how force affects the folding pathway itself. RNA unfolding under tension was found to display distinct differences from unfolding driven by temperature changes [39,41]. Folding pathways may also differ, in principle, at low and high force [42]. Furthermore, experimental interpretations in terms of a one-dimensional reaction coordinate (i.e., molecular extension) may sometimes represent a significant simplification, especially by neglecting dihedral angles [28•]. The effect of force on transition states has been a particular focus, with the apparent transition state location predicted to depend on both force and loading rate [20•,28•,35,40]. Models for extrapolating the kinetics to zero force and describing the loading-rate dependence of measurements have been improved, and constant-force results have now been related to those obtained at constant loading rate [20•].
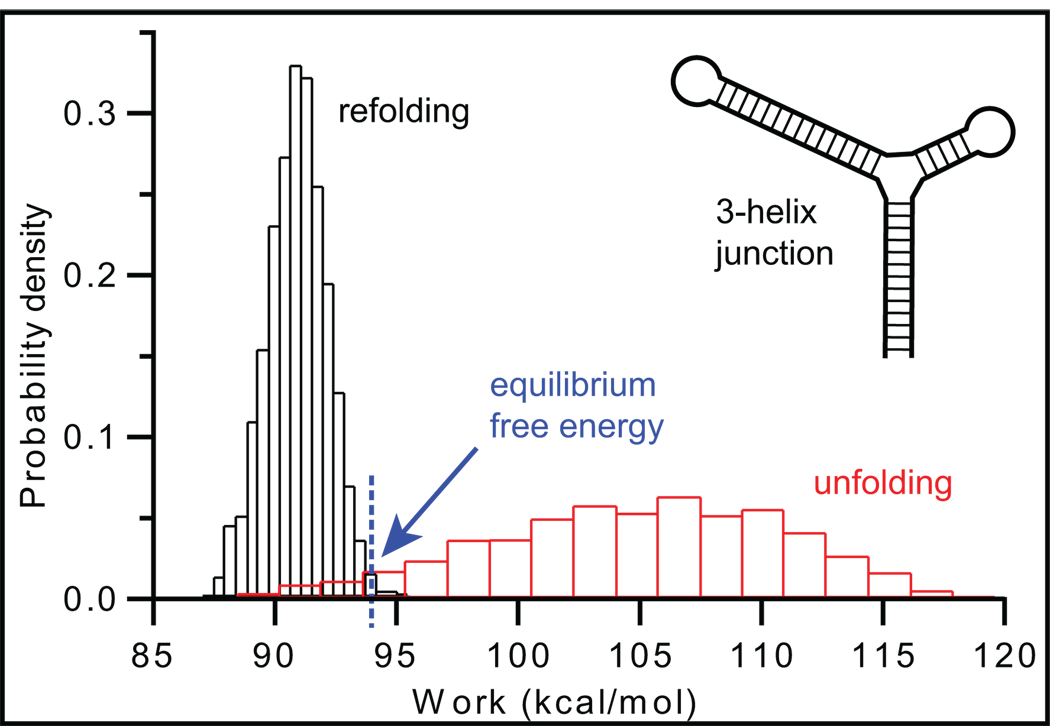
Fluctuation theorems, such as Jarzynski’s equality [22] and Crooks’ theorem [23], which derive equilibrium free energies from distributions of the non-equilibrium work performed on single molecules (Figure 2), are particularly noteworthy. These relations are exceedingly useful for SMFS, because slow folding rates often produce non-equilibrium measurements. First demonstrated on simple hairpins [9,10], fluctuation theorems have now been applied experimentally to estimate the free energy for pseudoknot [17] and riboswitch [26••] folding, as well as the salt-dependence of duplex stability [13]. A method for reconstructing the unfolding energy landscape based on the Jarzynski equality has also been proposed [43]. Although there is continuing debate about the general validity of such theorems [44,45] and they are challenging to implement experimentally (especially far from equilibrium [46]), they nevertheless represent an important addition to the RNA toolbox.
Figure 2.
Determining equilibrium free energies from non-equilibrium measurements using the Crooks fluctuation theorem. The point of intersection of the probability distributions for the irreversible work done when unfolding (red) and refolding (black) a 3-helix junction supplies the equilibrium free energy for the folding reaction. Adapted from [9].
