Abstract
More than half a century after the discovery of the molecular basis of Sickle Cell Disease (SCD), the causes of the phenotypic heterogeneity of the disease remain unclear. This heterogeneity manifests with different clinical outcomes such as stroke, vaso-occlusive episodes, acute chest syndrome, avascular necrosis, leg ulcers, priapism and retinopathy. These outcomes cannot be explained by the single mutation in the beta-globin gene alone but may be attributed to genetic modifiers and environmental effects. Recent advances in the post human genome sequence era have opened the door for the identification of novel genetic modifiers in SCD. Studies are showing that phenotypes of SCD seem to be modulated by polymorphisms in genes that are involved in inflammation, cell–cell interaction and modulators of oxidant injury and nitric oxide biology. The discovery of genes implicated in different phenotypes will help understanding of the physiopathology of the disease and aid in establishing targeted cures. However, caution is needed in asserting that genetic modifiers are the cause of all SCD phenotypes, because there are other factors such as genetic background of the population, environmental components, socio-economics and psychology that can play significant roles in the clinical heterogeneity.
Keywords: drepanocytose, genomics, hemoglobin, HbS, hemoglobinopathies, RBC
Introduction
Hemoglobinopathies are the world’s most widespread genetic diseases.1 The high frequency of hemoglobinopathies globally is of great concern to public health managers of various nations.2 Among the hemoglobinopathies, sickle cell disease is a genetic disorder in which the beta-chain of the human hemoglobin (Hb) gene is mutated, leading to an abnormal Hb. This mutation causes red blood cells (RBCs) to acquire a sickle shape under conditions of hypoxia, resulting in a very large range of phenotypes such as anemia, cell adhesion, vaso-occlusion, severe pain, stroke and organ failure. For the past several years, genetic studies and single-nucleotide polymorphisms (SNPs) genotyping on patients with various phenotypes showed involvements of SNPs of different genes.3,4 To date, over 100 SNPs that might be associated with specific phenotypes have been shown to be significantly involved in different genes that act as inflammatory mediators, modulators of oxidant injury, Nitric Oxide (NO) biology, vaso-regulatory molecules and cell adhesion factors. These studies show that SNPs in genes implicated in the transforming growth factor-beta/bone morphogenetic protein (TGF-beta/BMP) pathways and a few other genes such as Klotho gene (KL) are associated with several phenotypes of SCD.
Experiments showed that decreased NO bioavailability, due to scavenging of NO by cell free hemoglobin, a product of the hemolytic process associated with SCD, played a significant role in the vascular patho-biology of SCD.5 Thus, the emerging views of the pathogenesis of many complications of SCD involve complex interactions between sickle reticulocytes, neutrophils, monocytes, and the endothelium. It therefore follows that many factors that are also important in the pathways leading to inflammation, cellular adhesion, NO metabolism, vascular reactivity and coagulation are important in the patho-physiology of the disease and that variations in the expression of molecules in these pathways contribute to the heterogeneity of SCD.
The elucidation of genetic variants and mechanisms responsible for the phenotypic variability in SCD will have important clinical implications for genetic counseling and clinical management. Targeted follow up of patients and utilization of individual therapeutic strategies and preventive cures will produce better outcomes for patients with SCD.
In this review, we summarize some of the recent genomic findings focused on non globin genetic modifiers, shown to be potentially implicated in different clinical outcomes of SCD.
The beta-globin gene and the origin of sickle cell hemoglobin (HbS)
The main Hb function is to transport oxygen to tissues. Adult Hb is composed of a major HbA (alpha2beta2) and a minor HbA2 (alpha2delta2), with traces of fetal HbF (alpha2gamma2). Each of the different globin chains is controlled by distinct genes; two genes exist for the alpha and gamma chains and one for each of the other chains.2,6 HbS is caused by a single amino acid substitution of Glutamic Acid replaced by Valine at the sixth position of the beta-globin chain. This is due to a single nucleotide substitution, GAG —> GTG in codon 6 of the beta-globin gene on chromosome 11p15.5.7 This mutation alters the stability of the Hb and leads to the clinical disorder. The homozygous state of the sickle cell gene mutation (HbSS) results in sickle cell anemia (SCA). The replacement of the Glutamic Acid by a Valine causes Hb polymerization induced by deoxygenation.8 As the deoxy-HbS polymerizes and fibers align, the erythrocyte is transformed into a “sickle” shape. These deformed and rigid erythrocytes can obstruct normal blood flow in micro-circulation and thereby induce ischemia in tissues distal to the vascular blockage, which is the basis for many SCD complications.8,9 Thus, sickling is the direct consequence of the presence of HbS, but pleiotropic effects that follow depict the clinical outcome of the disease.
Since the clinical phenotypes of homozygous HbS are extremely variable, it is now clear that even if SCA is a monogenic disorder, at the phenotypic level it is most likely to be a multigenic disease. Among risk factors for early mortality are stroke, painful crisis and infection.10 There are many factors that might influence the outcome of these clinical presentations. These factors include, but are not limited to, environmental, psychological, cultural, and the socio-economical. However, the epigenetic mechanisms seem to play a primordial role in determining different SCD phenotypes.
Modulation of SCD by other disorders
Genetics is a major determinant of childhood survival in malaria endemic countries. The past few years have seen significant progress towards showing some of the best-known malaria resistance genes that determine the structure or function of RBCs, such as Gerbich blood group antigen negativity, polymorphisms of the complement receptor genes (most notably CR1), Southeast Asian ovalocytosis, pyruvate kinase deficiency; HbE, the sickle cell trait (SCT) and alpha-thalassemia.11 Despite conclusive evidence that these defects of the Hb molecule protect against severe and fatal Plasmodium falciparum malaria,12 the mechanisms underlying this protection are poorly understood.
Alpha-thalassemia is due to mutations of the alpha-globin genes (chromosome 16pter-p13.3) and it has been shown that the presence of alpha-thalassemia has a protective role against Plasmodium falciparum malaria infection. This could explain its high gene frequency in geographic malaria-endemic regions. However, several patients with SCA have coincidental alpha-thalassemia and the presence of both SCA and alpha-thalassemia mutations seems to act as a negative epistatic factor.13
Alpha-thalassemia reduces the concentration of HbS and therefore of HbS polymerization. Thus it is expected that this will prevent vaso-occlusive events that are consequences of hemolysis, including stroke, leg ulcer, priapism, and pulmonary hypertension. Complications more dependent on blood viscosity, such as painful episodes, acute chest syndrome (ACS) and avascular necrosis will usually be more prevalent when alpha-thalassemia coexists with SCD mutation.14,15 This is explained by patients with homozygous alpha-thalassemia and SCD having slightly lower levels of HbF than the non-thalassemic sickle cell patients. Preferential survival of F cells, a subpopulation of erythrocytes, occurs in SCA, with or without alpha-thalassemia, and the slight difference in HbF levels appears to reflect differences in numbers of circulating F cells. Thus, the change in the erythrocyte density profile in SCD with coexisting alpha-thalassemia, could explain the change in blood viscosity and the hematological improvement.16
Glucose-6-phosphate dehydrogenase (G6PD) deficiency (Chromosome Xq28) is commonly found in HbS populations. Although this deficiency does not appear to have a direct effect on the SCD phenotype,17 there are case reports of more severe hemolysis in patients with SCD and G6PD deficiency.18 Similarly, coinheritance of SCD and pyruvate kinase (Chromosome 1q21) deficiency can cause painful crisis,19 and co-inheritance of sherocytosis may cause recurrent acute splenic sequestration crisis.20,21 All these examples highlight the complexity of gene interactions.
Phenotype outcomes and potential modifier gene polymorphisms
The consequences of the sickle mutation and its downstream effects are clearly variable. Complications due to chronic hemolytic anemia, episodic vaso-occlusion with resultant painful episodes and chronic organ damage lead to very variable phenotypes of SCD. It is very difficult to determine the exact factors mediating the severity of the disease. It seems at least that all hematologists agree that they can definitely define the very mild or the “asymptomatic” patients as an obvious phenotype.22–24 Different authors have reported SCD as an inflammatory disease with endothelium involvement.25 Other studies have implicated the NO bio-availability, associated with the scavenging of NO by cell free Hb (product of hemolysis), in the vascular patho-biology of SCD.5
It is critical to carefully characterize phenotypes in order to study complex gene interactions. Studies of sickle cell patients from different populations will very likely yield important information due to the differences in genetic backgrounds of these populations and potential implications on the disease phenotype. In the past few years, many centers have focused on the study of genetic modifiers of SCD. Selected findings are summarized here. Table 1 reviews a list of SNPs reported to be significantly associated with different phenotypes of SCD.
Table 1.
