Abstract
Disruptions of beta-catenin and the canonical Wnt pathway are well documented in cancer. However, little is known of the non-canonical branch of the Wnt pathway. In this study, we investigate the transcript level patterns of genes in the Wnt pathway in squamous cell lung cancer using reverse-transcriptase (RT)-PCR. It was found that over half of the samples examined exhibited dysregulated gene expression of multiple components of the non-canonical branch of the WNT pathway. In the cases where beta catenin (CTNNB1) was not over-expressed, we identified strong relationships of expression between wingless-type MMTV integration site family member 5A (WNT5A)/frizzled homolog 2 (FZD2), frizzled homolog 3 (FZD3)/dishevelled 2 (DVL2), and low density lipoprotein receptor-related protein 5 (LRP5)/secreted frizzled-related protein 4 (SFRP4). This is one of the first studies to demonstrate expression of genes in the non-canonical pathway in normal lung tissue and its disruption in lung squamous cell carcinoma. These findings suggest that the non-canonical pathway may have a more prominent role in lung cancer than previously reported.
Keywords: WNT pathway, lung cancer, gene expression, NSCLC, non-canonical, squamous cell carcinoma
Background
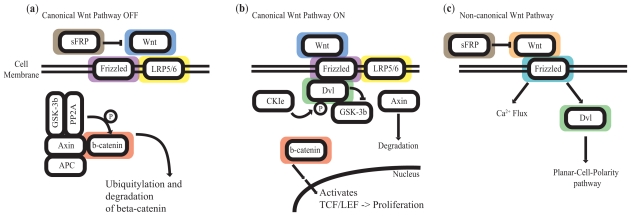
The Wnt pathway is integral to developmental biology. The canonical pathway determines β-catenin stability and influences the transcription of TCF/LEF target genes (Clevers, 2006). In the absence of Wnt ligands binding to frizzled receptors, the canonical Wnt pathway is turned off leading to the eventual degradation of β-catenin (Fig. 1A). Conversely, the binding of Wnt ligands promotes the formation of a tertiary complex between Wnt, Frizzled and LRP5/6, allowing β-catenin to shuttle into the nucleus and bind to TCF/LEF proteins, thus activating target gene transcription (Fig. 1B). The non-canonical pathway is β-catenin-independent and controls cell movements during morphogenesis. It is further subdivided into the Wnt/calcium pathway and the planar-cell-polarity (PCP) pathway (Fig. 1C) (Katoh, 2005; Veeman, Axelrod and Moon, 2003).
Figure 1.
Schematic representation of the canonical and non-canonical Wnt pathways. sFRPs are inhibitors of both the canonical and non-canonical branches of the Wnt pathway. (a) Canonical Wnt pathway in its off state. (b) Canonical Wnt pathway in its on state. (c) Non-canonical Wnt pathway. Color halos represent genes that were used in this study. Grey: SFRP1, SFRP2, SFRP3, SFRP4, SFRP5; Blue: WNT1, WNT3A; Purple: FZD1; Yellow: LRP5, LRP6; Red: CTNNB1; Orange: WNT5A, WNT11; Teal: FZD2, FZD3, FZD6; Green: DVL2.
The canonical Wnt pathway plays a critical role during the development of the lung (Eberhart and Argani, 2001; Mazieres et al. 2005). In the adult lung, the canonical Wnt pathway contributes to bronchial epithelial regeneration (Steel et al.). However, little is known about the non-canonical pathway in the adult lung. Furthermore, disruption of the canonical pathway branch is well documented in cancer (Clevers, 2006; Ilyas, 2005), but the involvement of the non-canonical branch of the Wnt pathway in cancer is virtually unknown. Disruptions have been reported for many canonical pathway components; for example, mutations in axin and APC are common in colorectal and hepatocellular cancers (Aust et al. 2002; Taniguchi et al. 2002). The consequence of disrupting the Wnt pathway is the constitutive activation of target genes, such as MYC, CCND1, VEGF, each contributing to the hallmarks of cancer (Hanahan and Weinberg, 2000).
Lung cancer is a highly aggressive disease and is the leading cause of cancer deaths worldwide (Minna, Roth and Gazdar, 2002). Identification of genes and pathways disrupted in lung cancer will improve our understanding of this disease. Recent studies have implicated the disruption of upstream Wnt components in lung cancer. For example, wingless-related MMTV integration site 1 (WNT1) and wingless-related MMTV integration site 2 (WNT2) are overexpressed in non-small cell lung cancer (NSCLC) (He, B et al. 2004; You et al. 2004); loss of wingless-related MMTV integration site family, member 7A (WNT7A) contributes to the progression of lung cancer through its inability to induce E-cadherin (Ohira et al. 2003); and DVL3 is reported to be overexpressed in NSCLC (Uematsu et al. 2003). However, disruption of downstream Wnt pathway components are not often reported in lung cancer (Shigemitsu et al. 2001; Ueda et al. 2001). Coordinated measurements of Wnt components expression will be necessary to define their involvement in lung cancer. In this study, we investigated the transcript level patterns of pathway components in normal lung tissue and lung squamous cell carcinoma (SCC) to determine if the expression of the non-canonical pathway is disrupted in lung cancer.