Complex systems: large RNAs and the interaction of folding with cellular processes
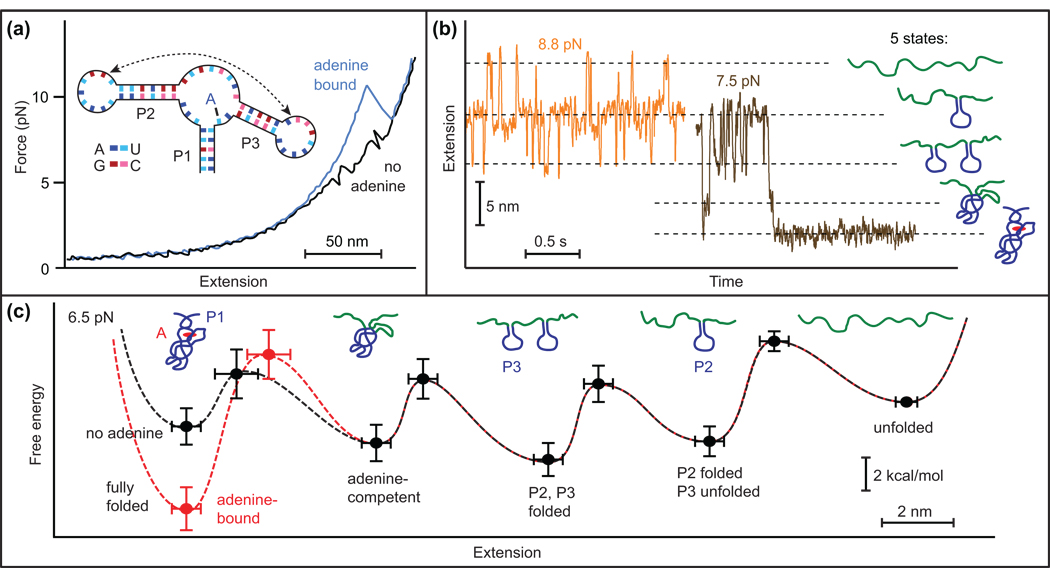
A few large RNA molecules were studied by SMFS early on, including the full-length ribozyme from T. thermophila [47] and the 16S rRNA from E. coli [8]. These studies observed complex FECs containing large numbers of unfolding events. This work illustrates a principal limitation of SMFS: because different structural elements (or combinations thereof) may lead to the same change in molecular extension, the assignment of particular unfolding events to specific substructures can be ambiguous. Additional measurements are often required to remove such ambiguities, including the use of mutations or anti-sense oligomers to block the formation of targeted structures or interactions [47]. The formation of unusually stable structures or interactions may also protect less stable ones from unfolding, further complicating the interpretation of FECs. This difficulty can sometimes be overcome by making constant-force measurements instead, as illustrated by recent work on folding and ligand binding in an adenine riboswitch aptamer [26••]. In that study, FECs displayed only a single transient intermediate, whereas constant-force trajectories revealed three distinct intermediate states, corresponding to various secondary and tertiary structures. High-resolution measurements allowed each intermediate to be assigned structurally, the sequence of folding events to be determined, the energetic effect of ligand binding to be measured, and the folding energy landscape to be reconstructed, providing an integrated picture of a molecule with multiple substructures and interactions (Figure 3).
Figure 3.
SMFS of folding in an adenine riboswitch aptamer. (a) Binding of the adenine ligand alters the shape of the FECs. (b) Constant-force trajectories obtained at two different forces indicate five distinct states in the folding, corresponding to the formation of specific structural elements of the aptamer, as indicated. (c) Energy landscape for the complete folding reaction, both with (red) and without (black) ligand. Adapted from [26••].
One of the most intriguing applications of SMFS has been to study how RNA folding interacts with processive nucleic acid enzymes, including polymerases, helicases, and ribosomes. During translocation, such enzymes often melt structures formed in their RNA templates. Such structures can, in turn, regulate enzyme progress, affecting the fundamental processes of transcription and translation. The mechanism of ribosomal frameshifting has been probed by SMFS measurements relating the mechanical stability of a frameshifting pseudoknot to the frameshift efficiency [48•]. Other studies have used the unfolding of RNA hairpins to probe the motion of helicases [49•,50] and ribosomes [51••]: folded hairpins held under moderate tension are melted by the enzyme as it translocates, revealing key motor characteristics such as the step size, processivity, and NTP dependence.