Review of polymorphisms reported to date to be significantly associated with different SCD phenotypes. (*) = protective.
| Phenotype | Gene | Chromosome | GeneID | SNP or allele | p value | Population | Ref | |
|---|---|---|---|---|---|---|---|---|
| HbF and HbF response to hydroxyurea | HBG2 | 11p15.5 | 3048 | −158 C->T (XmnI) | N/S | West African | 141 | |
| HBB | 11p15.5 | 3043 | rs7482144 (XmnI) | 4 × 10−7 | CSSCD | 142 | ||
| GPM6B | Xp22.2 | 2824 | rs1005589 | 0.047 | CSSCD, MSH | 143 | ||
| rs11095629 | 0.042 | CSSCD, MSH | 143 | |||||
| rs5978663 | 0.064 | CSSCD | 143 | |||||
| rs11095629 | 0.069 | CSSCD | 143 | |||||
| rs7890737 | 0.07 | CSSCD | 143 | |||||
| rs4830513 | 0.068 | CSSCD | 143 | |||||
| rs5979998 | 0.069 | CSSCD | 143 | |||||
| rs6654096 | 0.067 | CSSCD | 143 | |||||
| TOX | 8q12.1 | 9760 | rs10504269 | 0.054 | CSSCD, MSH | 143 | ||
| rs6997859 | 0.052 | CSSCD | 143 | |||||
| rs12155519 | 0.056 | CSSCD, MSH | 143 | |||||
| rs1947178 | 0.056 | CSSCD | 143 | |||||
| rs389349 | 0.039 | CSSCD, MSH | 143 | |||||
| rs4737532 | 0.058 | CSSCD | 143 | |||||
| rs851800 | 0.045 | CSSCD | 143 | |||||
| rs2726599 | 0.065 | CSSCD | 143 | |||||
| rs3109904 | 0.061 | CSSCD | 143 | |||||
| rs7821556 | 0.061 | CSSCD | 143 | |||||
| rs7817609 | 0.066 | CSSCD | 143 | |||||
| rs826730 | 0.064 | CSSCD | 143 | |||||
| rs3779999 | 0.063 | CSSCD | 143 | |||||
| rs1349115 | 0.066 | CSSCD | 143 | |||||
| rs2594953 | 0.065 | CSSCD | 143 | |||||
| rs10283344 | 0.063 | CSSCD | 143 | |||||
| rs12545204 | 0.066 | CSSCD | 143 | |||||
| rs380620 | 0.062 | CSSCD | 143 | |||||
| rs826729 | 0.045 | MSH | 48 | |||||
| rs765587 | 0.031 | MSH | 48 | |||||
| rs9693712 | 0.0098 | MSH | 48 | |||||
| rs172652 | 0.049 | MSH | 48 | |||||
| rs380620 | 0.016 | MSH | 48 | |||||
| rs2693430 | 0.049 | MSH | 48 | |||||
| rs12155519 | 0.037 | MSH | 48 | |||||
| HBE1 | 11p15.5 | 3046 | rs7130110 | 0.07 | CSSCD, MSH | 143 | ||
| rs3759070 | 0.060 | CSSCD | 143 | |||||
| HBG2 | 11p15.5 | 3048 | rs7482144 | 0.07 | CSSCD, MSH | 143 | ||
| AQP9 | 15q22.1–q22.2 | 366 | rs1867380 | 0.044 | CSSCD, MSH | 143 | ||
| MAP2K1 | 15q22.1–q22.33 | 5604 | rs4489951 | 0.051 | CSSCD | 143 | ||
| SMAD6 | 15q21–q22 | 4091 | rs1440372 | 0.042 | CSSCD | 143 | ||
| SMAD3 | 15q22.33 | 4088 | rs10518707 | 0.056 | CSSCD | 143 | ||
| rs8038623 | 0.043 | CSSCD | 143 | |||||
| KDR | 4q11–q12 | 3791 | rs6554233 | 0.07 | CSSCD | 143 | ||
| rs6828477 | 0.066 | CSSCD | 143 | |||||
| rs7654599 | 0.063 | CSSCD | 143 | |||||
| rs2305948 | 0.062 | CSSCD | 143 | |||||
| MAP3K7 | 6q16.1–q16.3 | 6885 | rs1145729 | 0.069 | CSSCD | 143 | ||
| rs157681 | 0.056 | CSSCD | 143 | |||||
| NOX3 | 6q25.1–q26 | 50508 | rs231944 | 0.068 | CSSCD | 143 | ||
| rs231945 | 0.066 | CSSCD | 143 | |||||
| rs9371889 | 0.067 | CSSCD | 143 | |||||
| rs6557420 | 0.066 | CSSCD | 143 | |||||
| NOS3 | 7q36 | 4846 | rs1008140 | 0.059 | CSSCD | 143 | ||
| rs743507 | 0.067 | CSSCD | 143 | |||||
| rs1808593 | 0.068 | CSSCD | 143 | |||||
| ASS | 9q34.1 | 445 | rs590086 | 0.066 | CSSCD | 143 | ||
| rs652313 | 0.067 | CSSCD | 143 | |||||
| rs12555797 | 0.069 | CSSCD | 143 | |||||
| rs543048 | 0.067 | CSSCD | 143 | |||||
| NOS1 | 12q24.2–q24.31 | 4842 | rs2682820 | 0.068 | CSSCD | 143 | ||
| rs3825102 | 0.067 | CSSCD | 143 | |||||
| rs1483757 | 0.066 | CSSCD | 143 | |||||
| rs816361 | 0.045 | CSSCD | 143 | |||||
| rs7977109 | 0.029 | MSH | 48 | |||||
| rs7309163 | 0.038 | MSH | 48 | |||||
| FLT1 | 13q12 | 2321 | rs7987291 | 0.064 | MSH | 48 | ||
| rs2387632 | 0.067 | CSSCD | 143 | |||||
| rs9513097 | 0.066 | CSSCD | 143 | |||||
| rs9508026 | 0.062 | CSSCD | 143 | |||||
| rs8002446 | 0.067 | CSSCD | 143 | |||||
| rs638889 | 0.065 | CSSCD | 143 | |||||
| rs2256849 | 0.066 | CSSCD | 143 | |||||
| rs670084 | 0.067 | CSSCD | 143 | |||||
| rs600640 | 0.065 | CSSCD | 143 | |||||
| rs9319428 | 0.047 | MSH | 48 | |||||
| rs2182008 | 0.003 | MSH | 48 | |||||
| rs8002446 | 0.011 | MSH | 48 | |||||
| rs3751395 | 0.039 | MSH | 48 | |||||
| rs9319428 | 0.047 | MSH | 48 | |||||
| rs2387634 | 0.037 | MSH | 48 | |||||
| ALOX5AP | 13q12 | 241 | rs4468448 | 0.064 | CSSCD | 143 | ||
| rs4769058 | 0.057 | CSSCD | 143 | |||||
| rs12019512 | 0.069 | CSSCD | 143 | |||||
| rs4445746 | 0.068 | CSSCD | 143 | |||||
| KL | 13q12 | 9365 | rs398655 | 0.062 | CSSCD | 143 | ||
| rs577912 | 0.068 | CSSCD | 143 | |||||
| rs7982726 | 0.069 | CSSCD | 143 | |||||
| rs685417 | 0.065 | CSSCD | 143 | |||||
| rs9527025 | 0.050 | CSSCD | 143 | |||||
| rs648202 | 0.067 | CSSCD | 143 | |||||
| PDE7B | 6q23-q24 | 27115 | rs2327669 | 0.041 | MSH | 48 | ||
| rs11154849 | 0.05 | MSH | 48 | |||||
| rs9376173 | 0.049 | MSH | 48 | |||||
| rs1480642 | 0.044 | MSH | 48 | |||||
| rs487278 | 0.017 | MSH | 48 | |||||
| HAO2 | 1p13.3–p13.1 | 51179 | rs10494225 | 0.0039 | MSH | 48 | ||
| MAP3K5 | 6q22.33 | 4217 | rs9376230 | 0.036 | MSH | 48 | ||
| rs9483947 | 0.034 | MSH | 48 | |||||
| rs2237262 | 0.03 | MSH | 29 | |||||
| ARG2 | 14q24.1–q24.3 | 384 | rs10483801 | 0.0075 | MSH | 48 | ||
| rs10483802 | 0.0038 | MSH | 48 | |||||
| NOS2A | 17q11.2–q12 | 4843 | rs1137933 | 0.031 | MSH | 48 | ||
| rs944725 | 0.02 | MSH | 48 | |||||
| HBS1L | 6q23–q24 | 10767 | rs52090901 | 1.10-5 | North European | 46 | ||
| rs9399137 | 1.10-45 | North European | 46 | |||||
| rs4895440 | 0.00031 | North European | 46 | |||||
| PDE7B | 6q23–q24 | 27115 | rs509342 | 0.03 | MSH | 29 | ||
| BTF | 6q22–q23 | 9774 | rs703193 | 0.05 | MSH | 29 | ||
| MAP7 | 6q23.3 | 9053 | rs2179288 | 0.003 | MSH | 29 | ||
| rs2076192 | 0.04 | MSH | 29 | |||||
| rs9977139 | 0.03 | MSH | 29 | |||||
| rs3778314 | 0.02 | MSH | 29 | |||||
| PEX7 | 6q21–q22.2 | 5191 | rs2012700 | 0.05 | MSH | 29 | ||
| rs3799476 | 0.01 | MSH | 29 | |||||
| rs1342645 | 0.01 | MSH | 29 | |||||
| Unknown | 6q22 | N/S | rs1342641 | 0.004 | MSH | 29 | ||
| Unknown | 6q22 | N/S | rs44450 | 0.01 | MSH | 29 | ||
| BCL11A | 2p16.1 | 53335 | rs11886868 | 10−20 | Sardinia | 144 | ||
| rs11886868 | 4.10−35 | CCSCD, Brazil | 142 | |||||
| rs4671393 | 2.10−42 | CCSCD, Brazil | 142 | |||||
| rs7557939 | 6.10−38 | CCSCD, Brazil | 142 | |||||
| HBS1L-MYB | 6q22-q24 | 10767–4602 | rs28384513 | 0.04 | CCSCD, Brazil | 142 | ||
| rs7776054 | 0.0009 | CCSCD, Brazil | 142 | |||||
| rs9399137, | 5. 10−11 | CCSCD, Brazil | 142 | |||||
| rs9389268 | 0.0002 | CCSCD, Brazil | 142 | |||||
| rs4895441 | 1.10−7 | CCSCD, Brazil | 142 | |||||
| rs6929404 | 0.0002 | North European | 46 | |||||
| Stroke | Large vessel | HLA-A (*) | 6p21.3 | 3105 | 0102 | 0.02 | CSSCD | 60 |
| 2612 | 0.007 | CSSCD | 60 | |||||
| 3301 | 0.04 | CSSCD | 60 | |||||
| IL-4R | 16p12.1–p11.2 | 3566 | rs1805015, S503P | 0.006 | CSSCD, STOP | 61, 63 | ||
| TNFa (*) | 6p21.3 | 7124 | rs1800629, −308G>A | 0.048 | CSSCD, STOP | 61, 63 | ||
| ADRB2 (*) | 5q31–q32 | 154 | rs1042714, Q27E | 0.033 | CSSCD | 145 | ||
| Stroke | Small vessel | HLA DPB1 (*) | 6p21 | 3115 | 0401 | 0.01 | CSSCD | 60 |
| 1701 | 0.02 | CSSCD | 60 | |||||
| VCAM1 | 1p32–p31 | 7412 | rs1041163, −1594T>C | 0.002 | CSSCD | 145 | ||
