Abstract
BACKGROUND
The molecular mechanisms that underlie colorectal cancer (CRC) include microsatellite instability (MSI), chromosomal instability, and the CpG island methylator phenotype. There is evidence to suggest that CRC incidence varies among different ethnic populations worldwide. The authors of this report hypothesized that environmental factors and lifestyle differences among various ethnic groups may differentially influence the epigenetic regulation of tumor suppressor genes in CRC.
METHODS
In the current study, microdissection and DNA extraction were performed on 128 samples of CRC from Israeli patients (85 Jews and 43 Arabs). MSI analysis, mutL homolog 1 (MLH1) and mutS homolog 2 (MSH2) protein expression levels, and MLH1 promoter methylation were investigated by combined bisulfite restriction analysis. The v-raf murine sarcoma viral oncogene homolog B1 (BRAF) valine-to-glutamic acid mutation at residue 600 was investigated by direct DNA sequencing.
RESULTS
High MSI (MSI-H), MLH1 methylation, and BRAF mutations were observed in 11.6%, 9.4%, and 23.5% of Jews, respectively, and in 16.2%, 17.6%, and 20.9% of Arabs, respectively (P value nonsignificant). MLH1 promoter methylation was observed in 22.6% of microsatellite-stable (MSS) tumors and in 53.8% of MSI-H tumors (P < .015). Extensive methylation (covering both 5′ and 3′ promoter regions) was present in all MSI-H tumors with loss of MLH1 expression. BRAF mutation was observed in 15.6% and 46.1% of MSS tumors and MSI-H tumors, respectively (P < .007). BRAF mutation was observed in 66%, 22.2%, and 14.7% of patients who had tumors with extensive MLH1 promoter methylation, methylation of the 5′ region alone, or without methylation, respectively (P < .006).
CONCLUSIONS
There was no difference in molecular signatures examined between Jewish and Arab patients with CRC in Israel. Extensive promoter methylation was associated with MLH1 inactivation, MSI, and BRAF mutation.
Keywords: microsatellite instability, mutL homolog 1, BRAF mutation, colorectal cancer, methylation
Colorectal cancer (CRC) is the second leading cause of cancer mortality in the Western countries and affects 5% to 6% of the population. In Israel, the rate of CRC differs significantly among different ethnic groups. The incidence is highest in Ashkenazi Jews (European-and American-born Jews), intermediate in Sephardic Jews (Asian- and African-born Jews), and lower in Israeli-born Jews.1 Israeli-born Arabs have the lowest incidence rate of CRC.2 The cause of these differences is not understood well and is believed to result from a combination of many genetic, epigenetic, and environmental factors, such as the I1307K adenomatous polyposis coli gene (APC) variant in Ashkenazi Jews3 and CpG island methylation that results in gene silencing.4
Microsatellite instability (MSI) is the hallmark of DNA mismatch-repair gene (MMR)-deficient CRCs. MSI has been observed in approximately 15% of sporadic colon cancers and in virtually all colorectal tumors arising in patients with hereditary nonpolyposis colorectal cancer (HNPCC) or Lynch syndrome (LS).5 Sporadic tumors that exhibit MSI have unique clinicopathologic characteristics, such as poor differentiation, mucinous histology, Crohn-like lymphoid infiltrate, preferential proximal location, and older age.6,7 The prognosis for patients who have sporadic tumors with high MSI (MSI-H) is better than that for patients who have microsatellite-stable (MSS) tumors, but they respond poorly to chemotherapy. 8,9 In contrast to HNPCC tumors, in which loss of MMR gene expression is caused by germline mutation, MSI in sporadic colon cancers is caused by epigenetic modification of the mutL homolog 1 gene (MLH1).4,7 The MLH1 gene has a large CpG island within its promoter region. Deng et al10 compared the methylated status of the MLH1 promoter and its protein expression in 24 cancer cell lines and observed that methylation of a small proximal region closer to the transcriptional start site (defined as Region C) in the promoter invariably was correlated with loss of gene expression; methylation outside of this region did not always correlates with the absence of MLH1 expression. Those results were confirmed by others in clinical samples and suggest that partial MLH1 promoter methylation limited to the upstream region of the MLH1 promoter is not critical for gene silencing.11,12 The mechanism of epigenetic modification of tumor suppressor genes in cancer remains unclear. It has been proposed that low consumption of folic acid, vitamin B6, and vitamin B12, high alcohol intake, and smoking may affect DNA methylation,13,14 but the critical factors in human cancer remain uncertain.
Recently, a v-raf murine sarcoma viral oncogene homolog B1 (BRAF) somatic mutation (valine-to-glutamic acid mutation at residue 600 [V600E]), has been identified in multiple human cancers. Davies et al15 reported a BRAF mutation in 66% of malignant melanomas and at lower frequency in other human malignancies, including CRC. BRAF is a serine/threonine kinase that is part of the Ras/Raf-extracellular signal-regulated kinase/mitogen activated protein kinase (MEK/MAPK) signaling pathway and is an essential component of intracellular signaling. In CRC, this BRAF mutation is associated strongly with sporadic MSI tumors and extensive MLH1 promoter methylation,12,16–18 but not with MMR-deficient tumors in patients with LS.19,20 The objective of this study was to investigate a possible relationship between MLH1 methylation in 2 different promoter regions, MSI status, and BRAF mutation in sporadic patients with CRC from different Israeli ethnic groups.