Methods
RNA isolation and cDNA synthesis
A total of 20 frozen squamous lung tumor with matched lung normal samples were obtained from St. Paul’s Hospital. Sections (10 μm) fixed in 70% ethanol were manually microdissected based on histopathologic evalution of hematoxylin and eosin stained sample sections by a lung pathologist. Dissected cells were homogenized in a guanidine thiocyanate lysis buffer and RNA was isolated using the RNeasy Mini Kit (Qiagen, Mississauga, ON, Canada). Matched normal lung tissue samples were homogenized in the presence of liquid nitrogen and RNA was extracted using Trizol reagent (Invitrogen, Burlington, ON, Canada). Purified total RNA (40 ng samples) was converted to cDNA using the Superscript II RNAse H reverse-transcriptase system (Invitrogen). Primer sequences and melting temperatures are described in Additional file 1. In addition, 10 frozen paired SCC samples were obtained for quantitative RT-PCR from Vancouver General Hospital. All samples for this study were collected with approval by the Review of Ethics Board of the Ministry of British Columbia.
Gene expression analysis
Expression levels were determined by gene-specific PCR (Additional file 1) and the β-actin gene was used for normalization. cDNA samples obtained from tissues known to express the Wnt pathway were used as positive controls (Clontech human multiple tissue cDNA Panels 1 and 2, BD Biosciences Clontech, Mississauga, ON, Canada). Forty nanograms of RNA were converted to cDNA as described above and 1/20 of the cDNA from each sample was used. PCR cycle conditions were as follow: one cycle of 95 °C, 1 min; 30–35 cycles of 95 °C, 30 s; 55 °C, 30 s (for β-actin); 72 °C, 30 s; and a final 10 min extension at 72 °C. PCR products were resolved by polyacrylamide gel electrophoresis, imaged by SYBR green staining (Roche, Laval, PQ, Canada) on a Molecular Dynamics Storm Phosphoimager model 860, and quantified using ImageQuant software (Molecular Dynamics, Piscataway, NJ, U.S.A.). To verify the absence of genomic DNA contamination in the cDNA, a ACTB primer was designed to yield a 597 bp fragment for genomic DNA amplification product and a 400 bp fragment for cDNA amplification.
For quantitative PCR, TaqMan primers (primer IDs in parentheses) for FZD3 (Hs00184043_m1), DVL2 (Hs00182901_m1), and CTNNB1 (Hs00170025_m1) were purchased from Applied Biosystems (Applied Biosystems, CA, U.S.A.). PCR was performed as recommended by Applied Bio-systems. All reactions were 25 μL in volume and performed in triplicate. To account for variations in template quantities, cycle threshold (Ct) values were normalized using the Ct values of ACTB. The efficiencies of all TaqMan primers were estimated using the raw data generated at each well as previously described (Liu and Saint, 2002; Weksberg et al. 2005).
Statistical analysis of gene expression levels
Gene expression levels of Wnt pathway components were determined by calculating the signal intensity ratio between each gene of interest and ACTB was calculated for all lung samples. For the negative control, cDNA template was omitted in the reaction.
For the expression level comparison between tumor and normal tissue, the intensity ratio of each gene in tumor was divided by the corresponding intensity ratio in the matched normal tissue samples. Correlation coefficient analysis was performed using the Matlab Statistics Toolbox (The Mathworks, Natick, MA).
Results and Discussion
Wnt pathway components representing the canonical and the non-canonical sub-paths were selected for expression analysis using RT-PCR in an effort to investigate the state of the pathways in normal lungs and their disruption in lung tumors. The genes representing the canonical pathway in this study include WNT1, wingless-related MMTV integration site family, member 3A (WNT3A), frizzled homolog 1 (FZD1), low density lipoprotein receptor-related protein 5 (LRP5), density lipoprotein receptor-related protein 6 (LRP6), and CTNNB1. The non-canonical components were represented by wingless-related MMTV integration site family, member 5A (WNT5A), wingless-related MMTV integration site family, member 11 (WNT11), frizzled homolog 2 (FZD2), frizzled homolog 3 (FZD3), and frizzled homolog 6 (FZD6) (Katoh, 2005; Pongracz and Stockley, 2006; Torres et al. 1996). In addition, representative members of the Dvl family and the sFRP family were also included in our analysis (Melkonyan et al. 1997; Schumann et al. 2000; Uematsu et al. 2003). It should be noted that the regulation of the wnt pathway is complex. Some of Wnt ligands may have the activation of both the non-canonical and canonical branches and as such, their effects are strongly dependent on the receptor.