| LDLR | 19p13.3 | 3949 | rs5742911 | 0.002 | CSSCD | 145 | ||
| Stroke | Vessel size N/S | ADCY9 | 16p13.3 | 115 | N/S | N/S | N/S | 59 |
| rs437115 | a3 | CSSCD | 66 | |||||
| rs2238432 | a98 | CSSCD | 66 | |||||
| rs2238426 | a3381 | CSSCD | 66 | |||||
| rs2072338 | a638 | CSSCD | 66 | |||||
| rs2283497 | a10 | CSSCD | 66 | |||||
| ANXA2 | 15q22.2 | 302 | hCV26910500 | a1.68 108 | CSSCD | 66 | ||
| BMP6 | 6p24.3 | 654 | rs267196 | a2.31 1016 | CSSCD | 66 | ||
| rs267201 | a1.92 10103 | CSSCD | 66 | |||||
| rs408505 | a4.06 10101 | CSSCD | 66 | |||||
| rs449853 | a2.20 1057 | CSSCD | 66 | |||||
| CCL2 | 17q11.2–q12 | 6347 | rs4586 | a844 | CSSCD | 66 | ||
| CSF2 | 5q31.1 | 1437 | rs25882 | a1.19 10198 | CSSCD | 66 | ||
| ECE1 | 1p36.12 | 1889 | rs212528 | a1.55 104 | CSSCD | 66 | ||
| rs212531 | a2.34 1080 | CSSCD | 66 | |||||
| ERG | 21q22.3 | 2078 | rs989554 | a62 | CSSCD | 66 | ||
| MET | 1q31.1 | 4233 | rs38850 | a68 | CSSCD | 66 | ||
| rs38859 | a1.58 1039 | CSSCD | 66 | |||||
| SELP | 1q24.2 | 6403 | rs2420378 | a1.90 1010 | CSSCD | 66 | ||
| rs3917733 | a2.84 1093 | CSSCD | 66 | |||||
| rs3753306 | a2.32 1065 | CSSCD | 66 | |||||
| TEK | 9p21 | 7010 | rs489347 | a2 | CSSCD | 66 | ||
| TGFBR3 | 1p22.1 | 7049 | N/S | N/S | N/S | 59 | ||
| rs284875 | a443992 | CSSCD | 66 | |||||
| rs2148322 | a68988 | CSSCD | 66 | |||||
| rs2765888 | a41968 | CSSCD | 66 | |||||
| rs2007686 | a1739 | CSSCD | 66 | |||||
| HLA DRB1 | 6p21.3 | 3123 | 0301 | 0.007 | African American | 146 | ||
| (*) | 0302 | 0.007 | African American | 146 | ||||
| 1501 | 0.019 | African American | 146 | |||||
| HLA DQB1 | 6p21.3 | 3119 | 0201 | 0.033 | African American | 146 | ||
| (*) | 0602 | 0.011 | African American | 146 | ||||
| VCAM1 (*) | 1p32–p31 | 7412 | rs3783613, G1238C | 0.04 | Jamaican, STOP | 4, 64 | ||
| AGT | 1p42–q43 | 183 | GT repeat | 0.05 | African American | 147 | ||
| Avascular Necrosis | MTHFR | 1p36.3 | 4524 | C677T | 0.006 | N/S | 76 | |
| BMP6 | 6p24–p23 | 654 | rs270393 | 0.009 | CSSCD | 81 | ||
| rs267196 | 0.001 | CSSCD | 81 | |||||
| rs267201 | 0.008 | CSSCD | 81 | |||||
| rs449853 | 0.012 | CSSCD | 81 | |||||
| rs1225934 | 0.001 | CSSCD | 81 | |||||
| TGFBR2 | 3p22 | 7048 | rs1019856 | 0.023 | CSSCD | 81 | ||
| rs934328 | <0.001 | CSSCD | 81 | |||||
| TGFBR3 | 1p22.1 | 7049 | rs284157 | <0.001 | CSSCD | 81 | ||
| EDN1 | 6p24.1 | 1906 | rs5369 | 0.001 | CSSCD | 81 | ||
| hCV7464888 | 0.001 | CSSCD | 81 | |||||
| ERG | 21q22.3 | 2078 | rs979091 | 0.014 | CSSCD | 81 | ||
| rs2836430 | 0.005 | CSSCD | 81 | |||||
| KL | 13q12 | 9365 | rs480780 | 0.001 | CSSCD | 81 | ||
| rs211235 | 0.001 | CSSCD | 81 | |||||
| rs2149860 | 0.021 | CSSCD | 81 | |||||
| rs685417 | 0.002 | CSSCD | 81 | |||||
| rs516306 | 0.019 | CSSCD | 81 | |||||
| rs565587 | 0.001 | CSSCD | 81 | |||||
| rs211239 | 0.001 | CSSCD | 81 | |||||
| rs211234 | 0.001 | CSSCD | 81 | |||||
| rs2238166 | 0.046 | CSSCD | 81 | |||||
| rs499091 | 0.020 | CSSCD | 81 | |||||
| rs576404 | 0.010 | CSSCD | 81 | |||||
| ECE1 | 1p36.12 | 1889 | rs212527 | <0.001 | CSSCD | 81 | ||
| ANXA2 | 15q22.2 | 302 | rs7163836 | 0.010 | CSSCD | 81 | ||
| hCV11770326 | <0.001 | CSSCD | 81 | |||||
| rs7170178 | <0.001 | CSSCD | 81 | |||||
| rs1033028 | 0.007 | CSSCD | 81 | |||||
| hCV26910500 | <0.001 | CSSCD | 81 | |||||
| hCV1571628 | 0.034 | CSSCD | 81 | |||||
| STARD13 | 13q12–q13 | 90627 | rs538874 | 0.015 | CSSCD | 81 | ||
| rs475303 | 0.029 | CSSCD | 81 | |||||
| rs648464 | 0.001 | CSSCD | 81 | |||||
| APRIN | 13q12.3 | 23047 | hCV3118898 | 0.001 | CSSCD | 81 | ||
| hCV11710292 | 0.014 | CSSCD | 81 | |||||
| Acute Chest Syndrome | NOS3 | 7q36 | 4846 | T-786C | 0.0051 | African American | 82 | |
| 0.021 | Guadeloupe | 85 | ||||||
| NOS1 | 12q24.2–q24.31 | 4842 | (AAT)n intron 20 | 0.005 | African American | 84 | ||
| ET-1 | 6p24.1 | 1906 | T8002C | 0.039 | Guadeloupe | 85 | ||
| Priapism | KL | 13q12 | 9365 | rs2249358 | b2.6 | CSSCD | 87 | |
| rs211239 | b1.7 | CSSCD | 87 | |||||
| rs211234 | b2.3 | CSSCD | 87 | |||||
| TGFBR3 | 1p22.1 | 7049 | rs7526590 | 0.00058 | N/S | 88 | ||
| AQP1 | 7p14 | 358 | rs10244884 | 0.00068 | N/S | 88 | ||
| ITGAV | 2q31–q32 | 3685 | rs3768780 | 0.00090 | N/S | 88 | ||
| F13A1 | 6p25.3–p24.3 | 2162 | hcv1860621 | 0.00156 | N/S | 88 | ||
| Leg Ulcers | TGFBR3 | 1p22.1 | 7049 | rs2038931 | 0.0387 | CSSCD | 148 | |
| TGFBR2 | 3p22 | 7048 | rs1019856 | 0.0170 | CSSCD | 148 | ||
| BMP6 | 6p24–p23 | 654 | rs270393 | 0.0362 | CSSCD | 148 | ||
| TEK | 9p21 | 7010 | rs603085 | 0.0108 | CSSCD | 148 | ||
| rs671084 | 0.0251 | CSSCD | 148 | |||||
| KL | 13q12 | 9365 | rs685417 | 0.0186 | CSSCD | 148 | ||
| rs516306 | 0.0076 | CSSCD | 148 | |||||
| rs2149860 | 0.0480 | CSSCD | 148 | |||||
| APRIN | 13q12.3 | 23047 | hCV3118898 | 0.0153 | CSSCD | 148 | ||
| BMPR1B | 4q22–q24 | 658 | rs1560909 | 0.0263 | CSSCD | 148 | ||
| rs7661539 | 0.0389 | CSSCD | 148 | |||||
| MAP3K7 | 6q16.1–q16.3 | 6885 | rs157702 | 0.0080 | CSSCD | 148 | ||
| SMURF1 | 7q22.1 | 57154 | rs219825 | 0.0389 | CSSCD | 148 | ||
| SMAD9 | 13q12–q14 | 4093 | rs9576135 | 0.0167 | CSSCD | 148 | ||
| Unknown | 18q21.1 | N/S | rs736839 | 0.0004 | CSSCD | 148 | ||
| MAP2K1 | 15q22.1–q22.33 | 5604 | rs8036023 | 0.0414 | CSSCD | 148 | ||
| Gallstone | UGT1A | 2q37 | 7361 | Promoter (TA)n | 0.0015 | see text | 106 | |
| Infection and Bacteriamia | MPO | 17q23.1 | 4353 | G463A | 0.0112 | Brazilian | 111 | |
| IGF1R | 15q26.3 | 3480 | rs1319868 | 0.0059 | CSSCD | 108 | ||
| rs1567811 | 0.0586 | CSSCD | 108 | |||||
| rs8041224 | 0.0133 | CSSCD | 108 | |||||
| rs2872060 | 0.0321 | CSSCD | 108 | |||||
| BMP6 | 6p24–p23 | 654 | rs270387 | 0.0401 | CSSCD | 108 | ||
| rs267188 | 0.0047 | CSSCD | 108 | |||||
| rs408505 | 0.0006 | CSSCD | 108 | |||||
| rs449853 | 0.02 | CSSCD | 108 | |||||
| TGFBR3 | 1p22.1 | 7049 | rs2765888 | 0.0251 | CSSCD | 108 | ||
| BMPR1A | 10q22.3 | 657 | rs6586039 | 0.0248 | CSSCD | 108 | ||
| hCV1663921 | 0.0115 | CSSCD | 108 | |||||
| SMAD6 | 15q21–q22 | 4091 | rs5014202 | 0.0324 | CSSCD | 108 | ||
| SMAD3 | 15q22.33 | 4088 | rs10518707 | 0.0114 | CSSCD | 108 | ||
| Unknown | 1p22.1 | N/S | rs6662385 | 0.0222 | CSSCD | 108 | ||
| HLA-E | 6p21.3 | 3133 | 0101 | 0.003 | African | 110 | ||
| Pulmonary Hypertension | ACVRLI | 12q11–q14 | 94 | rs3847859 | 0.003 | African American | 115 | |
| rs706814 | 0.009 | African American | 115 | |||||
| ADCY6 | 12q12–q13 | 112 | rs9804777 | 0.017 | African American | 115 | ||
| ADRB1 | 10q24–q26 | 153 | rs1801253 | 0.006 | African American | 115 | ||
| BMP6 | 6p24–p23 | 654 | rs267192 | 0.014 | African American | 115 | ||
| rs267196 | 0.013 | African American | 115 | |||||
| rs267201 | 0.019 | African American | 115 | |||||
| BMPR2 | 2q33–q34 | 659 | rs17199249 | 0.018 | African American | 115 | ||
| rs35711585 | 0.025 | African American | 115 | |||||
| CR1 | 1q32 | 1378 | rs6663530 | 0.024 | African American | 115 | ||
| FY | 1q21–q22 | 2532 | rs3027045 | 0.035 | African American | 115 | ||
| KL | 13q12 | 9365 | rs1888057 | 0.031 | African American | 115 | ||