MATERIALS AND METHODS
Patients
This study included consecutive patients with sporadic CRC from Rabin Medical Center, Beilinson Hospital, Tel Aviv University, Israel who underwent curative surgery and had specimens available for research between the years 2000 and 2003. The exclusion criteria were patients with CRC who belonged to families with familial adenomatous polyposis or LS according to detailed family history, Amsterdam criteria, or Bethesda guidelines.21 Demographic, clinical, and pathologic data were computed from the patients’ files, surgical reports, and pathology reports. Staging was assessed according to the TNM classification. The study was approved by the institutional review board (IRB) of Rabin Medical Center (RMC IRB no. 4087) and Baylor University Medical Center (IRB no. 005-134).
DNA Extraction
For DNA extraction, we used formalin-fixed, paraffin-embedded tissue slides. Hematoxylin and eosin-stained slides were reviewed, and areas of tumors were marked to exclude normal tissue. Ten-micron slides were microdissected using a sterile scalpel blade under a dissecting microscope. Genomic DNA was extracted using QIAmp DNA Mini Kits (Qiagen, Valencia, Calif), according to the manufacturer’s instructions.
MSI Analysis
MSI testing was done by using 5 quasimonomorphic repeat markers (BAT25, BAT26, NR21, NR24, and NR27) and pentaplex polymerase chain reaction (PCR).22 The pentaplex PCR was performed in a 25 µL volume containing 12.5 µL of HotStarTaq Master Mix (Qiagen), 0.4 µM of each primer pair, and approximately 50 ng of genomic DNA; then, the slides were amplified by PCR under the following conditions: denaturation at 94°C for 15 minutes, 35 cycles of denaturation at 95°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 30 seconds. This was followed by an extension step at 72°C for 10 minutes in a PTC 200 DNA Engine Thermocyler (MJ Research, Inc., Waltham, Mass). The PCR products were loaded onto an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, Calif), and the results were analyzed using Gene Mapper Analysis software. MSI-H was defined when ≥3 markers were mutated, and MSS was defined when <3 markers were unstable.22
Immunohistochemistry for MLH1 and MSH2
Immunohistochemical staining was performed on the paraffin-embedded CRC tissues. Five-micrometer-thick tissue sections were mounted on positively charged slides. Tissue sections were deparaffinized with xylene and rehydrated through a gradient alcohol series. The sections sub-sequently were immersed in 10 mmol/L citrate buffer, pH 6.0, and heated in an autoclave at 121°C for 5 minutes for nonenzymatic antigen retrieval. Dual endogenous enzyme block solution (Dako Cytomation Inc., Carpinteria, Calif) was applied to the tissue sections for 10 minutes to block the endogenous peroxidase activity. The primary antibodies were mouse monoclonal antibodies against MLH1 protein (clone G168-15; dilution, 1:100) and mutS homolog 2 (MSH2) protein (clone G219-1129; dilution, 1:100). Both antibodies were purchased from PharMingen (San Diego, Calif). The sections were incubated for 90 minutes at room temperature, followed by incubation with Dako EnVision-labeled polymer (Dako Cytomation Inc.) for 30 minutes. Staining was developed by reaction with diaminobenzidine chromogen for 5 to 10 minutes and then counterstained for 5 minutes with hematoxylin. A negative control was run without the primary antibody. The normal staining pattern for both MLH1 and MSH2 monoclonal antibodies is nuclear. Tissues with positive nuclear staining in the tumor cells were scored as positive. Tissue specimens with complete loss of nuclear staining were scored as negative. The non-neoplastic colonic tissues, stroma, and infiltrating lymphocytes normally show positive nuclear staining, and this served as an internal positive control for the immuno-histochemistry.
Combined Bisulfite Restriction Analysis
Methylation status of the 5′ and 3′ regions of the MLH1 promoter was analyzed quantitatively by combined bisulfite restriction analysis (COBRA) on bisulfite-modified DNA that was obtained using the EpiTect bisulfite kit (Qiagen). The modified DNA was used as a template for PCR using the following primers: 5′ region: forward, YGGGTAAGTYGTTTTGAYGTAGA; reverse, TATA CCTAATCTATCRCCRCCTCA; 3′-region: forward, GGGAGGGATGAAGAGATTTAGT; reverse, ACCTTCAACCAATCACCTCAAT. PCR for both regions was carried out in a volume of 25 µL containing 12.5 µL of HotStarTaq Master Mix (Qiagen), 0.4 µM of each primer set, and approximately 50 ng of bisulfite-modified DNA using the following amplification profile: denaturation at 94°C for 15 minutes, 39 cycles of denaturation at 95°C for 30 seconds, annealing at 58°C for the 5′ region or at 54°C for the 3′ region for 30 seconds, and extension at 72°C for 30 seconds. This was followed by an extension step at 72°C for 10 minutes in a PTC 200 DNA Engine Thermocyler (MJ Research, Inc.). Appropriate negative and positive controls were included with each PCR. Normal colonic mucosal DNA artificially methylated by SssI was used as a positive control, whereas untreated normal colonic mucosa was used as an unmethylated control. A water blank was used as negative control in every PCR reaction. The PCR products of the 5′ region were digested with HhaI (New England Biolabs, Beverly, Mass) at 37°C for 16 hours, and PCR products of the 3′ region were digested with RsaI (New England Biolabs) at 37°C for 16 hours. The HhaI restriction enzyme cleaves GĈGC sites, and RsaI cleaves GT̂AC sites if it is retained through bisulfite-mediated deamination. Thus, enzymatic cleavage indicates methylation, whereas lack of digestion indicates the absence of methylation. Digested PCR products were identified by ethidium bromide staining on 2.5% agarose gels and quantified using a Gel Logic 200 Imaging System (Eastman Kodak Company, Rochester, NY). The methylation density was computed as a ratio of the density of digested and undigested bands.