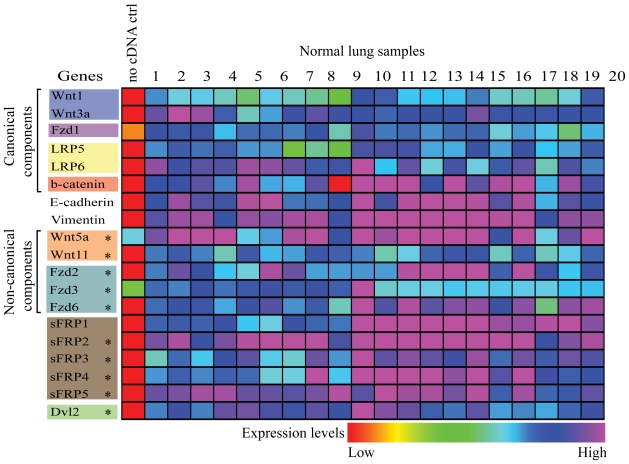
Expression profiles of the Wnt components in 20 normal lung samples are shown (Fig. 2). Analysis of the canonical Wnt pathway genes suggests their transcription in normal lung. Notably, the non-canonical Wnt components, WNT5A, WNT11, FZD2, FZD3, and FZD6, are also present in the normal lung. This is one of the first reports of non-canonical pathway expression in adult human non-malignant lung tissue (Pongracz and Stockley, 2006; Winn et al. 2005). In addition, dishevelled 2, dsh homolog (DVL2) and members of the sFRP family are also expressed in the normal lung (Fig. 2). Although the role of DVL2 is not entirely clear in humans, it has been shown to activate the PCP signaling pathway in a series of experiments involving HEK293T cell and Xenopus models (Habas, Kato and He, 2001). As for the sFRP family, not all members serve the same functions. For example, sFRP2 enables the breast cancer cell line MCF-7 to resist TNF-induced apoptosis while sFRP1 sensitizes the cells to TNF-induced apoptosis (Melkonyan et al. 1997). The gene expression data on normal lung tissue provide a baseline for comparison against those of NSCLC.
Figure 2.
Expression profiles of 19 genes in 20 normal lung samples. Raw data was shifted by adding a constant to get rid of negative values. A trimmed mean was calculated (excluding the lower and upper 2% values) and a scaling factor was calculated as 500 divided by the trimmed mean. Each raw value was then multiplied by the scaling factor to create a new distribution centered at 500. The value displayed is the log10 of the scaled data. *represent expression of genes that have not been reported in normal lung in literature.
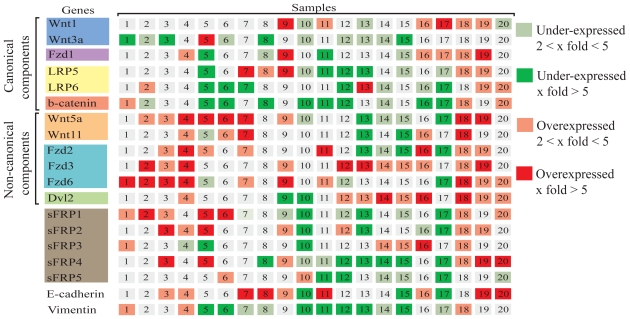
To investigate which Wnt pathway components are disrupted in lung tumors, a pairwise comparison between tumour and matched normal lung samples was performed on the Wnt pathway genes (Fig. 3). A comparison of the components in the canonical and non-canonical pathway shows that the non-canonical pathway may be involved in a subset of tumor cases. For example, patient 4 (Fig. 4A) shows high level up-regulation of all non-canonical components while there is minimal disruption of the transcription levels of canonical components. In contrast, patient 12 (Fig. 4B) shows high level down-regulation of canonical components with minimal disruptions of the non-canonical components. In fact, the twenty samples have varying patterns of expression changes in the Wnt pathway components (Additional file 2). We only observed overexpression of CTNNB1 in three out of 20 samples, and this observation held true in an independent set of ten cases by quantitative PCR (Fig. 5). This is not surprising as CTNNB1 activity is determined by protein stability and nuclear localization (Blache et al. 2004; He, TC et al. 1998; Korinek et al. 1997; Mann et al. 1999; Morin et al. 1997). However, it is remarkable that 11 out of 20 samples showed overexpression of multiple non-canonical components. These findings strongly suggest the involvement of the non-canonical pathway in lung SCC.
Figure 3.
Expression data of the 19 genes in a pairwise comparison between lung tumors and their matched normals. Colored spots represent expression fold changes of genes by dividing tumor intensity ratio by the normal intensity ratio. Only 2 fold changes are displayed for the 20 tumor-normal pairs.
Figure 4.
Comparison of pairwise (tumor versus matched normal) expression profiles between two patients. (a) Patient 7 show high level disruption of non-canonical WNT components and low to no change in expression of canonical WNT components. (b) Patient 10 shows high level disruption of canonical WNT components and low to no change in expression of non-canonical WNT components.
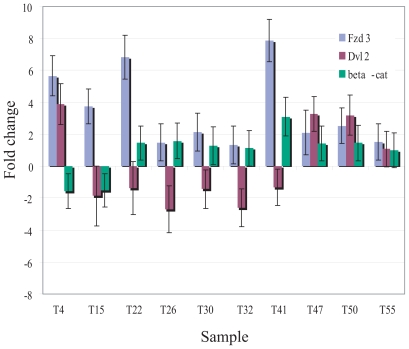
Figure 5.
Differential expression of FZD3, DVL2 and CTNNB1 between 10 lung squamous cell carcinoma and matched normals (Samples T21–T30). Results are generated by real-time RT-PCR using TaqMan gene specific primers from Applied Biosystems.