| LCAT | 16q22.1 | 3931 | rs5923 | 0.033 | African American | 115 | ||
| hcv2846928 | 0.014 | African American | 115 | |||||
| LTA4H | 12q22 | 4048 | rs10492226 | <0.001 | African American | 115 | ||
| rs1978331 | 0.022 | African American | 115 | |||||
| SELP | 1q22–q25 | 6403 | rs2235302 | 0.014 | African American | 115 | ||
| rs6131 | 0.019 | African American | 115 | |||||
| SERPINCI | 1q23–q25.1 | 462 | rs2227617 | 0.001 | African American | 115 | ||
| SLC12A6 | 15q13 | 9990 | rs426634 | 0.028 | African American | 115 | ||
| TGFBR3 | 1p22.1 | 7049 | rs10874940 | 0.002 | African American | 115 | ||
| rs17443164 | 0.045 | African American | 115 | |||||
| Glomerular | BMPR1B | 4q22–q24 | 658 | rs2240036 | 0.0434 | CSSCD | 94 | |
| Filtration | rs4145993 | 0.0352 | CSSCD | 94 | ||||
| Rate | rs17022863 | 0.011 | CSSCD | 94 | ||||
| rs1434549 | 0.0109 | CSSCD | 94 |
Abbrevations: CSSCD, Cooperative Study of Sickle Cell Disease; MSH, Multicenter Study of Hydroxyurea in SCA; STOP, Stroke Prevention Trial in Sickle Cell Anemia study.
Notes:
Bayes factor of the model associating the SNP to stroke versus the model of independence (large Bayes factor implies that there is very strong evidence for the associations).66
Odd ratio.87 In Bold is the highest significance SNP for each gene.
Fetal Hb and hydroxyurea response
HbF concentration is high at birth and declines slowly during the first three decades of life and the final level in adults is variable. It has been well established that in most cases, HbF has an ameliorative effect on SCD patients because the gamma-chains of HbF do not polymerize with HbS. This potential anti-sickling effect resulted in the development of a major therapeutic product for amelioration of SCD effects. To date, the only drug that is therapeutically used in this way is the S-phase specific chemotherapeutic agent, hydroxyurea (HU) which increases HbF production.26
Among SCA patients, HbF concentrations vary from 0.1% to 30% with an average of about 8%. Many laboratories have made efforts to elucidate the reasons behind differences in HbF levels between patients.27–29 Understanding the mechanisms controlling expression of HbF (gamma-globin gene) may help in the design of better therapeutic strategies in the cure of SCA severity. Unfortunately, some inconsistencies have been reported in patients with very severe disease that have an HbF level of nearly 20% and in some older SCA patients that often have very low levels of HbF.24,30,31 Moreover, the different effect of HbF on the clinical course and the differences in response to HU treatment among patients with similar HbF levels may be related to heterogeneity in the cellular distribution of HbF or to the effect of other modifying genes.29,32
Several genetic determinants have been shown to contribute to the heterogeneity of baseline HbF levels in SCD. Haplotypes of the beta-globin gene cluster are among the most extensively studied.33 High HbF levels are associated with Senegal and Asian-Indian haplotypes, which confer a general milder clinical and hematological phenotype compared with the other African haplotypes (Benin, Bantu and Cameroon). Individuals with Bantu haplotypes have the lowest HbF level and the most severe phenotype.34 Individuals with the Benin haplotype usually have intermediate features.34 The common feature of the Senegal and Asian-Indian haplotype is the presence of a C->T polymorphism in the promoter of the G-gamma-gene [-158(C->T)] detectable with the XmnI restriction enzyme.35–37 Other studies suggested that the beta-globin gene cluster haplotype, independently of the HbF levels, is correlated with survival of SCA patients treated with HU.38 Further research is necessary to understand how that occurs. Modifier genes could be interacting with the beta-globin-like cluster. HbS haplotypes and variations in cis-acting elements associated with different haplotypes are only partially responsible for the variation seen in HbF levels among SCD patients.39–41 This has prompted the search for transacting regulatory elements controlling HbF levels. Among these elements was cited a quantitative trait locus (QTL) on chromosome Xp22 that was associated with the F cell number production.39–41 Together with the −158 (C->T) polymorphism in the G-gamma promoter, this locus is estimated to account for half of the variation of HbF level in SCD.3 Other QTLs associated with HbF level have been described on chromosome 6q22.3–23.2 in a single extended Asian-Indian family.29,42,43 Another QTL on chromosome 8q was described in an Asian family with beta-thalassemia. Beta-thalassemia is due to different mutations on the beta-globin gene and the severity of the disease depends on the nature of the mutation. This QTL seems to interact with the −158 (C->T) polymorphism.44,45 Some studies showed the presence of SNPs in intronic regions of genes in the 6q QTL (Table 1). SNPs in several genes were found to be associated with the HbF levels suggesting that these genes are regulatory elements and modulators of HbF.23 These genes are phosphodiesterase 7B (PDE7B), microtubule-associated protein 7 (MAP7), mitogen-activated protein kinase kinase kinase 5 (MAP3K5), BCL2-associated transcription factor 1 (BTF) and peroxisomal biogenesis factor 7 (PEX7).29 They are also associated with the p38-MAPK pathway with the activation of gamma-globin gene expression by histone deacetylase inhibitors in K562 cells. Another interesting study46 screened two panels of 824 and 1,217 individuals of twin pairs from Northern European origin. They selectively chose v-myb myeloblastosis viral oncogene homolog (MYB) and HBS1-like (HBS1L) candidate genes in the 6q23 QTL interval. They identified three linkage disequilibrium (LD) blocks, which span a nearly contiguous segment of 79 kb long, starting 188 bp upstream from HBS1L exon 1 and ending 45 kb upstream of MYB. Among the 12 markers exhibiting the strongest evidence of association, one, rs52090909, is located in the 5′ UTR of exon 1a of HBS1L. The other strongly associated markers are either in intron 1a (rs9376090, rs9399137, rs9402685 and rs11759553), or directly upstream of the 5’ UTR of HBS1L exon 1a (rs4895440, rs4895441, rs9376092, rs9389269, rs9402686, rs11154792 and rs9483788). A test of linkage in the European dizygotic twins showed that the 6q23 QTL was completely accounted for by the markers in the trait-associated blocks. Based on measured haplotype analysis, they estimate that 17.6% of the trait variance is attributed to the markers in the three HBS1L-MYB blocks. An additional 11.6% of the trait variance is influenced by the XmnI variant on chromosome 11. As the overall heritability of the cellular HbF trait in Europeans is 89%,47 it is suggested that additional genetic or other familial factors contribute substantially (residual heritability = 59.8%) to the trait variation. They suggest that genetic variants that are associated with high F cell levels are also strongly correlated with increased expression of HBS1L in cultured erythroid cells.