Direct Sequencing of the BRAF Gene
The BRAF exon 15, which contains the V600E mutation hotspot, was amplified using the following primers: forward, TGCTTGCTCTGATAGGAAAATGA; reverse, TGGATCCAGACAACTGTTCAAA. PCR was carried out in a volume of 25 µL in the following sequence: denaturation at 94°C for 15 minutes, 39 cycles of denaturation at 95°C for 30 seconds, annealing at 54°C for 30 seconds, and extension at 72°C for 30 seconds. This was followed by an extension step at 72°C for 10 minutes. Three micro-liters of PCR product were run on an agarose gel to verify the adequacy of the reaction, and the rest of the PCR product was purified by using the QIAquick PCR purification kit (Qiagen). Direct sequencing was performed using the BigDye version 1.1 cycle-sequencing kit (Applied Biosystems) in a capillary automated sequencer (ABI PRISM 3100 Genetic Analyzer; Applied Biosystems). RKO cells with the V600E BRAF mutation were used as a positive control. Water was used as a negative control for every PCR.
RESULTS
Patients and Pathologic Staging
Microdissection and DNA extraction were performed on 128 CRC specimens. The mean age at CRC diagnosis was 67.6 ± 12.3 years among all patients; 60 patients (46.8%) were men. Eighty-five patients (66.4%) were Jews, and 43 patients (33.6%) were Israeli-born Arabs. In 29.7% of patients, the tumor was located proximal to the splenic flexure. Clinical, epidemiological, and pathologic data (stage and grade) are shown in Table 1.
Table 1.
Patient Characteristics (n=128)
| Characteristic | No. of Patients (%) |
|---|---|
| Average ± SD age at CRC diagnosis, y | 67.6± 12.3 |
| Sex | |
| Men | 60 (46.8) |
| Women | 68 (53.2) |
| Origin | |
| Israeli Jews | 85 (66.4) |
| Ashkenazi | 39 (45.8) |
| Sephardic | 29 (34.1) |
| Israeli-born Jews | 17 (20.1) |
| Israeli Arabs | 43 (33.6) |
| Tumor site | |
| Left sided | 90 (70.3) |
| Right sided | 38 (29.7) |
| Tumor grade | |
| Well differentiated | 26 (20.3) |
| Moderately differentiated | 70 (54.6) |
| Poorly differentiated | 32 (25.1) |
| Tumor stage | |
| A/B | 56 (54.9) |
| C/D | 46 (45.1) |
| Undetermined | 26 |
CRC indicates colorectal cancer; SD, standard deviation.
Microsatellite Instability Status and Immunohistochemistry Staining
MSI testing was performed in all patients. Overall, 13 patients (10.2%) had MSI-H tumors, and 115 patients (89.8%) had MSS tumors. A proximal tumor location and poor differentiation were observed in 69.2% and 53.9% of MSI-H tumors, respectively, versus 26.1% and 22.6% of MSS tumors, respectively (P = .008 and P = .05, respectively). There were no differences in age, sex, or origin between MSI-H tumors and MSS tumors (Table 2).
Table 2.
Microsatellite Instability and Clinicopathologic Characteristics in Patients With Colorectal Cancer
| No. of Patients (%) | |||
|---|---|---|---|
| Characteristic | MSS | MSI | P |
| No. of patients | 115 (89.8) | 13 (10.2) | |
| Age, y | |||
| Mean ± SD | 67.1 ± 12.4 | 72.2 ± 10.6 | .16 |
| Range | 38–92 | 52–85 | |
| Sex | |||
| Men | 52 (45.2) | 8 (61.6) | .26 |
| Women | 63 (54.8) | 5 (38.4) | |
| Tumor location | |||
| Proximal | 30 (26.1) | 9 (69.2) | .008 |
| Distal | 85 (79.3) | 4 (30.8) | |
| Tumor grade | |||
| Well or moderately differentiated |
89 (77.4) | 6 (46.1) | .05 |
| Poorly differentiated | 26 (22.6) | 7 (53.9) | |
| Ethnicity | |||
| Jews | 77 (91.6) | 8 (9.4) | |
| Ashkenazi | 32 (82.1) | 7 (17.9) | .1 |
| Sephardim | 28 (96.6) | 1 (3.4) | |
| Israeli-born Jews | 17 (100) | 0 (0) | |
| Israeli-born Arabs | 38 (88.4) | 5 (11.6) | |
MSS indicates microsatellite stable; MSI-H, high microsatellite instability; SD, standard deviation.