Based on the expression patterns of CTNNB1, it appears not all tumors solely involve the canonical pathway. We next investigated which particular non-canonical components are involved in the samples without CTNNB1 overexpression. As some of the components affect both the canonical and non-canonical pathway, we selected only genes belonging to one or the other, namely those listed in Table 1. The expression of each gene was categorized as +1 for up-regulation, -1 for down-regulation, and 0 for unchanged, with a 2-fold expression difference deemed change. The genes were paired and a percentage was calculated for each pair of genes based on the number of times they showed the same category of expression. In other words, the percentage is an indication of how similar the expression changes are for a given set of genes. The table of gene comparisons with the corresponding percentages is shown in Table 1. Gene pairs that were less than 50% concordant in expression change were eliminated from further analysis. For the remaining gene pairs, a Spearman correlation was calculated. Eleven gene pairs showed statistically significant correlation with three gene pairs showing greater than 65% concordance: LRP5 and secreted frizzled-related protein 4 (SFRP4), WNT5A and FZD2, and FZD3 and DVL2. We also investigated the frequency of discordant expression changes but, there were no gene pairs that were significantly related (data not shown).
Table 1.
Pairwise expression correlation of genes in WNT pathway.
| Gene Pairs | (%) | R | pval | |
|---|---|---|---|---|
| Wnt1 | Wnt11 | 53 | 0.22 | 0.39 |
| B-catenin | sFRP5 | 53 | 0.04 | 0.87 |
| B-catenin | Wnt3a | 59 | 0.14 | 0.59 |
| B-catenin | Lrp6 | 53 | 0.41 | 0.11 |
| sFRP5 | Wnt3a | 53 | −0.02 | 0.95 |
| sFRP5 | Lrp6 | 53 | 0.35 | 0.17 |
| sFRP5 | sFRP4 | 53 | 0.31 | 0.22 |
| Wnt3a | sFRP1 | 59 | 0.06 | 0.81 |
| Wnt3a | sFRP4 | 59 | 0.45 | 0.07 |
| Fzd1 | Lrp5 | 53 | 0.48 | 0.05 |
| Fzd1 | sFRP4 | 53 | 0.45 | 0.07 |
| Fzd3 | sFRP2 | 53 | 0.3 | 0.24 |
| Fzd3 | Dvl2 | 77* | 0.6 | 0.01 |
| Lrp5 | sFRP4 | 71* | 0.49 | 0.04 |
| sFRP1 | sFRP4 | 59 | 0.49 | 0.04 |
| sFRP1 | Wnt5a | 59 | 0.67 | 0 |
| sFRP2 | Wnt5a | 59 | 0.69 | 0 |
| sFRP2 | Dvl2 | 53 | 0.05 | 0.86 |
| sFRP2 | Fzd6 | 53 | 0.31 | 0.22 |
| sFRP2 | Fzd2 | 53 | 0.46 | 0.07 |
| sFRP3 | Wnt11 | 59 | 0.28 | 0.28 |
| sFRP4 | Wnt5a | 59 | 0.78 | 0 |
| Wnt5a | Fzd6 | 53 | 0.55 | 0.02 |
| Wnt5a | Fzd2 | 65* | 0.7 | 0 |
| Wnt5a | Wnt11 | 53 | 0.48 | 0.05 |
| Fzd6 | Fzd2 | 53 | 0.48 | 0.05 |
| Fzd2 | Wnt11 | 53 | 0.43 | 0.08 |
denote gene pairs that are over 65% similar in the 17 samples
Abbrevations: R:Spearman correlation coefficient; pval:p-value of spearman correlation coefficient.
The first pair of genes showing high concordance is WNT5A and FZD2 (65%) with a correlation coefficient of 0.7 ( p < 0.01). FZD2 and WNT5A are coordinately increased in 5 samples and decreased in 4 samples. The relationship between WNT5A and FZD2 is novel in human lung but their association has been documented in other animal models. For example, previous studies in zebrafish models suggest that Fzd2 induces intra-cellular release of Ca2+ via Wnt5a activation. The release of Ca2+ involves the activation of the phosphatidylinositol pathway in a G-protein-dependent manner (Kuhl et al. 2000; Sheldahl et al. 1999; Slusarski, Corces and Moon, 1997) which in turn activates CamKII and PKC. The implications of PKCs have been reported in various types of cancer. For example, human small cell lung cancer (SCLC) cells have shown to exhibit rapid growth due to over-expression of PKCɛ and similarly, breast cancer cells displayed an enhanced rate of proliferation due to PKCα transfection (Hofmann, 2004).
The next pair, the non-canonical components, FZD3 and DVL2 are similar in 77% of the 17 tumor samples with a corresponding correlation coefficient of 0.6 ( p ≤ 0.01). We discovered that the expression levels of both FZD3 and DVL2 are up-regulated in 7 out of 17 tumor samples and unchanged in 6 tumor samples where the expression of CTNNB1 is down or unchanged. FZD3 and DVL2 have independently been reported to be involved in the non-canonical pathway. The patterns of expression of FZD3 and DVL2 do not seem to affect the expression levels of CTNNB1. Although the Dvl family has been shown to be able to activate the canonical and non-canonical pathway, DVL2 alone does not display a high frequency of coordinate expression change with CTNNB1 in this study. Likewise, FZD3 alone does not seem to affect the expression of CTNNB1 as well, which agrees with the majority of studies done on this gene. Quantitative RT-PCR was performed on FZD3 and DVL2 on an independent set of 10 lung SCC samples and the results confirmed that FZD3 is up-regulated in 7 out of 10 samples as shown in Figure 5. However, DVL2 is only up-regulated in 3 out of 10 samples. When we applied the same concordance analysis onto these 10 samples, 9 samples showed reduced or unchanged expression of CTNNB1. Nearly half of these samples show that FZD3 and DVL2 have the same pattern of expression. FZD3 and DVL2 are increased in 67% and 33% of the samples, respectively. These results are consistent to what was observed in the first panel of lung tumors of 58% and 41%, respectively. Limited knowledge exists of the involvement of FZD3 and DVL2 in cancer. FZD3 is reported to be down-regulated in ovarian cancer (Tapper et al. 2001) but up-regulated in chronic lymphocytic leukemia (Lu et al. 2004). Although DVL2 has never been directly linked to cancer, its associations with Rho GTPases have been reported. Rho family of proteins are involved in a number of essential cellular processes such as cell growth, lipid metabolism, cytoskeleton architecture, membrane trafficking, transcriptional regulation, and apoptosis (Aznar and Lacal, 2001), with many of those processes disrupted in cancer.