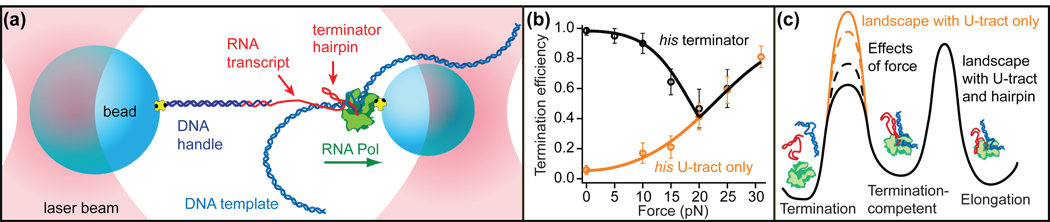
The interaction between RNA transcript structure and the process of transcription has been another focus of study. By applying force to RNA molecules transcribed in situ (Figure 4a), the effects of RNA structures on the polymerase motion can be probed, as can the effects of the polymerization process on the folding of the transcript. The structure of mRNA was shown to have little or no effect [52] on the “ubiquitous” pauses that are found in single-molecule records of RNA polymerase motion [53,54]. However, force applied to terminator hairpins was found to exert a strong effect on transcriptional termination (Figure 4b). SMFS measurements of the termination efficiency for various terminator hairpin constructs led to a quantitative model for termination efficiency (Figure 4c) [55••]. The folding of a riboswitch aptamer during transcription has also been compared to its folding after transcription, in order to discern any differences arising from competition between the kinetics of folding and transcription [26••]. No co-transcriptional effects of note were found, but only the one such comparison has been made to date, and there are many opportunities for further exploration.
Figure 4.
Effect of co-transcriptional folding/unfolding of the histidine (his) terminator hairpin upon transcription. Adapted from [55••]. (a) Force is applied to an RNA transcript during transcription, biasing the structure (folded/unfolded) of the terminator hairpin as the motion of polymerase is continuously measured. (b) Efficiency of transcription is found to depend on force through two mechanisms that work in conjunction: (i) shearing of the RNA/DNA hybrid at a U-rich tract (yellow), most evident at high force, and (ii) folding/unfolding of the intrinsic terminator hairpin (black), most evident at low force. (c) Model of the energy landscape for termination, where the terminator hairpin lowers the barrier to termination. Force applied to the RNA can raise this barrier in the presence of the hairpin by causing the hairpin to unfold (dashed black line), but can lower the barrier in the absence of the hairpin by shearing the RNA/DNA hybrid at the U-tract (dashed yellow line).
Conclusions
Significant advances in instrumentation, theory, and modeling have greatly enhanced the utility of SMFS as a probe of RNA folding, especially for determining elusive properties such as the structure or energetics of intermediate and transition states. Most work to date has involved relatively simple RNA structures. The information gained from these systems, however, is now enabling a detailed study of more complex RNAs, yielding an integrated picture of folding landscapes and dynamics. Efforts to obtain additional information about conformational rearrangements by combining SMFS with other modalities, particularly single-molecule fluorescence, are being actively pursued and now beginning to bear fruit [56,57•]. Some issues about the interpretations of SMFS measurements remain to be addressed, however, such as why SMFS findings occasionally differ from other types of measurement, e.g., fluorescence studies of hairpin folding suggesting the existence of an ensemble of collapsed intermediate states prior to duplex formation [58,59]. Looking forward, one of the key contributions anticipated from SMFS is a deeper examination of how RNA structures interact with enzymes. Such measurements hold great promise for new insights into the molecular mechanisms for the regulation of transcription and translation.
Acknowledgements
We acknowledge the support of the U.S. National Institutes for Health (grants GM57035 and GM66275) and the National Institute for Nanotechnology (National Research Council of Canada).
References and recommended reading
• of special interest
•• of outstanding interest
- 1.Brion P, Westhof E. Hierarchy and dynamics. Annu Rev Biophys Biomol Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 2.Draper DE, Grilley D, Soto AM. Ions and RNA folding. Annu Rev Biophys Biomol Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 3.Chen SJ. RNA folding: conformational statistics, folding kinetics, and ion electrostatics. Annu Rev Biophys. 2008;37:197–214. doi: 10.1146/annurev.biophys.37.032807.125957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bustamante C, Chemla YR, Forde NR, Izhaky D. Mechanical Processes in Biochemistry. Annu Rev Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- 5.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Woodside MT, Behnke-Parks WM, Larizadeh K, Travers K, Herschlag D, Block SM. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc Natl Acad Sci USA. 2006;103:6190–6195. doi: 10.1073/pnas.0511048103. A high-precision, systematic study of how hairpin folding under tension depends on the stem length, loop length, and G:C content. The transition state for folding a hairpin with an unpatterned stem sequence is found to involve the formation of just a few basepairs.