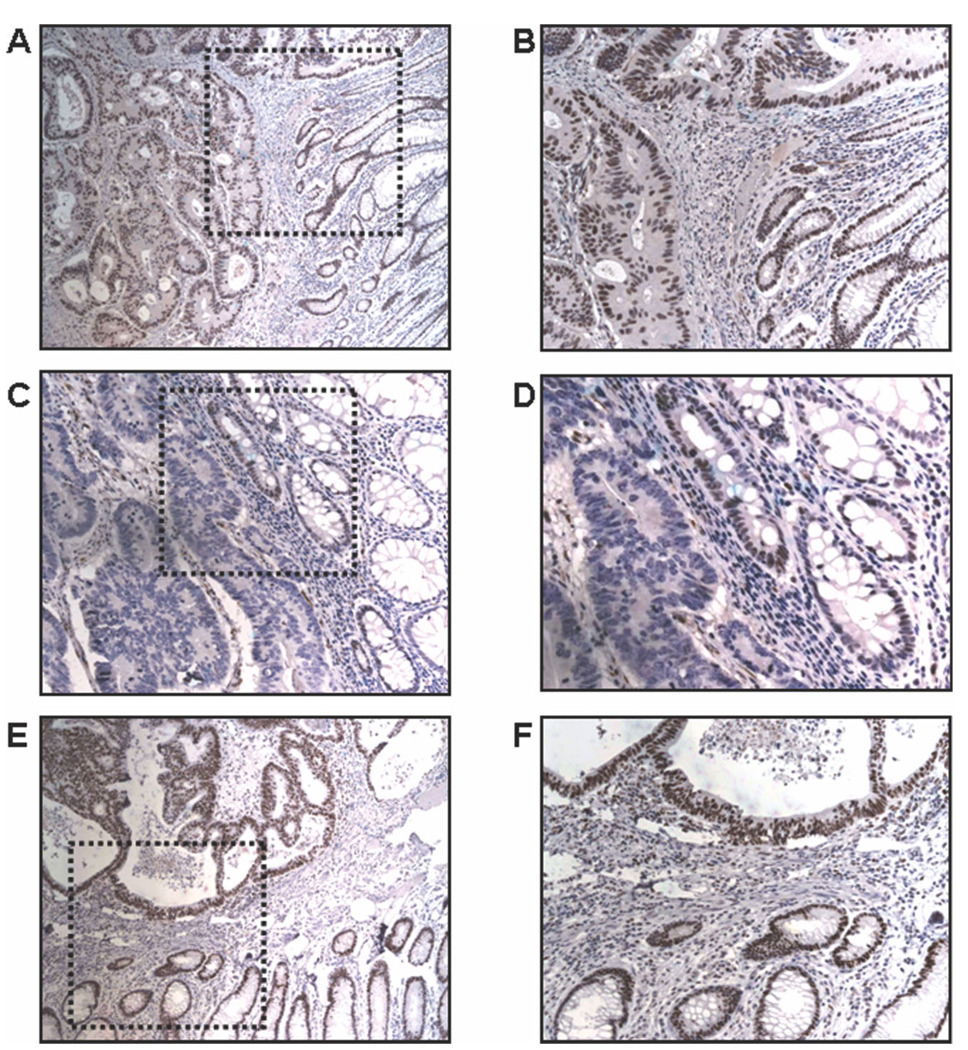
Immunohistochemistry staining for MLH1 and MSH2 protein expression was performed for all 13 MSI-H tumors and for 70 (60.8%) MSS tumors. In 12 MSI-H tumors (92.3%), loss of MLH1 expression was demonstrated; and, in 1 tumor, there was lack of expression of both MLH1 protein and MSH2 protein (Fig. 1). All MSS tumors were expressed both MLH1 protein and MSH2 protein.
FIGURE 1.
Immunohistochemistry staining for protein expression of the mutL homolog 1 gene MLH1 and the mutS homolog 2 gene MSH2. (A,B) Immunohistochemistry results for MLH1 protein expression from a microsatellite-stable (MSS) tumor (original magnification, ×100 in A, ×200 in B). MLH1 expression can be seen as nuclear brown staining in normal and tumor tissue. (C,D) Immunohistochemistry results for MLH1 expression from a tumor with high microsatellite instability (original magnification, ×100 in C, ×200 in D) There is nuclear staining in normal mucosa and absent nuclear staining in the tumor tissue. Inflammatory cells and non-neoplastic cells (internal control) are positively stained. (E,F) MSH2 protein expression in an MSS tumor (original magnification, ×100 in E, ×200 in F). There is normal nuclear staining in the normal mucosa and the tumor tissue.