Lastly, the LRP5 (of the canonical pathway) and SFRP4 pair is concordant in 71% of the samples with a corresponding correlation coefficient of 0.49 (p = 0.04). Interestingly, relationships between LRPs and sFRPs have not been previously reported. A total of 6 out of the 17 samples show coordinate down-regulation of LRP5 and SFRP4 in lung tumors. LRP5 is a single transmembrane co-receptor that forms an active complex with the Fzd protein and an incoming Wnt ligand, to activate the canonical Wnt signaling pathway. As for SFRP4, although this protein exhibits the same domain architecture as other sFRP family members, its expression behaviour is different from its other family members. In contrast to the other sFRP members, SFRP4 has been shown to be up-regulated where there is positive expression of CTNNB1 (Feng Han et al. 2006) in a study involving human colorectal carcinoma. In vitro studies have also shown that overexpression of SFRP4 does not lead to reduced expression of CTNNB1 (Suzuki et al. 2004). Although the mechanisms behind the activation of the canonical pathway by sFRP4 in these studies still needs more investigation, past and present evidence suggests that the sFRP genes may have more complex roles in addition to their pre-defined roles as Wnt antagonists.
Conclusions
Based on the results in this study, the non-canonical pathway is active in normal lung. Activation of the non-canonical pathway in development has been associated with the control of specific morphogenetic movements during and following vertebrate gastrulation. This is one of the first reports to show activity of the non-canonical pathway in the human adult lung at the gene expression level. Previous studies of lung tumors have mainly focused on the canonical components. However, tumor gene expression analysis in this study shows that in fact, the non-canonical pathway may provide an alternative explanation to the proliferation of lung cancer cells. Further investigation at the protein level and phosphorylation state of CTNNB1 will provide a more comprehensive understanding of the biological impact of changes in the non-canonical components. We suggest that the non-canonical pathway may have a more prominent role in lung cancer than previously reported and future studies of the WNT pathway should encompass both the canonical and the non-canonical branches.
Supplement Material
Table S1.
Primer sequences and conditions for RT-PCR analysis.
| Gene name | Primer sequence | MgCl2 (mM) | Cycles | Tm (°C) |
|---|---|---|---|---|
| DVL2 | 5′-aatcccagcgagttctttgt-3′ 5′-caatctcctgtatggcagca-3′ |
1 | 35 | 58.3 |
| FZD1 | 5′-tacacgaggctcaccaacag-3′ 5′-gagcctgcgaaagagagttg-3′ |
1 | 35 | 52.3 |
| FZD2 | 5′-catcgaggccaactctcagt-3′ 5′-gtgccgatgaacaggtacac-3′ |
1.5 | 35 | 52 |
| FZD3 | 5′-tgagtgttcgaagctcatgg-3′ 5′-ttaactctcggggacaccaa-3′ |
1.5 | 30 | 60.9 |
| FZD6 | 5′-caggcaggcagtgtatctga-3′ 5′-accacctccctgctcttttc-3′ |
2 | 30 | 58 |
| LRP5 | 5′-cccgtcacaggtacatgtact-3′ 5′-gaacgagccgtccaggtt-3′ |
1 | 30 | 55 |
| LRP6 | 5′-ttccaggaatgtctcgaggt-3′ 5′-ggttcaaaattgcagggaag-3′ |
1 | 35 | 51 |
| SFRP1 | 5′-gagctccagtttgcatttgg-3′ 5′-tagggtgctctcctcaaaca-3′ |
1 | 35 | 58 |
| SFRP2 | 5′-gacctgaagaaatcggtgct-3′ 5′-atgcgcttgaactctctctg-3′ |
1 | 35 | 60 |
| SFRP3 | 5′-tgttaccagagcctctttgc-3′ 5′-gagaatgcccaaaaggcata-3′ |
2 | 35 | 64 |
| SFRP4 | 5′-gtttccaaagcggagacttc-3′ 5′-atggcttgtgatggcttaca-3′ |
2 | 35 | 62.1 |
| SFRP5 | 5′-actggagggtgttttcacga-3′ 5′-ctcccctgcctactttctga-3′ |
2 | 35 | 63.4 |
| WNT1 | 5′-acagagccacgagtttggat-3′ 5′-gaggcaaacgcatctttgag-3′ |
1 | 35 | 55 |
| WNT3A | 5′-agagctgctggtctcatttg-3′ 5′-aggaaagcggaccatttctc-3′ |
2 | 35 | 58 |
| WNT5A | 5′-tggaccatgtgtggtgtctc-3′ 5′-gtgcagcactgtccagattt-3′ |
2 | 35 | 60.9 |
| WNT11 | 5′-gaagccaccaggaacagaag-3′ 5′-gccctgaaaggtcaagtctg-3′ |
2 | 31 | 64 |
| CADH | 5′-agccatgggcccttggag-3′ 5′-ccagaggctctgtgcaccttc-3′ |
1 | 40 | 50 |
| VIM | 5′-tggcacgtcttgaccttgaa-3′ 5′-ggtcatcgtgatgctgagaa-3′ |
1 | 35 | 55 |
| CTNNB1 | 5′-gagcctgccatctgtgctct-3′ 5′-acgcaaaggtgcatgatttg-3′ |
1 | 35 | 60 |
Pairwise expression profile analysis (tumor versus matched normal) of non-canonical and canonical Wnt pathway components in 20 SCC samples. Each tumor and normal pair is represented as an individual case, numbered from Case 1 to Case 20. For each gene, color gradient shading represents magnitude of over and underexpression.