- 7.Liphardt J, Onoa B, Smith SB, Tinoco I, Jr, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 8.Harlepp S, Marchal T, Robert J, Léger JF, Xayaphoummine A, Isambert H, Chatenay D. Probing complex RNA structures by mechanical force. Eur Phys J E. 2003;12:605–615. doi: 10.1140/epje/e2004-00033-4. [DOI] [PubMed] [Google Scholar]
- 9.Collin D, Ritort F, Jarzynski C, Smith SB, Tinoco JI, Bustamante C. Verification of the Crooks fluctuation theorem and recovery of RNA folding free energies. Nature. 2005;437:231–234. doi: 10.1038/nature04061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liphardt J, Dumont S, Smith SB, Tinoco IJ, Bustamante C. Equilibrium information from nonequilibrium measurements in an experimental test of Jarzynski's equality. Science. 2002;296:1832–1835. doi: 10.1126/science.1071152. [DOI] [PubMed] [Google Scholar]
- 11.Li PTX, Collin D, Smith SB, Bustamante C, Tinoco IJ. Probing the mechanical folding kinetics of TAR RNA by hopping, force-jump, and force-ramp methods. Biophys J. 2006;90:250–260. doi: 10.1529/biophysj.105.068049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li PTX, Bustamante C, Tinoco IJ. Real-time control of the energy landscape by force directs the folding of RNA molecules. Proc Natl Acad Sci USA. 2007;104:7039–7044. doi: 10.1073/pnas.0702137104. Demonstrates how the folding pathway of RNAs can be manipulated by modulating the relaxation rate of the applied force.
- 13.Vieregg J, Cheng W, Bustamante C, Tinoco IJ. Measurement of the effect of monovalent cations on RNA hairpin stability. J Am Chem Soc. 2007;129:14966–14973. doi: 10.1021/ja074809o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Woodside MT, Anthony PC, Behnke-Parks WM, Larizadeh K, Herschlag D, Block SM. Direct measurement of the full, sequence-dependent folding landscape of a nucleic acid. Science. 2006;314:1001–1004. doi: 10.1126/science.1133601. Shows that the folding energy landscape of hairpins can be controlled systematically via the stem sequence, and demonstrates a high-resolution method to measure the energy landscape along the entire reaction coordinate with an optical trap.
- 15. Li PTX, Bustamante C, Tinoco IJ. Unusual mechanical stability of a minimal RNA kissing complex. Proc Natl Acad Sci USA. 2006;106:15847–15852. doi: 10.1073/pnas.0607202103. Investigates the mechanical folding of a minimal kissing loop complex, resolving the intermediate and transition states for the unusually stable loop-loop interactions.
- 16.Chen G, Wen JD, Tinoco IJ. Single-molecule mechanical unfolding and folding of a pseudoknot in human telomerase RNA. RNA. 2007;13:2175–2188. doi: 10.1261/rna.676707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green L, Kim CH, Bustamante C, Tinoco IJ. Characterization of the mechanical unfolding of RNA pseudoknots. J Mol Biol. 2007;375:511–528. doi: 10.1016/j.jmb.2007.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li PTX, Vieregg J, Tinoco IJ. How RNA unfolds and refolds. Annu Rev Biochem. 2008;77 doi: 10.1146/annurev.biochem.77.061206.174353. [DOI] [PubMed] [Google Scholar]
- 19.Greenleaf WJ, Woodside MT, Block SM. High-resolution, single-molecule measurements of biomolecular motion. Annu Rev Biophys Biomol Struct. 2007;36:171–190. doi: 10.1146/annurev.biophys.36.101106.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dudko OK, Hummer G, Szabo A. Intrinsic rates and activation free energies from single-molecule pulling experiments. Phys Rev Lett. 2006;96:108101. doi: 10.1103/PhysRevLett.96.108101. Presents an improved theoretical framework to extract kinetic rates at zero force, transition states and activation energies from single molecule experiments. Also relates results obtained at constant force to results obtained at constant loading rate.