Studies in 137 SCA patients treated with HU showed an association between SNPs and the change in HbF level after 2 years of treatment with HU. Using a candidate gene approach, they found SNPs in TOX (thymocyte selection-associated high mobility group box) gene within the 8q12.1 linkage peaks, and SNPs within the 6q22.33 region on mitogen-activated protein kinase kinase kinase 5 (MAP3K5) gene. They reported SNPs in other genes such as hydroxyacid oxidase 2 (HAO2), nitric oxide synthase 1 (NOS1), fms-related tyrosine kinase 1 (FLT1), arginase, type II (ARG2), nitric oxide synthase 2A (NOS2A) and phosphodiesterase 7B (PDE7B) that were associated with the HbF response to HU.48
The interactions of NO with Hb result in the formation of a vast biological sink of NO activity that modulates microvascular tone throughout the body. NO reacts with oxygenated Hb and reduced Hb to produce nitrate plus met-Hb and iron-nitrosyl-Hb, respectively; NO has several mechanisms of action, but it appears to play a major role in both the regulation of vascular muscle tone at the cellular level as well as in platelet aggregation (clumping).49 In SCD, especially in SCA disease, the rapid release of cell-free Hb may exceed the normal clearance mechanisms such as by binding to haptoglobin, and thus the excess free Hb consumes NO and impairs its regulatory role.5
Using a newly developed Bayesian modeling approach, the same team used two independent groups of patients, one group of 1,518 adults and children from the Cooperative Study of Sickle Cell Disease (CSSCD), and validated the results in a second independent group of 211 adults from the Multicenter Study of Hydroxyurea (MSH) in SCA.23 The study screened about 850 SNPs in 320 candidate genes. They found SNPs in the aquaporin 9 (AQP9), mitogen-activated protein kinase kinase 1 (MAP2K1), SMAD family member 3 (SMAD3), kinase insert domain receptor (KDR), mitogen-activated protein kinase kinase kinase 7 (MAP3K7), NADPH oxidase 3 (NOX3), argininosuccinate synthetase 1 (ASS), NOS1, arachidonate 5-lipoxygenase-activating protein (ALOX5AP), nitric oxide synthase 3 (NOS3), KL and SMAD family member 6 (SMAD6) (15q21.22) suggesting that these genes are regulatory elements in this region and novel modulators of HbF. The SNP rs1867380 in AQP9 is a functional SNP with an amino acid change located in the last exon of the gene. The minimum allele frequency observed for the minor allele A is 22% in the CSSCD set, and 19% in the MSH set which is consistent with 20% estimated in the African American panel of 23 samples reported in dbSNP. HbF levels are slightly lower in subjects who are homozygous for the minor allele A, while heterozygous subjects have a highly variable distribution of HbF. They also reported SNPs in TOX (8q21.1), hemoglobin, epsilon 1 (HBE1) and hemoglobin, gamma G (HBG2) genes, coinciding the beta-globin gene-like cluster (11p15.4) and SNPs in the glycoprotein M6B (GPM6B) gene (Xq22.2 QLT). Other SNPs in different genes have been reported in the CSSCD study only with a difference of association between age groups. Table 1 and Figure 1 summarizes all the SNPs described.23
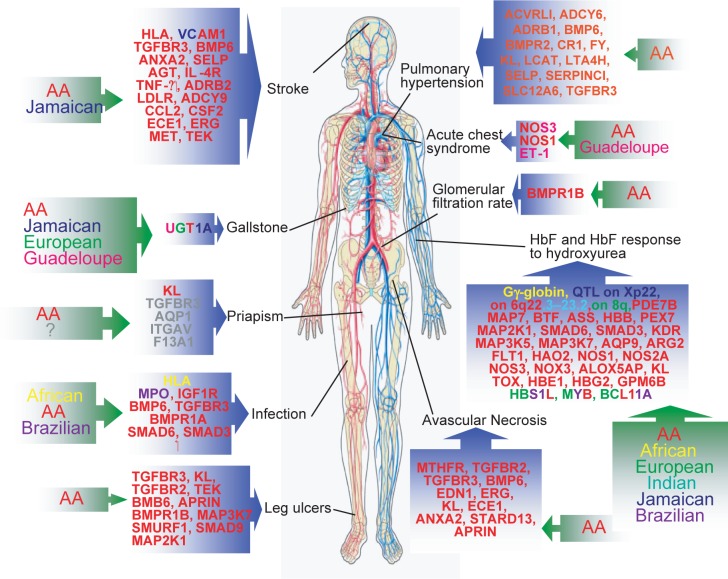
Figure 1.
Schematics of genes where SNPs were reported to be significantly implicated on different phenotypes of SCD and on which population it was described. (AA= African American).
Although the significance of all these data seems attractive, a genome-wide screening using new high throughput technologies is suggested to find how all the polymorphisms in genes regulating HbF expression, HU metabolism and erythroid progenitor proliferation might modulate patient response to HU. Also, it is important to note that, some SNPs and genes found to be associated in the CSSCD population were not found to be associated in the MSH population. Because these two populations were of African American origins, it is possible that some modifier genes may not have originated from an African ancestry. This shows the importance of studying different isolated populations composed of large cohorts of individuals to conduct association studies.
Multiple genes are very likely to affect the response of HU and HbF levels. Their interactions and the predictive values of their polymorphisms will help to elucidate their complex mechanisms.
Stroke
Stroke is a devastating complication of SCD, which occurs in about 11% of patients under 20 years of age.50,51 Among these patients, stroke is predominantly ischemic and results from the involvement of medium sized to large intracranial arteries. Ischemic stroke alone is considered in the general population as a multigenic disorder.52–54 In many cases it results from gene-gene and gene-environment interactions. In the last few years, genetic determinants have been shown to influence the risk of stroke and many SNPs in different genes have been found to be associated with ischemic stroke.55 It was reported that alpha-thalassemia is a protective factor56 and increased levels of HbF are associated with reduced risks for other complications.57
Studies have shown that increased TCD (Trans Cranial Doppler) velocities of blood flow (>200 cm/sec) in major intracranial arteries, is a predictor of stroke risk in children with SCD.58 The value of TCD as a predictor of stroke risk has been validated in the randomized STOP (Stroke Prevention in Sickle Cell Anemia) trial.58 As reviewed in Driscoll et al,59 genetic association studies in SCA have examined stroke phenotype and have shown the possibility of inherited modulation of this phenotype.59 Recent studies have found association of increase stroke risk with several Human leucocyte antigen (HLA) genotypes, especially association of certain phenotypes with distinct subtypes such as large vessel or small vessel stroke.60,61 HLA DRB1*0301 and *0302 alleles increased the susceptibility of stroke while the DRB1*1501 protected against stroke.62 DRB1*0201, in LD with DRB1*0301, was associated with stroke and DQB1*0602, in LD with DRB1*1501, was protective from stroke. Using the CSSCD patient database, HLA genotyping was performed in 36 patients with large vessel stroke and 35 with small vessel stroke.60,62 A total of 160 patients with a negative magnetic resonance imaging scan served as controls. In the small vessel stroke group, HLA DPB1*0401 was associated with increased stroke risk, whereas DPB1*1701 conferred protection from stroke. The DPB1*0401 allele was associated with susceptibility and DPB1*1701 was associated with a trend toward protection from stroke in the small vessel stroke group. In the large vessel stroke subgroup, HLA-A*0102 and *2612 conferred susceptibility, whereas the allele *3301 protected from stroke. These results suggest that specific HLA alleles influence stroke risk and appear to contribute differently to small vessel and large vessel stroke subtypes. Therefore, different pathologic processes may be involved in the development of stroke in children with SCA.60 Separation of ischemic stroke into subtypes based on presumed mechanisms may help clarify the contribution of HLA to stroke risk in SCA. These results need to be confirmed in studies with a larger number of patients, and using different populations’ if possible.
In a study of seven candidate genes in 42 children with large vessel cerebral artery stenosis, SNPs in the transforming growth factor-β receptor 3 (TGFBR3) and adenine cyclase 9 (ADCY9) were associated with this phenotype, when compared with 71 controls.3,59
Other studies, investigating candidate genes in about 230 children from the CSSCD population found associations of stroke with several genes including interleukin-4 receptor (IL-4R) S503P, tumor necrosis factor super-family, member 2 (TNFalpha) −308G and adrenergic beta-2 receptor (ADRB2) Q27E polymorphisms with large vessel stroke and vascular cell adhesion molecule 1 (VCAM1) −1594T>C, low density lipoprotein receptor (LDLR) NcoI polymorphisms with small vessel stroke.61,63 According to this study the combination of TNFalpha-308 GG homozygosity and IL-4R S503P polymorphism conferred a particularly strong risk for large vessel stroke. Another study reported polymorphism VCAM1 G1238C as protective against symptomatic stroke.64 Independent studies from the STOP population confirmed the association of large vessel stroke in SCD with both TNFalpha −308G and IL-4R S503P polymorphisms63 as well as the association of VCAM1 G1238C.65
Another study66 analyzed 108 SNPs in 39 candidate genes in 1,398 individuals from the CSSCD African American population with SCD. Thirty-one SNPs in 12 genes were found to interact with HbF to modulate the risk of stroke. This network of interactions includes three genes in the TGF-beta pathway and selectin P (SELP), which is associated with stroke in the general population.67 The researchers used the Bayesian networks modeling to capture the relationship between genotypes and phenotypes that can be used to compute the probability that a new individual with a particular genotype, will have the phenotype of interest.68 The network dissects the genetic basis of stroke into 11 genes whose variants have a direct effect on the disease that is modulated by HbF levels, and 9 genes whose variants are indirectly associated with stroke. For example, the cluster of five SNPs in endothelin 1 (EDN1) is associated with SNPs in annexin A2 (ANXA2) and bone morphogenetic protein 6 (BMP6). Both EDN1 and BMP6 are on chromosome 6 (at 6p24.1 and 6p24.3, respectively), and their association suggests that this chromosomal region may be associated with an increased risk of stroke. ANXA2 has a regulatory role in cell surface plasmin generation; EDN1 might be a potent vaso-constrictor and mitogen secreted in response to hypoxia, supporting the hypothesis that EDN1 antagonists may be useful in the prevention and treatment of sickle vaso-occlusive crises. They suggest that these model variants in BMP6 are the strongest risk factors, whereas variants in EDN1 are associated with stroke through BMP6 and ANXA2 but are not as relevant for the risk prediction. Stroke is also directly associated with variants in TGFBR3 and indirectly associated with variants in transforming growth factor, beta receptor II (TGFBR2), which have essential, non-redundant roles in TGF-betạ signaling. BMP6 is part of the TGF-beta super-family, and the simultaneous association of three genes with functional roles in TGF-betạ signaling suggests that this pathway might be involved with increased risk of stroke. This conjecture is further supported by the association of stroke with colony stimulating factor 2 (CSF2), a protein necessary for the survival, proliferation and differentiation of leukocyte progenitors. Using this method, they can compute the probability distribution of the phenotype (stroke) given the genotype of any SNP and, conversely, compute the conditional distribution of any genotype given values of other variables in the network. The overall predictive accuracy reported is 98.2%.66 In this way, the model is able to describe the determinant effects of genetic variants on stroke, to predict the odds for stroke of new individuals given their genotypes and to find the most probable combination of genetic variants leading to stroke. This method is very promising in that it can be used to find relationships between genotypes and phenotypes of all the SCD symptoms. Other genes involved according to this study are ADCY9, chemokine (C-C motif) ligand 2 (CCL2), endothelin converting enzyme 1 (ECE1), v-ets erythroblastosis virus E26 oncogene homolog (ERG), hepatocyte growth factor receptor (MET) and TEK tyrosine kinase (TEK).