Acknowledgements
The authors thank Timon P. H. Buys, Bradley P. Coe, Jonathan J. Davies, William W. Lockwood, and Teresa Mastracci for critical discussion. This work was supported by funds from Genome Canada/ British Columbia, Canadian Institutes of Health Research, and NIDCR grant R01 DE15965-01. RC is supported by scholarships from the Canadian Institutes of Health Research and Michael Smith Foundation for Health Research.
Footnotes
Competing Interests
The authors declare that they have no competing interests.
Authors’ Contributions
EHLL and RC designed and performed experiments and wrote manuscript.
AL performed experiments.
RTN and CM performed statistical analysis. JY and KGE isolated specimens.
JE performed pathology review.
SL and WLL are principle investigators of this project.
References
- Aust DE, Terdiman JP, Willenbucher RF, Chang CG, Molinaro-Clark A, Baretton GB, Loehrs U, Waldman FM. The APC/ beta-catenin pathway in ulcerative colitis-related colorectal carcinomas: a mutational analysis. Cancer. 2002;94(5):1421–7. doi: 10.1002/cncr.10334. [DOI] [PubMed] [Google Scholar]
- Aznar S, Lacal JC. Rho signals to cell growth and apoptosis. Cancer Lett. 2001;165(1):1–10. doi: 10.1016/s0304-3835(01)00412-8. [DOI] [PubMed] [Google Scholar]
- Blache P, van de Wetering M, Duluc I, Domon C, Berta P, Freund JN, Clevers H, Jay P. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J. Cell. Biol. 2004;166(1):37–47. doi: 10.1083/jcb.200311021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-Catenin Signaling in Development and Disease. Cell. 2006;127(3):469–80. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Eberhart CG, Argani P. Wnt signaling in human development: beta-catenin nuclear translocation in fetal lung, kidney, placenta, capillaries, adrenal, and cartilage. Pediatr Dev. Pathol. 2001;4(4):351–7. doi: 10.1007/s10024001-0037-y. [DOI] [PubMed] [Google Scholar]
- Feng Han Q, Zhao W, Bentel J, Shearwood AM, Zeps N, Joseph D, Iacopetta B, Dharmarajan A. Expression of sFRP-4 and beta-catenin in human colorectal carcinoma. Cancer Lett. 2006;231(1):129–37. doi: 10.1016/j.canlet.2005.01.026. [DOI] [PubMed] [Google Scholar]
- Habas R, Kato Y, He X. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell. 2001;107(7):843–54. doi: 10.1016/s0092-8674(01)00614-6. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- He B, You L, Uematsu K, Xu Z, Lee AY, Matsangou M, McCormick F, Jablons DM. A monoclonal antibody against Wnt-1 induces apoptosis in human cancer cells. Neoplasia. 2004;6(1):7–14. doi: 10.1016/s1476-5586(04)80048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281(5382):1509–12. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- Hofmann J. Protein kinase C isozymes as potential targets for anti-cancer therapy. Curr. Cancer Drug Targets. 2004;4(2):125–46. doi: 10.2174/1568009043481579. [DOI] [PubMed] [Google Scholar]
- Ilyas M. Wnt signalling and the mechanistic basis of tumour development. J. Pathol. 2005;205(2):130–44. doi: 10.1002/path.1692. [DOI] [PubMed] [Google Scholar]
- Katoh M. WNT/PCP signaling pathway and human cancer (review) Oncol. Rep. 2005;14(6):1583–8. [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275(5307):1784–7. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Kuhl M, Sheldahl LC, Malbon CC, Moon RT. Ca(2+)/ calmodulin-dependent protein kinase II is stimulated by Wnt and Frizzled homologs and promotes ventral cell fates in Xenopus. J. Biol. Chem. 2000;275(17):12701–11. doi: 10.1074/jbc.275.17.12701. [DOI] [PubMed] [Google Scholar]
- Liu W, Saint DA. A new quantitative method of real time reverse transcription polymerase chain reaction assay based on simulation of polymerase chain reaction kinetics. Anal. Biochem. 2002;302(1):52–9. doi: 10.1006/abio.2001.5530. [DOI] [PubMed] [Google Scholar]