- 21.Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarzynski C. Nonequilibrium equality for free energy differences. Phys Rev Lett. 1997;78:2690–2693. [Google Scholar]
- 23.Crooks GE. Entropy production fluctuation theorem and the nonequilibrium work relation for free energy differences. Phys Rev E. 1999;60:2721–2726. doi: 10.1103/physreve.60.2721. [DOI] [PubMed] [Google Scholar]
- 24.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 25.García-García C, Draper DE. Electrostatic interactions in a peptide-RNA complex. J Mol Biol. 2003;331:75–88. doi: 10.1016/s0022-2836(03)00615-6. [DOI] [PubMed] [Google Scholar]
- 26. Greenleaf WJ, Frieda KL, Foster DAN, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. The most in-depth SMFS study of RNA folding to date. Characterizes folding and ligand binding in a riboswitch aptamer using both equilibrium and non-equilibrium methods, identifies multiple intermediate states, and reconstructs the energy landscape quantitatively.
- 27.Greenleaf WJ, Woodside MT, Abbondanzieri EA, Block SM. Passive all-optical force clamp for high-resolution laser trapping. Phys Rev Lett. 2005;95:208102. doi: 10.1103/PhysRevLett.95.208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hyeon C, Thirumalai D. Forced-unfolding and force-quench refolding of RNA hairpins. Biophys J. 2006;90:3410–3427. doi: 10.1529/biophysj.105.078030. Coarse-grained simulations are used to investigate systematically the effects of force on RNA folding and how SMFS measurements work.
- 29.Manosas M, Wen JD, Li PTX, Smith SB, Bustamante C, Tinoco JI, Ritort F. Force unfolding kinetics of RNA using optical tweezers. II. Modeling experiments. Biophys J. 2007;92:3010–3021. doi: 10.1529/biophysj.106.094243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen JD, Manosas M, Li PTX, Smith SB, Bustamante C, Ritort F, Tinoco IJ. Force unfolding kinetics of RNA using optical tweezers. I. Effects of experimental variables on measured results. Biophys J. 2007;92:2996–3009. doi: 10.1529/biophysj.106.094052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moffitt JR, Chemla YR, Izhaky D, Bustamante C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc Natl Acad Sci USA. 2006;103:9006–9011. doi: 10.1073/pnas.0603342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manosas M, Ritort F. Thermodynamic and kinetic aspects of RNA pulling experiments. Biophys J. 2005;88:3224–3242. doi: 10.1529/biophysj.104.045344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreuzer HJ, Payne SH, Livadaru L. Stretching a macromolecule in an atomic force microscope: statistical mechanical analysis. Biophys J. 2001;80:2505–2514. doi: 10.1016/S0006-3495(01)76222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vieregg JR, Tinoco IJ. Modelling RNA folding under mechanical tension. Mol Phys. 2006;104:1343–1352. doi: 10.1080/00268970500525986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manosas M, Collin D, Ritort F. Force-dependent fragility in RNA hairpins. Phys Rev Lett. 2006;96:218301. doi: 10.1103/PhysRevLett.96.218301. [DOI] [PubMed] [Google Scholar]
- 36.Cocco S, Marko JF, Monasson R, Sarkar A, Yan J. Force-extension behavior of folding polymers. Eur Phys J E. 2003;10:249–263. doi: 10.1140/epje/i2002-10113-2. [DOI] [PubMed] [Google Scholar]
- 37.Liu F, Ou-Yang Z. Unfolding single RNA molecules by mechanical force: a stochastic kinetic method. Phys Rev E. 2004;70:40901–40904. doi: 10.1103/PhysRevE.70.040901. [DOI] [PubMed] [Google Scholar]
- 38.Gerland U, Bundschuh R, Hwa T. Force-induced denaturation of RNA. Biophys J. 2001;81:1324–1332. doi: 10.1016/S0006-3495(01)75789-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hyeon C, Thirumalai D. Mechanical unfolding of RNA hairpins. Proc Natl Acad Sci USA. 2005;102:6789–6794. doi: 10.1073/pnas.0408314102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hyeon C, Thirumalai D. Mechanical unfolding of RNA: from hairpins to structures with internal multiloops. Biophys J. 2007;92:731–743. doi: 10.1529/biophysj.106.093062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hyeon C, Thirumalai D. Multiple probes are required to explore and control the rugged energy landscape of RNA hairpins. J Am Chem Soc. 2008;130:1538–1539. doi: 10.1021/ja0771641. [DOI] [PubMed] [Google Scholar]
- 42.Best RB, Paci E, Hummer G, Dudko OK. Pulling direction as a reaction coordinate for the mechanical unfolding of single molecules. J Phys Chem B. 2008;112:5968–5976. doi: 10.1021/jp075955j. [DOI] [PubMed] [Google Scholar]