Other studies attempted to associate stroke risk with SNPs in the angiotensinogen (AGT), cystathionine-beta-synthase (CBS), cholesteryl ester transfer protein (CETP), integrin, beta 3 (ITGB3), apolipoprotein C-III (APOC3), 5,10-methylenetetrahydrofolate reductase (MTHFR), serpin peptidase inhibitor, clade E (PAI1), intercellular adhesion molecule 1 (ICAM1) and thrombospondin receptor (CD36) genes,64,69 but due to the few patients studied, replication of these results in a larger patient sample is necessary.
To further define the genetic basis of stroke, the association of SNPs in candidate genes of different functional classes with the likelihood of having a stroke was examined. A total of 113 patients with SCA and a confirmed history of, or incident complete, non-hemorrhagic stroke, documented by imaging studies were compared with 493 control patients. Polymorphisms in four candidate genes, KL, TGFBR3, ANXA2 and BMP6, were associated with stroke.70 These genes play roles in the TGF-beta/BMP pathway, cell adhesion and NO biology. KL (13q12) encodes a membrane protein and regulates many vascular functions including vascular endothelial growth factor expression and NO release by the endothelium.
No association with cerebro-vascular disease was found when multiple coagulation factors were measured in children transfused for stroke, at risk for stroke and in untransfused controls.71–73 Polymorphisms in low-affinity Fc leucocyte receptors were not associated with stroke.64 Hypoxia-induced cellular activation and release of adhesive and inflammatory mediators might be related to stroke and other vaso-occlusive complications, as suggested by a study of cell adhesion molecules in children with mild sleep hypoxia who had a higher vaso-occlusive episodes and increased markers of cell adhesion and activation.74
To examine the interaction among genes and their variant SNPs and to develop a prognostic model for stroke in SCA, a Bayesian network was developed to analyze 235 SNPs in 80 candidate genes in 1398 unrelated subjects with SCA. SNPs on 11 genes and four clinical variables, including alpha-thalassemia and HbF, interacted in a complex network of dependency to modulate the risk of stroke. This network of interactions included three genes, BMP6, TGFBR2, TGFBR3 with a functional role in the TGF-beta pathway and one gene (SELP) associated with stroke in the general population. The model was validated in a different population by predicting the occurrence of stroke in 114 unrelated individuals with 98% accuracy, predicting the correct outcome for all seven stroke subjects, and for 105 of 107 non-stroke subjects. This gave a 100% true positive rate and 98. 14% true negative rate, and an overall predictive accuracy of 98%.66 As traditional analytical methods are often inadequate for the discovery of the genetic basis of complex traits in large association studies, Bayesian networks are emerging as a promising approach. The predictive accuracy of this stroke model is a step toward the development of prognostic tests which will allow a better identification of patients at risk for stroke. The presence among the risk factors of genes already associated with stroke in the general population, such as SELP, suggests that some genetic factors predisposing to stroke may be shared by both SCA patients and stroke victims in general.
Avascular necrosis/Osteonecrosis
In the CSSCD study it was shown that alpha-thalassemia, age, high hematocrit and frequent vaso-occlusive events, are risk factors for the development of avascular necrosis (AVN) of the femoral head in SCD.75 Kutlar et al investigated the frequency of the MTHFR C677T gene polymorphism in the sickle cell population as it is associated with elevated serum homocysteine levels and resultant vascular complications in relation with AVN. They discovered that MTHFR C677T was present in 16% of the sickle cell patients. There was a strong association of the presence of MTHFR C677T with AVN in 35.6% of the AVN patients having the MTHFR polymorphism as opposed to only 12.9% of sickle cell patients without AVN.76 This association has been confirmed in a Brazilian population,77 however, this was not confirmed in the high Hb F population of Kuwaiti sickle cell patients, suggesting that different genetic factors may be operative in different populations.78 Some other smaller studies in different patient populations have also failed to show an association between this MTHFR polymorphism and the risk of AVN in SCD.72,79,80 A recent study81 analyzed 442 patients with AVN and 455 SCD controls from the CSSCD cohort. They studied SNPs in 66 candidate genes and found significant association of AVN with seven SNPs in BMP6, TGFBR2, TGFBR3, EDN1, ERG, KL, ECE1, ANXA2, StAR-related lipid transfer domain containing 13 (STARD13) and PDS5, regulator of cohesion maintenance, homolog B (APRIN) genes. The precise mechanism(s) whereby variation in these genes causes AVN is not yet understood.
Acute chest syndrome
Increased susceptibility to ACS has been associated with the T-786C SNP in the NOS3 gene in females.82 Gender specific disease modifications in the endothelial NOS3 have been proposed as explanation for these modifications.83 It is suggested that the differences between sexes could possibly be explained by modulation of NOS3 activity through circulating estrogen, resulting in differences of NO formation in pulmonary vascular endothelium between females and males. Exhaled NO is significantly lower in healthy females compared with males, and in the case of relative NO deficiency, such as in SCD and CF, NOS3 variants associated with altered response to female hormones may then be relevant for the pathophysiology of the disease.83 Low exhaled levels of NO were also seen in patients with ACS compared with controls, and this was associated with the number of AAT repeats in intron 20 of NOS1.84 Another recent study shows that endothelin 1 (ET-1) T8002C and NOS3 C-786 alleles are associated with both an increased and a decreased risk of ACS in SCA patients.85 It was also proposed that Secretory phospholipase A(2) (sPLA2) may be related to the severity of the ACS and capable of predicting its onset.62 sPLA2, is found in low concentration in normal plasma, however, its levels are increased in response to inflammation and were found to be very high in the ACS.86
Priapism
Priapism, a persistent, usually painful, erection that lasts for more than four hours and occurs without sexual stimulation, occurs in 30%–45% of male patients with SCD. The possible influence of genetic risk factors on the incidence of priapism is not well understood. A study made on 44 candidate genes, in 148 patients with SCA and a confirmed history of priapism, versus 529 control SCA patients who never developed priapism, revealed a polymorphic association with the KL gene.87 Another recent study examined genetic polymorphisms in 199 unrelated, adult (>18 years) male patients with HbSS and HbS/beta (0)-thalassemia, 83 (42%) with a reported history of priapism. Candidate genes for association with priapism were identified based on their involvement in adhesion, coagulation, inflammation and cell signaling. They also examined genes involved in nitric oxide biology (NOS2, NOS3, SLC4A1), as well as polymorphisms in the KL gene. They reported strong evidence of association found for SNPs in the TGFBR3, aquaporin 1 (AQP1), integrin, alpha V (ITGAV), and the coagulation factor XIII, A1 polypeptide (F13A1). Associations with TGFBR3, AQP1, and ITGAV remained significant after adjusting for multiple testing, using the Benjamini-Hochberg procedure. These data suggest that genes involved in the TGF-β pathway, coagulation, cell adhesion and cell hydration pathways may be important in risk for priapism.88 In this study however, associations with the SNPs in the KL gene as reported before, were not significant.