MLH1 Promoter Methylation
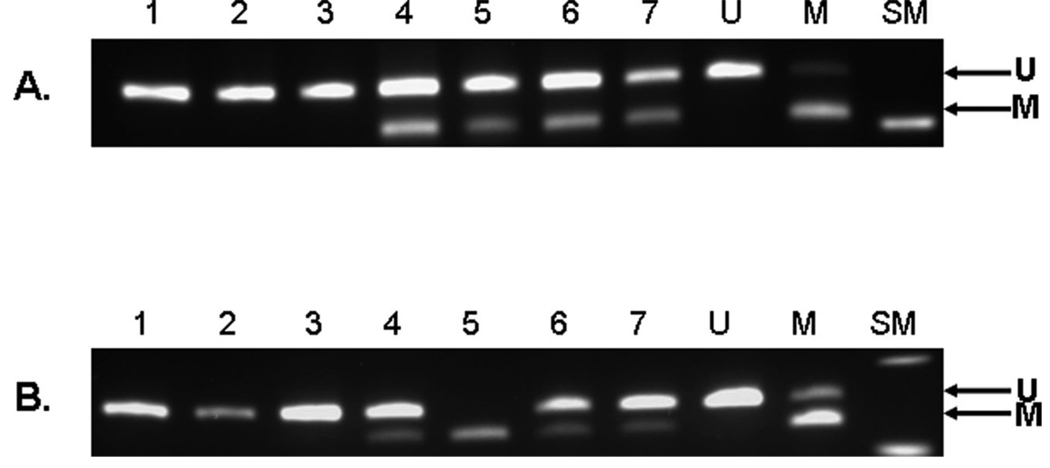
The methylation status of both the 5′ region and the 3′ region of the MLH1 promoter was assessed using COBRA (Fig. 2). Ninety-five patients (74.2%) had unmethylated tumors, 27 patients (21.1%) had tumors with methylation of the 5′ region only, and 6 patients (4.7%) had tumors with methylation of both the 5′ region and the 3′ region (extensive methylation). Specific sex and ethnic group origin was not associated with MLH1 promoter methylation (Table 3). Patients who had tumors with extensive methylation were older than patients who either had tumors with 5′-region methylation only or had unmethylated tumors. Patients who had tumors with extensive methylation also had significantly more MSI-H tumors, most of their tumors were poorly differentiated, and they had a trend toward more proximal location. There were no significant differences between patients who had tumors with methylation of the 5′-region and the patients who had tumors without promoter methylation in any clinical or pathologic parameter that we examined (Table 3).
FIGURE 2.
Methylation analysis of the mutL homolog 1 gene (MLH1) promoter in the 5′ and 3′ regions. Combined bisulfite restriction analysis for the MLH1 promoter region: (A) shows the 5′ region after digestion with the restrictive enzyme HhaI and (B) is the 3′ region after digestion with the restriction enzyme RsaI. The upper band is the unmethylated product and the lower band is a methylated product. Lanes 1–3, unmethylated samples; lanes 4–7, methylated samples; U, unmethylated control; M, methylated control; SM, size marker.
Table 3.
MLH1 Promoter Methylation and Clinicopathologic Characteristics in Patients With Colorectal Cancer
|
MLH1 Methylation Status: No. of Patients (%) |
||||
|---|---|---|---|---|
| Characteristic | Unmethylated | 5′-Region Alone | Extensive | P |
| All patients | 95 (74.2) | 27 (21.1) | 6 (4.7) | |
| Mean age ± SD, y | 66.3 ± 12.8 | 69.8 ± 11 | 79.8 ± 3.6 | .02 |
| Sex | ||||
| Men | 48 (50.5) | 9 (33.3) | 3 (50) | .28 |
| Women | 47 (49.5) | 18 (66.7) | 3 (50) | |
| Tumor location | ||||
| Proximal | 25 (26) | 9 (33) | 4 (66.6) | .09 |
| Distal | 70 (74) | 18 (67) | 2 (33.4) | |
| Tumor grade | ||||
| Well or moderately differentiated | 66 (80) | 19 (70.4) | 1 (16.7) | .02 |
| Poorly differentiated | 19 (20) | 8 (29.6) | 5 (83.3) | |
| Ethnicity | ||||
| Jews | 61/85 (71.8) | 20/85 (23.5) | 4 (4.7) | |
| Ashkenazi | 25/39 (64.1) | 10/39 (25.6) | 4 (10.3) | .63 |
| Sephardim | 24/29 (82.7) | 5/29 (17.3) | 0 (0) | |
| Israeli-born Jews | 12/17 (70.5) | 5/17 (29.6) | 0 (0) | |
| Israeli-born Arabs | 34/43 (79.1) | 7/43 (16.2) | 2 (4.7) | |
| MSI status | ||||
| MSS | 83 (93.7) | 26 (96.3) | 0 (0) | .001 |
| MSI-H | 6 (6.3) | 1 (3.7) | 6 (100) | |
| MLH1 expression | ||||
| Positive | 57 (92) | 14 (93.3) | 0 (0) | .001 |
| Negative | 5 (8) | 1 (6.7) | 6 (100) | |
| BRAF mutation | ||||
| Wild type | 81(85.3) | 21 (77.8) | 2 (33.4) | .006 |
| V600E | 14 (14.7) | 6 (22.2) | 4 (66.4) | |
MLH1 indicates mutL homolog 1 gene; SD, standard deviation; MSS, microsatellite stable; MSI-H, high microsatellite instability; BRAF, v-raf murine sarcoma viral oncogene homolog B1; V600E, valine-to-glutamic acid mutation at residue 600.