- 43.Hummer G, Szabo A. Free energy surfaces from single-molecule force spectroscopy. Acc Chem Res. 2005;38:504–513. doi: 10.1021/ar040148d. [DOI] [PubMed] [Google Scholar]
- 44.Cohen EGD, Mauzerall D. A note on the Jarzynski equality. J Stat Mech: Theory Exp. 2004;2004:P07006. [Google Scholar]
- 45.Vilar JMG, Rubi JM. Failure of the work-hamiltonian connection for free-energy calculations. Phys Rev Lett. 2008;100:20601. doi: 10.1103/PhysRevLett.100.020601. [DOI] [PubMed] [Google Scholar]
- 46.Gore J, Ritort F, Bustamante C. Bias and error in estimates of equilibrium free-energy differences from nonequilibrium measurements. Proc Natl Acad Sci USA. 2003;100:12564–12569. doi: 10.1073/pnas.1635159100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Onoa B, Dumont S, Liphardt J, Smith SB, Tinoco IJ, Bustamante C. Identifying kinetic barriers to mechanical unfolding of the T. thermophila ribozyme. Science. 2003;299:1892–1895. doi: 10.1126/science.1081338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hansen TM, Reihani SNS, Oddershede LB, Sorensen MA. Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proc Natl Acad Sci USA. 2007;104:5830–5835. doi: 10.1073/pnas.0608668104. The resistance of frameshifting pseudoknots to mechanical unfolding measured by SMFS is correlated with frameshifting efficiency measured in bulk assays.
- 49. Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco IJ, Pyle AM, Bustamante C. RNA translocation and unwinding mechanism of HCV NS 3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. The NS3 helicase is shown to translocate through RNA hairpins by melting them using an inchworm-type mechanism.
- 50.Cheng W, Dumont S, Tinoco IJ, Bustamante C. NS3 helicase actively separates RNA strands and senses sequence barriers ahead of the opening fork. Proc Natl Acad Sci USA. 2007;104:13954–13959. doi: 10.1073/pnas.0702315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wen JD, Lancaster L, Hodges C, Zeri AC, Yoshimura SH, Noller HF, Bustamante C, Tinoco IJ. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. The most complex biological system studied using SMFS to date. The translocation of a single ribosome through mRNA hairpin structures is observed directly, revealing single-codon steps. Sequence-dependent pausing is also characterized.
- 52.Dalal RV, Larson MH, Neuman KC, Gelles J, Landick R, Block SM. Pulling on the nascent RNA during transcription does not alter kinetics of elongation or ubiquitous pausing. Mol Cell. 2006;23:231–239. doi: 10.1016/j.molcel.2006.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing Is Independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- 54.Adelman K, La Porta A, Santangelo TJ, Lis JT, Roberts JW, Wang MD. Single molecule analysis of RNA polymerase elongation reveals uniform kinetic behavior. Proc Natl Acad Sci USA. 2002;99:13538–13543. doi: 10.1073/pnas.212358999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Larson MH, Greenleaf WJ, Landick R, Block SM. Applied force reveals mechanistic and energetic details of transcription termination. Cell. 2008;132:971–982. doi: 10.1016/j.cell.2008.01.027. A co-transcriptional, single-molecule assay is used to quantify the contribution to transcription termination efficiency of two critical elements of the RNA transcript: terminator hairpins and associated U-rich tracts.
- 56.Lang MJ, Fordyce PM, Engh AM, Neuman KC, Block SM. Simultaneous, coincident optical trapping and single-molecule fluorescence. Nat Methods. 2004;1:133–139. doi: 10.1038/nmeth714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hohng S, Zhou R, Nahas MK, Yu J, Schulten K, Lilley DMJ, Ha T. Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the Holliday junction. Science. 2007;318:279–283. doi: 10.1126/science.1146113. Single-molecule fluorescence and force spectroscopy are combined in a technical tour-de-force to study the dynamics of Holliday junctions.
- 58.Ma H, Wan C, Wu A, Zewail AH. DNA folding and melting observed in real time redefine the energy landscape. Proc Natl Acad Sci USA. 2007;104:712–716. doi: 10.1073/pnas.0610028104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jung J, Ihly R, Scott E, Yu M, Van Orden A. Probing the complete folding trajectory of a DNA hairpin using dual beam fluorescence fluctuation spectroscopy. J Phys Chem B. 2008;112:127–133. doi: 10.1021/jp076248t. [DOI] [PubMed] [Google Scholar]