Leg ulcer
Cutaneous leg ulcers are common in SCA. In the United States, 2.5% of patients with all common genotypes of SCD have leg ulcers.89 In Jamaica, >40% of patients90 and between 1.5% and 13.5% of SCD patients in Africa were reported to have leg ulcers.91–93
In a study made on the CSSCD cohort, and after screening 215 SNPs in more than 100 candidate genes, associations were found with SNPs in KL, TEK and several genes in the TGF-beta/BMP signaling pathway by genotypic association analyses. KL directly or indirectly promotes endothelial NO production and the TEK receptor tyrosine kinase is involved in angiogenesis. The TGF-beta/BMP signaling pathway modulates wound healing and angiogenesis, among its other functions. They suggest that hemolysis-driven phenotypes, such as leg ulcers, could be improved by agents that reduce sickle erythrocyte density or increase NO bioavailability.94 Among the genes reported to be significantly associated are: TGFBR3, TGFBR2, BMP6, TEK, KL, APRIN, bone morphogenetic protein receptor, type IB (BMPR1B), SMAD specific E3 ubiquitin protein ligase 1 (SMURF1), MAP3K7, SMAD family member 9 (SMAD9) and MAP2K1.94
Gallstones, cholelithiasis and bilirubin levels
Twin and family linkage studies suggested a genetic component to the development of gallstones.95–97 Pigment stones are the predominant variety in SCD due to chronic hemolysis. Homozygosity for the promoter 7(TA) repeat in the UDP glucuronosyltransferase 1 family, polypeptide A cluster (UGT1A) gene promoter has been associated with unconjugated hyperbilirubinaemia and Gilbert syndrome.98,99 The normal UGT1A promoter contains an A(TA)nTAA nucleotide-sequence motif with 6 (TA) dinucleotide repeats. A shorter 5(TA) repeat and longer 8(TA) allele have been described in persons of African descent100 but without clinical correlation. UGT1A promoter polymorphisms act as an important genetic modifier of hepatobiliary disease in SCA, influencing baseline bilirubin levels and the incidence of cholecystectomy.101 This suggests that symptomatic cholelithiasis is more common in carriers of this genotype.101,102 When treated with HU, children with the 6/6 UGT1A genotype had normal bilirubin levels compared with individuals with the 6/7 or 7/7 genotypes. A recent study suggested that the 7/7 and 7/8 genotypes were risk factors for symptomatic gallstones only in older subjects with SCD.103
Carriers of the 7/7 genotype had bilirubin levels greater than 51,3 μmol/l despite full-dose hydroxyurea therapy,104 suggesting that this polymorphism may influence the ability of hydroxyurea to prevent gallstone formation. In this study, they analyzed the effect of the UGT1A genotype on the response to HU therapy, which decreases hemolysis in children with SCA with significant increase in Hb concentration, decreases in reticulocytes and lower serum total bilirubin.105 They analyzed the reduction in serum bilirubin that occurs with decreased hemolysis in association with HU therapy.104 In a large cohort of children with SCA, from the Duke Pediatric Sickle Cell program, taking HU therapy at the maximum tolerated dose demonstrated significant reductions in hemolysis independent of UGT1A promoter polymorphism genotype, but there were no HU-related decreases in serum bilirubin levels. Children with the wild-type 6/6 UGT1A genotype demonstrated normalized bilirubin levels with HU therapy, but children with the heterozygous 6/7 or abnormal 7/7 genotypes did not. Children with the abnormal 7/7 genotype, which confers the phenotype of Gilbert syndrome, had bilirubin levels greater than 3 mg/dL despite full-dose HU therapy. These data indicate that the UGT1A promoter polymorphism is a powerful non-globin genetic modifier in SCA that influences serum bilirubin both at baseline and on HU therapy. UGT1A promoter polymorphisms may therefore influence the ability of HU to prevent gallstone formation in patients with SCA.104 Similar findings were confirmed in various cohort studies including Jamaica,103 Guadeloupe106 and Greece107 which makes this modifier gene consistent with different populations.
Infection and bacteremia
Infection and bacteremia are common in SCD and severe bacterial infections are the major causes of morbidity and mortality in SCA. A recent study on the CSSCD cohort, showed significant associations with SNPs in insulin-like growth factor 1 receptor (IGF1R) and genes of the TGF-beta/BMP pathway (BMP6, TGFBR3, bone morphogenetic protein receptor, type IA (BMPR1A), SMAD6 and SMAD3) suggesting that both could play important roles in immune function in SCA and their polymorphisms may help identify a “bacteremia-prone” phenotype.108
Another study on 144 sub-Saharan African SCD patients, 73 of whom had at least one severe bacterial infection history and 71 had none, showed a bi-allelic polymorphism (Arg107Gly) distribution of a human leukocyte antigen–E (HLA-E) locus. The HLA-E*0101/E*0101 genotype was more frequent among the group with infections than their counterparts (47% vs. 21%). This genetic association is of relevance for the involvement of HLA-E molecules in host response to pathogens.109,110 Another study involving a small group of subjects from a Brazilian cohort with SCD identified a G463A polymorphism in the myeloperoxidase (MPO) gene.111
Albuminuria and glomerular filtration rate
The glomerular filtration rate (GFR) in SCA is supra-normal in childhood but falls quickly with age, often resulting in renal failure. The renal failure is common among older patients with homozygous SCA and contributes to death of adult patients.112 The risk factors underlying these observations are unclear. Studies have correlated the high GFR with macroalbuminuria and proteinemia.113,114 In a study of 1,140 patients with SCA, SNPs of 70 candidate genes of the TGF-β/BMP pathway were screened for association with GFR. They found 4 SNPs (rs2240036, rs4145993, rs17022863, rs1434549) in BMPR1B, to be significantly associated. Three haplotypes in this gene were also associated with GFR. The TGF-beta/BMP pathway has been also associated with the development of diabetic nephropathy, which has some features in common with sickle cell nephropathy. These results suggest that, as with other sub-phenotypes of SCD, renal function may be genetically modulated.94
Pulmonary hypertension
Genetic studies concerning this phenotype are very limited. One recent study measured with a tricuspid regurgitation jet of >2.5 m/sec, showed evidence of association on genes of the TGF-beta superfamily, including activin A receptor type II–like 1 (ACVRL1), bone morphogenetic protein receptor 2 (BMPR2), and BMP6, the beta-1 adrenergic receptor (ADRB1).115
Gene polymorphisms have been found associated with eNOS and ACE genes in other diseases dealing with pulmonary hypertension-like asthma and chronic obstructive pulmonary disease116,117 emphasizing the NO pathway again as contributing candidate genes for severity.
Sickle cell adhesion
Sickle (SS) RBCs, unlike unaffected (AA) RBCs, adhere avidly to components of the vascular wall, and this abnormal adhesion is believed to contribute to the painful vaso-occlusive crises that occur in patients with SCA.118 It was reported that up-regulation of intracellular cyclic adenosine monophosphate (cAMP)-dependent protein kinase A (PKA) by epinephrine significantly increased sickle but not normal erythrocyte adhesion to both primary and immortalized endothelial cells.119 This suggests that adrenergic hormones such as epinephrine may initiate or exacerbate vaso-occlusion and thus contribute to the association of vaso-occlusive events with physiologic stress.119 In an other study, the same group showed that the alphavbeta3 integrin is the extra-cellular ligand involved in the adhesion of SS RBCs through the LW blood group antigen glycoprotein (a member of the ICAM subfamily).120 They conclude that LW appears to be the SS RBC receptor that mediates binding to at least one endothelial integrin, the alphavbeta3. Furthermore, stress hormones, such as epinephrine, may contribute to vaso-occlusion by activating LW to mediate adhesion at least in part through cAMP/PKA-dependent signaling pathways.
A recent study suggested that polymorphisms in the beta2-adrenergic receptors (ADRB2) and adenylate cyclase 6 (ADCY6) may influence SCD severity through the signalling pathway of RBC adhesion to laminin.121 In this study, they found that SNP rs1042713 in ADRB2 and SNP rs3730070 of ADCY6 polymorphisms were associated with elevated adhesion (P = 0.037 and P = 0.0002 respectively). These two genes are activated by adrenaline and may affect SS RBC adhesion to laminin. They theorize that these two genetic polymorphisms of signalling pathways on SS RBC adhesion may lead to pathophysiological consequences as well as disease variability in SCD. Future investigations are needed to confirm a role for adrenaline-mediated in vivo activation of SS RBC adhesion in vaso-occlusion.
Painful episodes
Acute painful episodes are a major complication of SCD and can be predictive of early death in adults. The only genetic modulations known to date about pain in SCA are the protective role of HbF and the deleterious or neutral role of alpha-thalassemia.122 A major limitation of the polymorphism study is how painful episodes are defined. The detection of genomic variations leading to painful episodes will be very important for SCD patients. The finding of new candidate genes will open the possibilities to use them as biomarkers to diagnose the severity of the disease.
Other phenotypes
Other complications can be associated with genetic predisposition. Although only few studies exist on the risk factors of some of these phenotypes, many of them remain to be studied. Some potential sub-phenotypes that can be considered genetically associated with the disease, and thus needing further genetic association studies are: Mortality, dactylitis, aplastc crisis, acute splenic sequestration, chronic hypersplenism, megaloblastic change, iron and folate deficiency, bone pain, pregnancy complications, osteomyelitis, acute pulmonary sequestration, pulmonary fat embolism, deep jaundice, iris atrophy and eye complications and chronic transfusion complications.3,4,123 All these complications, may or may not be directly or indirectly associated with a gene polymorphism, but further studies should be considered.
The “no complication” phenotype
Due to the advances in alternative controls and therapies such as HU treatment, transfusions, prophylaxis and antibiotics to prevent infection, new born screening programs, prevention and gene counseling, and improving of socio-economical features, the survival rate of SCA patients is clearly increasing.57,124 Determining the severity of the disease is very difficult. Isolating one specific sub-phenotype for genetic association screening requires very careful monitoring of the patient’s complications throughout their life. The only clear phenotype that we can surely characterize is the “no complication” phenotype. Indeed, some patients that carry the SS mutated hemoglobin have very mild or no complication throughout their life. We can imagine that these SS individuals carry a gene polymorphism that “protects” them from anemia and vaso-occlusive events. In many of the studies above, they used these patients as negative controls to find polymorphisms in different sub-phenotypes. Some patients with the SS mutation can survive until their 6th and 7th decade (Serjeant, personal communication).24 Further studies on gene polymorphisms in these individuals can reveal interesting findings.