BRAF Mutation Analysis
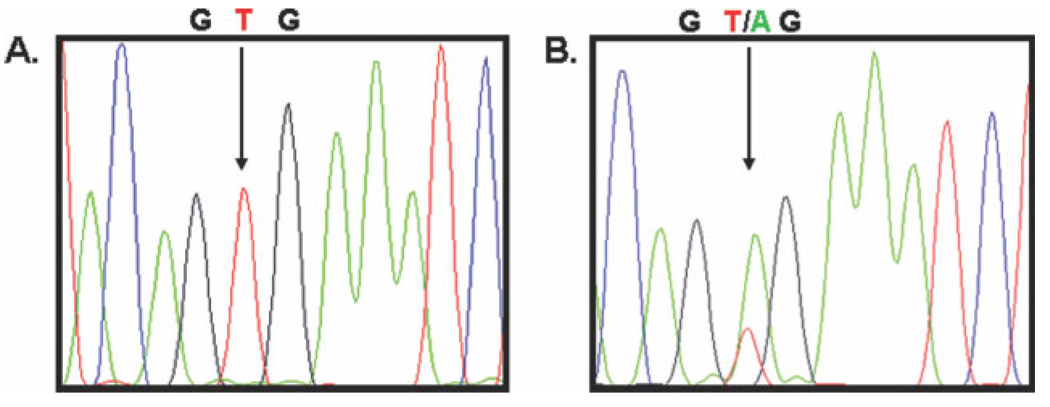
Direct sequencing for the BRAF hotspot mutation V600E was performed in all 128 CRC samples. Twenty-four samples (18.7%) harbored BRAF mutations, and 104 samples (82.3%) had wild-type alleles (Fig. 3 and Table 4). Age, sex, and ethnic origin were not associated with BRAF mutations. However, tumors with BRAF mutations more often were located proximally in the colon and more frequently had MSI-H status. Patients who had tumors with extensive promoter methylation, compared with patients who had tumors with methylation of the 5′ region only and patients who had tumors without methylation, harbored more BRAF mutations (66.6% vs 22.2% and 14.7%, respectively; P = .006). It is noteworthy that, among the patients who had MSI-H tumors without MLH1 promoter methylation, only 1 of 6 tumors had BRAF mutations.
FIGURE 3.
Direct sequencing for a v-raf murine sarcoma viral oncogene homolog B1 (BRAF) mutation. Representative sequence chromatographs from (A) a wild-type BRAF gene and (B) a BRAF gene with the V600E valine-to-glutamic acid mutation at residue 600 in exon 15. G indicates guanine; T, thymine; A, adenine.
Table 4.
BRAF Mutation and Clinicopathologic Characteristics in Patients With Colorectal Cancer
|
BRAF Mutation: No. of Patients (%) |
|||
|---|---|---|---|
| Characteristic | Wild-Type Allele | Mutant Allele | P |
| All patients | 104 (82.3) | 24 (18.7) | |
| Age, y | |||
| Mean ± SD | 67.1 ± 12.7 | 70.04 ± 0.4 | .29 |
| Range | 38–92 | 52–85 | |
| Sex | |||
| Men | 47 (45.2) | 13 (54.2) | .42 |
| Women | 57 (54.8) | 11 (45.8) | |
| Tumor location | |||
| Proximal | 78 (75) | 12 (50) | .016 |
| Distal | 26 (25) | 12 (50) | |
| Histologic grade | |||
| Well or moderately differentiated |
78 (75) | 18 (74.9) | .997 |
| Poorly differentiated | 26 (25) | 6 (25.1) | |
| Ethnicity | |||
| Jews | 70 (82.4) | 15 (17.6) | |
| Ashkenazi | 31 (79.4) | 8 (20.6) | |
| Sephardim | 25 (86.2) | 4 (13.8) | .87 |
| Israeli-born Jews | 14 (82.3) | 3(17.7) | |
| Israeli-born Arabs | 34 (79.1) | 9 (20.9) | |
| MSI status | |||
| MSS | 97 (84.4) | 18 (15.6) | |
| MSI-H | 7 (53.9) | 6 (46.1) | .008 |
BRAF indicates v-raf murine sarcoma viral oncogene homolog B1; SD, standard deviation; MSS, microsatellite stable; MSI-H, high microsatellite instability.
DISCUSSION
Our objective for the current study was to investigate whether there are differences in the molecular characteristics of sporadic CRC among patients from different Israeli ethnic groups. The major group in our patient population consisted of Jews who were born in Eastern Europe and America (Ashkenazi Jews) or in Asia and North Africa (Sephardic Jews) who immigrated to Israel. In addition, there are Arabic individuals who live in Israel, are diverse ethnically, live separately, maintain an Arabic lifestyle, and have unique environmental and dietary exposures. The incidence of CRC varies among these groups, but the cause for these differences is not known.