- Lu D, Zhao Y, Tawatao R, Cottam HB, Sen M, Leoni LM, Kipps TJ, Corr M, Carson DA. Activation of the Wnt signaling pathway in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 2004;101(9):3118–23. doi: 10.1073/pnas.0308648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann B, Gelos M, Siedow A, Hanski ML, Gratchev A, Ilyas M, Bodmer WF, Moyer MP, Riecken EO, Buhr HJ, Hanski C. Target genes of beta-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc. Natl. Acad. Sci. U.S.A. 1999;96(4):1603–8. doi: 10.1073/pnas.96.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazieres J, He B, You L, Xu Z, Jablons DM. Wnt signaling in lung cancer. Cancer Lett. 2005;222(1):1–10. doi: 10.1016/j.canlet.2004.08.040. [DOI] [PubMed] [Google Scholar]
- Melkonyan HS, Chang WC, Shapiro JP, Mahadevappa M, Fitzpatrick PA, Kiefer MC, Tomei LD, Umansky SR. SARPs: a family of secreted apoptosis-related proteins. Proc. Natl. Acad. Sci. U.S.A. 1997;94(25):13636–41. doi: 10.1073/pnas.94.25.13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minna JD, Roth JA, Gazdar AF. Focus on lung cancer. Cancer Cell. 2002;1(1):49–52. doi: 10.1016/s1535-6108(02)00027-2. [DOI] [PubMed] [Google Scholar]
- Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275(5307):1787–90. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Ohira T, Gemmill RM, Ferguson K, Kusy S, Roche J, Brambilla E, Zeng C, Baron A, Bemis L, Erickson P, Wilder E, Rustgi A, Kitajewski J, Gabrielson E, Bremnes R, Franklin W, Drabkin HA. WNT7a induces E-cadherin in lung cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2003;100(18):10429–34. doi: 10.1073/pnas.1734137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongracz JE, Stockley RA. Wnt signalling in lung development and diseases. Respir Res. 2006;7(15) doi: 10.1186/1465-9921-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann H, Holtz J, Zerkowski HR, Hatzfeld M. Expression of secreted frizzled related proteins 3 and 4 in human ventricular myocardium correlates with apoptosis related gene expression. Cardiovasc. Res. 2000;45(3):720–8. doi: 10.1016/s0008-6363(99)00376-4. [DOI] [PubMed] [Google Scholar]
- Sheldahl LC, Park M, Malbon CC, Moon RT. Protein kinase C is differentially stimulated by Wnt and Frizzled homologs in a G-protein-dependent manner. Curr. Biol. 1999;9(13):695–8. doi: 10.1016/s0960-9822(99)80310-8. [DOI] [PubMed] [Google Scholar]
- Shigemitsu K, Sekido Y, Usami N, Mori S, Sato M, Horio Y, Hasegawa Y, Bader SA, Gazdar AF, Minna JD, Hida T, Yoshioka H, Imaizumi M, Ueda Y, Takahashi M, Shimokata K. Genetic alteration of the beta-catenin gene (CTNNB.1) in human lung cancer and malignant mesothelioma and identification of a new 3p21.3 homozygous deletion. Oncogene. 2001;20(31):4249–57. doi: 10.1038/sj.onc.1204557. [DOI] [PubMed] [Google Scholar]
- Slusarski DC, Corces VG, Moon RT. Interaction of Wnt and a Frizzled homologue triggers G-protein-linked phosphatidylinositol signalling. Nature. 1997;390(6658):410–3. doi: 10.1038/37138. [DOI] [PubMed] [Google Scholar]
- Steel MD, Puddicombe SM, Hamilton LM, Powell RM, Holloway JW, Holgate ST, Davies DE, Collins JE. Beta-catenin/T-cell factor-mediated transcription is modulated by cell density in human bronchial epithelial cells. Int. J. Biochem. Cell. Biol. 2005;37(6):1281–95. doi: 10.1016/j.biocel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, Toyota M, Tokino T, Hinoda Y, Imai K, Herman JG, Baylin SB. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat. Genet. 2004;36(4):417–22. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- Taniguchi K, Roberts LR, Aderca IN, Dong X, Qian C, Murphy LM, Nagorney DM, Burgart LJ, Roche PC, Smith DI, Ross JA, Liu W. Mutational spectrum of beta-catenin, AXIN1, and AXIN2 in hepatocellular carcinomas and hepatoblastomas. Oncogene. 2002;21(31):4863–71. doi: 10.1038/sj.onc.1205591. [DOI] [PubMed] [Google Scholar]
- Tapper J, Kettunen E, El-Rifai W, Seppala M, Andersson LC, Knuutila S. Changes in gene expression during progression of ovarian carcinoma. Cancer Genet. Cytogenet. 2001;128(1):1–6. doi: 10.1016/s0165-4608(01)00386-7. [DOI] [PubMed] [Google Scholar]