From the molecular genetic point of view, CRC is not a homogenous disease. Three main pathways have been described in the multistep carcinogenesis of CRC: chromosomal instability,23 MSI,4,5 and CpG island methylator phenotype (CIMP).16,24,25 With regard to CIMP, Weisenberger and colleagues recently identified additional methylation markers for a more accurate classification of CIMP and observed a strong relation between CIMP and BRAF mutation in MSI-H tumors.26 Epigenetic modification of multiple genes is an acquired event in certain cancers, and different environmental factors have been proposed to affect DNA methylation and result in the methylator phenotype.13,14 Recently, Hitchins and colleagues described the possibility of inheriting a cancer-associated MLH1 germline epimutation, but this must be rare.27
The prevalence of CRC differs among different ethnic groups within the United States, and African Americans have a 10% higher incidence of CRC compared with Caucasian Americans.28 However, in Africa, the prevalence of CRC is substantially lower than in the United States. This observation suggests that combinations of genetic and environmental factors play important roles in the development of CRC.
Israeli Arabs have a lower incidence of CRC compared with Israeli Jews2; however, there is not enough information on the molecular characterization of CRCs in these groups to account for the observed differences. In the current study, we report that there were no significant differences in the frequency of MSI, MLH1 promoter methylation, or BRAF mutation between Arabic and Jewish patients with sporadic CRC in Israel. Our results support previous findings by Shpitz and colleagues, who investigated the mutator phenotype in 125 Arab patients and 210 Jewish patients with sporadic CRC over a 20-year period.29 Those authors did not observe any significant difference in MLH1 or MSH2 expression by immunohistochemistry, but they did not perform any MSI analyses.
We also demonstrated that partial methylation of the MLH1 promoter in the upstream region (the 5′ region) is not sufficient for MLH1 silencing. Extensive methylation of the MLH1 promoter (the 5′ and 3′ regions) is essential for MLH1 inactivation and for developing MSI-H in CRC. Tumors with isolated methylation of the 5′ region were not different from unmethylated tumors in any parameter that we examined. Conversely, patients who had tumors with extensive methylation of both the 5′ and 3′ regions of the MLH1 promoter were significantly older, had MSI-H, had a proximal tumor location, had poor tumor differentiation, and harbored BRAF mutations. In our work, all 6 tumors that had extensive methylation of the MLH1 promoter were MSI-H tumors. Our results are similar to those from other studies, which observed that only full methylation of the MLH1 gene, including methylation of a small region proximal to the transcription start site (Region C), is critical for MLH1 inactivation. 10–12
Recently, BRAF mutation was described in a small percentage of MSS tumors but in 50% of MSI-H tumors, particularly in tumors with methylation of the MLH1 promoter.12,17 To our knowledge, a BRAF mutation has never been described in tumors with germline mutations of the MLH1 gene.19,20 The link between BRAF mutations and methylation suggests the possibility of using this marker in the discrimination between MSI-H sporadic tumors (the result of MLH1 promoter methylation) and MSI-H tumors with germline mutations (LS).19,20,30 We identified BRAF mutations in 15.6% of MSS tumors and in 46.1% of MSI-H tumors. Within the group of MSI-H tumors, 66.6% of tumors with extensive methylation of the MLH1 promoter harbored BRAF mutations versus only 1 of 6 unmethylated MSI-H tumors (16%). Our results are similar to the results of other studies,18,26 which demonstrated that mutations in BRAF are associated with extensive methylation of the MLH1 promoter.
The differences in CRC incidence among ethnic groups in Israel may have a genetic basis, an environmental basis, or an interaction between both factors. Dietary practices and lifestyle issues differ between these groups. We initially had hypothesized that the differences in CRC incidence were linked to nongenetic issues, which might create detectable differences in the molecular phenotypes of the tumors and perhaps provide insights into possible prevention strategies. However, the tumors in our study had indistinguishable differences in the mutational ‘fingerprints’ that we measured; there appear to be more similarities than differences in the way that CRC develops among different ethnic groups in Israel.
In conclusion, we did not observed any significant differences in MSI status, MLH1 promoter methylation, or BRAF mutation status in Israeli patients with sporadic CRC from different ethnic groups. Full methylation of the MLH1 promoter is needed for MLH1 inactivation and development of MSI. BRAF mutation is associated with MSI-H tumors that have extensive methylation of the promoter region.
Footnotes
Conflict of Interest Disclosures
The authors made no disclosures.
References
- 1.Barchana M, Liphshitz I, Rozen P. Trends in colorectal cancer incidence and mortality in the Israeli Jewish ethnic populations. Fam Cancer. 2004;3:207–214. doi: 10.1007/s10689-004-9546-y. [DOI] [PubMed] [Google Scholar]
- 2.Fireman Z, Sandler E, Kopelman Y, Segal A, Sternberg A. Ethnic differences in colorectal cancer among Arab and Jewish neighbors in Israel. Am J Gastroenterol. 2001;96:204–207. doi: 10.1111/j.1572-0241.2001.03476.x. [DOI] [PubMed] [Google Scholar]
- 3.Laken SJ, Petersen GM, Gruber SB, et al. Familial colorectal cancer in Ashkenazim due to a hypermutable tract in APC. Nat Genet. 1997;17:79–83. doi: 10.1038/ng0997-79. [DOI] [PubMed] [Google Scholar]
- 4.Herman JG, Umar A, Polyak K, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aaltonen LA, Peltomaki P, Leach FS, et al. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 6.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 7.Lynch HT, Smyrk TC, Watson P, et al. Genetics, natural history, tumor spectrum, and pathology of hereditary non-polyposis colorectal cancer: an updated review. Gastroenterology. 1993;104:1535–1549. doi: 10.1016/0016-5085(93)90368-m. [DOI] [PubMed] [Google Scholar]
- 8.Carethers JM, Smith EJ, Behling CA, et al. Use of 5-fluorouracil and survival in patients with microsatellite-unstable colorectal cancer. Gastroenterology. 2004;126:394–401. doi: 10.1053/j.gastro.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 9.Niv Y. Microsatellite instability and MLH1 promoter hypermethylation in colorectal cancer. World J Gastroenterol. 2007;13:1767–1769. doi: 10.3748/wjg.v13.i12.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng G, Chen A, Hong J, Chae HS, Kim YS. Methylation of CpG in a small region of the hMLH1 promoter invariably correlates with the absence of gene expression. Cancer Res. 1999;59:2029–2033. [PubMed] [Google Scholar]
- 11.Miyakura Y, Sugano K, Konishi F, et al. Extensive methylation of hMLH1 promoter region predominates in proximal colon cancer with microsatellite instability. Gastroenterology. 2001;121:1300–1309. doi: 10.1053/gast.2001.29616. [DOI] [PubMed] [Google Scholar]
- 12.Nagasaka T, Sasamoto H, Notohara K, et al. Colorectal cancer with mutation in BRAF, KRAS, and wild-type with respect to both oncogenes showing different patterns of DNA methylation. J Clin Oncol. 2004;22:4584–4594. doi: 10.1200/JCO.2004.02.154. [DOI] [PubMed] [Google Scholar]
- 13.Diergaarde B, Braam H, van Muijen GN, Ligtenberg MJ, Kok FJ, Kampman E. Dietary factors and microsatellite instability in sporadic colon carcinomas. Cancer Epidemiol Biomarkers Prev. 2003;12:1130–1136. [PubMed] [Google Scholar]
- 14.Samowitz WS, Albertsen H, Sweeney C, et al. Association of smoking, CpG island methylator phenotype, and V600E BRAF mutations in colon cancer. J Natl Cancer Inst. 2006;98:1731–1738. doi: 10.1093/jnci/djj468. [DOI] [PubMed] [Google Scholar]
- 15.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 16.Goel A, Nagasaka T, Arnold CN, et al. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132:127–138. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 17.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934–934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 18.Koinuma K, Shitoh K, Miyakura Y, et al. Mutations of BRAF are associated with extensive hMLH1 promoter methylation in sporadic colorectal carcinomas. Int J Cancer. 2004;108:237–242. doi: 10.1002/ijc.11523. [DOI] [PubMed] [Google Scholar]
- 19.McGivern A, Wynter CV, Whitehall VL, et al. Promoter hypermethylation frequency and BRAF mutations distinguish hereditary non-polyposis colon cancer from sporadic MSI-H colon cancer. Fam Cancer. 2004;3:101–107. doi: 10.1023/B:FAME.0000039861.30651.c8. [DOI] [PubMed] [Google Scholar]
- 20.Domingo E, Laiho P, Ollikainen M, et al. BRAF screening as a low-cost effective strategy for simplifying HNPCC genetic testing. J Med Genet. 2004;41:664–668. doi: 10.1136/jmg.2004.020651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suraweera N, Duval A, Reperant M, et al. Evaluation of tumor microsatellite instability using 5 quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 2002;123:1804–1811. doi: 10.1053/gast.2002.37070. [DOI] [PubMed] [Google Scholar]
- 23.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 24.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toyota M, Ohe-Toyota M, Ahuja N, Issa JP. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci USA. 2000;97:710–715. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 27.Hitchins MP, Wong JJ, Suthers G, et al. Inheritance of a cancer-associated MLH1 germ-line epimutation. N Engl J Med. 2007;356:697–705. doi: 10.1056/NEJMoa064522. [DOI] [PubMed] [Google Scholar]
- 28.Ghafoor A, Jemal A, Cokkinides V, et al. Cancer statistics for African Americans. CA Cancer J Clin. 2002;52:326–341. doi: 10.3322/canjclin.52.6.326. [DOI] [PubMed] [Google Scholar]
- 29.Shpitz B, Millman M, Ziv Y, Klein E, Grankin M, Gochberg S, et al. Predominance of younger age, advanced stage, poorly differentiated and mucinous histology in Israeli Arab patients with colorectal cancer. Anticancer Res. 2006;26:533–537. [PubMed] [Google Scholar]
- 30.Samowitz WS, Albertsen H, Herrick J, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–845. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]