- Torres MA, Yang-Snyder JA, Purcell SM, DeMarais AA, McGrew LL, Moon RT. Activities of the Wnt-1 class of secreted signaling factors are antagonized by the Wnt-5A class and by a dominant negative cadherin in early Xenopus development. J Cell Biol. 1996;133:1123–37. doi: 10.1083/jcb.133.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M, Gemmill RM, West J, Winn R, Sugita M, Tanaka N, Ueki M, Drabkin HA. Mutations of the beta- and gamma-catenin genes are uncommon in human lung, breast, kidney, cervical and ovarian carcinomas. Br. J. Cancer. 2001;85(1):64–8. doi: 10.1054/bjoc.2001.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu K, He B, You L, Xu Z, McCormick F, Jablons DM. Activation of the Wnt pathway in non small cell lung cancer: evidence of dishevelled overexpression. Oncogene. 2003;22(46):7218–21. doi: 10.1038/sj.onc.1206817. [DOI] [PubMed] [Google Scholar]
- Veeman MT, Axelrod JD, Moon RT. A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev. Cell. 2003;5(3):367–77. doi: 10.1016/s1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- Weksberg R, Hughes S, Moldovan L, Bassett AS, Chow EW, Squire JA. A method for accurate detection of genomic microdeletions using real-time quantitative PCR. BMC Genomics. 2005;6(180) doi: 10.1186/1471-2164-6-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn RA, Marek L, Han SY, Rodriguez K, Rodriguez N, Hammond M, Van Scoyk M, Acosta H, Mirus J, Barry N, Bren-Mattison Y, Van Raay TJ, Nemenoff RA, Heasley LE. Restoration of Wnt-7a expression reverses non-small cell lung cancer cellular transformation through frizzled-9-mediated growth inhibition and promotion of cell differentiation. J Biol Chem. 2005;280:19625–34. doi: 10.1074/jbc.M409392200. [DOI] [PubMed] [Google Scholar]
- You L, He B, Xu Z, Uematsu K, Mazieres J, Mikami I, Reguart N, Moody TW, Kitajewski J, McCormick F, Jablons DM. Inhibition of Wnt-2-mediated signaling induces programmed cell death in non-small-cell lung cancer cells. Oncogene. 2004;23(36):6170–4. doi: 10.1038/sj.onc.1207844. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Primer sequences and conditions for RT-PCR analysis.
| Gene name | Primer sequence | MgCl2 (mM) | Cycles | Tm (°C) |
|---|---|---|---|---|
| DVL2 | 5′-aatcccagcgagttctttgt-3′ 5′-caatctcctgtatggcagca-3′ |
1 | 35 | 58.3 |
| FZD1 | 5′-tacacgaggctcaccaacag-3′ 5′-gagcctgcgaaagagagttg-3′ |
1 | 35 | 52.3 |
| FZD2 | 5′-catcgaggccaactctcagt-3′ 5′-gtgccgatgaacaggtacac-3′ |
1.5 | 35 | 52 |
| FZD3 | 5′-tgagtgttcgaagctcatgg-3′ 5′-ttaactctcggggacaccaa-3′ |
1.5 | 30 | 60.9 |
| FZD6 | 5′-caggcaggcagtgtatctga-3′ 5′-accacctccctgctcttttc-3′ |
2 | 30 | 58 |
| LRP5 | 5′-cccgtcacaggtacatgtact-3′ 5′-gaacgagccgtccaggtt-3′ |
1 | 30 | 55 |
| LRP6 | 5′-ttccaggaatgtctcgaggt-3′ 5′-ggttcaaaattgcagggaag-3′ |
1 | 35 | 51 |
| SFRP1 | 5′-gagctccagtttgcatttgg-3′ 5′-tagggtgctctcctcaaaca-3′ |
1 | 35 | 58 |
| SFRP2 | 5′-gacctgaagaaatcggtgct-3′ 5′-atgcgcttgaactctctctg-3′ |
1 | 35 | 60 |
| SFRP3 | 5′-tgttaccagagcctctttgc-3′ 5′-gagaatgcccaaaaggcata-3′ |
2 | 35 | 64 |
| SFRP4 | 5′-gtttccaaagcggagacttc-3′ 5′-atggcttgtgatggcttaca-3′ |
2 | 35 | 62.1 |
| SFRP5 | 5′-actggagggtgttttcacga-3′ 5′-ctcccctgcctactttctga-3′ |
2 | 35 | 63.4 |
| WNT1 | 5′-acagagccacgagtttggat-3′ 5′-gaggcaaacgcatctttgag-3′ |
1 | 35 | 55 |
| WNT3A | 5′-agagctgctggtctcatttg-3′ 5′-aggaaagcggaccatttctc-3′ |
2 | 35 | 58 |
| WNT5A | 5′-tggaccatgtgtggtgtctc-3′ 5′-gtgcagcactgtccagattt-3′ |
2 | 35 | 60.9 |
| WNT11 | 5′-gaagccaccaggaacagaag-3′ 5′-gccctgaaaggtcaagtctg-3′ |
2 | 31 | 64 |
| CADH | 5′-agccatgggcccttggag-3′ 5′-ccagaggctctgtgcaccttc-3′ |
1 | 40 | 50 |
| VIM | 5′-tggcacgtcttgaccttgaa-3′ 5′-ggtcatcgtgatgctgagaa-3′ |
1 | 35 | 55 |
| CTNNB1 | 5′-gagcctgccatctgtgctct-3′ 5′-acgcaaaggtgcatgatttg-3′ |
1 | 35 | 60 |
Pairwise expression profile analysis (tumor versus matched normal) of non-canonical and canonical Wnt pathway components in 20 SCC samples. Each tumor and normal pair is represented as an individual case, numbered from Case 1 to Case 20. For each gene, color gradient shading represents magnitude of over and underexpression.