Discussion
Differences between populations
The beta-globin gene is located on the short arm of Chromosome 11. Variations in non-coding nucleotides flanking the globin genes have been used to track the origin of the sickle cell mutation and its flow to other geographical regions in the world. Using restriction endonucleases, haplotype blocks of 8 SNPs spanning ∼30,000 bases of the beta-globin locus could be determined and was used to study the ancestral origin of the sickle cell mutation in individuals from different parts of Africa and Asia. In Africa, the HbS gene was associated with four haplotypes representing regions where independent mutations occurred. These haplotypes include the Benin, the Senegal, the Bantu (Central African Republic) and the Cameroon.36,125 A fifth Asian beta-locus haplotype was reported in Saudi Arabia and India.126 In America, Jamaica and Brazil, the African ethnic groups were subject to a considerable admixture of different haplotype groups. Calculations suggest that the HbS-globin gene mutation first appeared approximately 70,000∼150,000 years ago.127 As described above in the Fetal Hb and hydroxyurea response section, the Senegal and Asian-Indian haplotypes have a general milder clinical and hematological phenotype compared with the other African haplotypes (Benin, Bantu and Cameroon). Individuals with Bantu haplotypes have the most severe phenotype and individuals with the Benin haplotype usually have intermediate features.34
Global frequency and distribution
While the mutation prevalence is the highest in the Mediterranean, Africa and Asia, the migration of the populations from these areas has increased globally.128 SCD is now endemic throughout Europe, the Americas and Australia.1
Comprehensive control programs in recent years have succeeded in limiting the numbers of new births and prolonging life in affected individuals. Such programs have been successful in a minority of countries and have little global impact. Over 300,000 infants with major syndromes are born every year and the majority dies undiagnosed, untreated or under-treated. Countries may be divided into three general categories according to the services available: A. Endemic Mediterranean countries. In these countries, long-established prevention programs have achieved 80 to 100% prevention through specialized clinics which provide optimum treatment. B. Areas of the developed, industrialized world where prevalence is increasing because of migration. These countries have the means to provide adequate control but have problems in reaching immigrant groups with different cultural background. C. Countries of the developing world where the provision of services is hampered by economic difficulties, other health priorities due to high infant mortality from infectious diseases, and religious/cultural constraints.129
Cooperative studies
In Table 1 the majority of polymorphism discoveries on sub-phenotypes have been made on the CSSCD cohort. This shows the importance of establishing organized cooperative research centers and new born screening projects that help not only for gene polymorphism discoveries of modifier genes, but also enables us to understand the exact mechanism leading to different phenotypes. To conduct such genetic studies, it is very important to have a large number of patients included in each study. The relevance of the findings strongly depends on the statistical significance of the genetic results. This significance is directly associated with the number of individuals included in the study, which defines the power of the results. Independent studies, using different techniques and approaches (as well as different laboratories) must be done, even if the screening will be conducted on the same population. The findings will be additive and will confirm previous results. Confirmation of modifier gene action will enhance the discovery of a potential cure for that phenotype, not only for SCD but also for other diseases sharing similar phenotypes.
The National Institutes of Health (NIH) has established funds to monitor clinical trials and human genetic studies. However, genetic studies require a large number of patient DNA and clinical information. To collect information on patients and protect the personal privacy of each individual, the institutional review board (IRB), which is an independent committee of physicians, statisticians, community advocates and others, insure that the clinical trial or the genetic studies conducted by the investigators are ethical, and all the rights of the studied participants are protected. In the past however, minorities and populations from developing countries were exploited and abused in several clinical trials which has created resentment and suspicion of researchers among these populations. Regaining the trust of minorities and establishment of collaborative studies with developing countries to participate in large scale genetic screenings remains a problematic and challenging task (US DHHS 1994). Recruiting volunteer participants for medical research is critical to developing medical innovation toward improved patient health. A sophisticated and clear collaborative policy will definitely help regain trust in medical research, the backbone of medical cure.
Comparative studies between populations
SCD is one of more than 10,000 human diseases that are caused by defects in single genes. Screening for a diverse group of serious and common disorders in the prenatal setting presents a great challenge.
Although it is well known that 85%–95% of human genetic variation is due to variation among individuals within a population, whereas 5%–15% is attributable to variation among populations,130,131 it remains unclear whether similar levels of within-versus among-population components of variation will extend to higher-level phenotypes such as gene-expression levels. Understanding patterns of gene-expression variation within and among human populations will provide important insights into the molecular basis of phenotypic diversity and the interpretation of patterns of expression variation in disease. Finding that a single locus has a strong signal in two different populations and in two separate studies, is very important in validating biomarkers for the phenotype. Such results may be combined to maximize the amount of available information that can be extracted from these expensive and laborious experiments.
One example of the importance of comparing the population SNPs is the MTHFR gene described above and its implication in the avascular necrosis. This gene has been found associated with this phenotype in a large number of patients of the CSSCD population but could not be confirmed in the Kuwaiti population.78 This suggests that different genetic factors may be operative in different populations. On the other hand the UGT1A gene promoter (TA) repeat polymorphism has been confirmed in different populations, making this gene a potentially reliable biomarker that can be used as a diagnostic predictor.
Importance of collecting environmental information including diet and traditions
The differences between populations do not only refer to a pure genetic paradigm, but also refer to geographic, social, dietetic, traditional and socio-economical matter. The outcome of the hemoglobin sickling on patients eating different diet will be different. Even though very few studies has been made on water intake and it’s real patho-physiological influence on the phenotype, doctors advise children with SCA to drink water and fluids and avoid dehydration. Very few studies have been done on the effect of diet on SCD patients. Some studies done in our laboratory, showed correlations between the metabolic demands of increased erythropoiesis and cardiac energy consumption with the excess protein and energy metabolism in children with SCA.132 The same group conducted studies using a sickle cell mouse model and showed a direct role of the protein intake on the outcome of the disease.133
SCD is a blood disorder. Blood does not only carry oxygen and carbon dioxide, but also has primordial roles in distribution of water, metabolites, energy, hormones and enzymes to different parts of the body. It is logical that the diet, environmental factors and lifestyle in general, as well as psychological and socio-economical factors will definitely influence the outcome of the disease. It is important to note that, when we genetically screen patients for polymorphisms, we take into consideration the environmental factors that could influence the outcome of the disease.
Importance of conducting genetic studies in endemic malaria populations
For thousands of years, in sub-Saharan Africa, malaria transmission resulted in natural selection which resulted in preferential killing of healthy and SCA children while individuals with SCT were protected. This resulted in high frequency of SCD patients in malaria endemic regions. The global frequency of hemoglobinopathies shows a remarkably consistent match of hemoglobinopathies with malaria endemic region. The eradication of malaria in some countries, such as in the Mediterranean countries, has contributed to the lower frequency of HbS in these regions. Interestingly, SCD is a genetic disorder while malaria is due to a parasitic infection. It is therefore important to continue the fight to completely eradicate malaria in all the countries.
Susceptibility/resistance to Plasmodium falciparum malaria has been correlated with polymorphisms in more than 30 human genes on patients from Africa and south-east Asia and India.134
Genome-wide association analysis based on 10,000 single-nucleotide polymorphisms in a Ghanaian population identified several regions which showed evidence for linkage disequilibrium to parasitological and clinical phenotypes. Among them a prominent signal on Chromosome 10p15 obtained with malaria fever episodes135 and on Chromosome 5q31–q33.136 Several human gene SNPs have been described to be linked with the severity of the malaria such as: CYBB gene,137 the gp91 phox subunit of the NADPH oxidase,137 the TNF-enhancer and gene for FcgammaRIIa,134 IL12B136 as well as the haptoglobin haplotype polymorphism.138 These studies show a relationship between red blood cell efficiency and the severity of the malaria as observed in the SCA.
Benefits and limitations
It is extremely important to utilize new high throughput technologies now available commercially to determine how best to translate such progress into improved patient care. Nowadays, a single DNA chip microarray can be used to screen a whole genome-wide SNPs panel per individual. New technologies have enabled genome-wide association studies to be conducted with hundreds of thousands of genotyped SNPs. Several different first-generation genome-wide panels of SNPs have been commercialized. The total amount of common genetic variation is still unknown; however, the coverage of commercial panels can be evaluated against reference population samples genotyped by the International HapMap project. Less information is available about coverage in samples from other populations. The high cost of these technologies is a major limitation not only for the gene polymorphism studies, but also for the practical exploitation of the biomarker-based therapies. But, despite the high cost, the new technologies became more “per-sample” cost effective and can allow the determination of a whole patient’s genotype in one single experiment. It also gives us the copy number of the gene expression. This advancement of biomedical technologies enables the efficient and reliable screening of patients with different phenotypes and also appears to be the key for establishing biomakers that will allow better diagnosis and treatment of patients that have a susceptibility SNP implicated in the clinical manifestation of the disease. These technologies have been proven to be effective in revealing associations between SNPs and many disease phenotypes, even in non monogenic disorders such as infectious diseases and in different populations including Caucasians, Asians and Africans.135,139
On the other hand, a recent twin study, introduced a controversy in the paradigm of epistatic genetic control of disease.140 This study compared 9 pairs of identical twins of a Jamaican cohort and showed that twins presented similarities in the prevalence and degree of splenomegaly, susceptibility to priapism, and in onset of menarche, but other clinical complications were discordant in prevalence and severity. These findings suggest that physical growth and many hematological characteristics are subject to genetic influences, but that non-genetic factors contribute to the variance in disease manifestations. Thus, in these studies, even though very limited in the sample size, a better understanding of phenotype/genotype correlation in SCD can be acquired.
Coordinating efforts and cooperative studies with different laboratories and hospitals around the world, setting up a common database that is freely shared, and helping developing countries to set up new born screening centers for collecting data will facilitate the search for a cure for SCD.
These kind of genomic investigations are very important for the detection of genomic variations leading to different phenotypes in SCD patients. However, functional studies will be required to correlate the results with the phenotypes. These studies are limited to the extent to which it is a genetic study using new genomic tools and the complexity of genetic interactions and environmental factors require further gene expression confirmations such RNA and protein levels.
The understanding of genomic variations in SCD patients will be key in the use of possible biomarkers for diagnostic of the severity of SCD and that is critical for targeted therapeutics delivery and personalized treatments.
Acknowledgments
– NIH/Fogarty International Center (FIC) postdoctoral training grant in Genomics and Hemoglobinopathies (NIH-FIC-1R90-HG004151–01).
– NIH-RCMI (RR033062).
Footnotes
Disclosure
The authors report no conflicts of interest.
References
- 1.Angastiniotis M, Modell B. Global epidemiology of hemoglobin disorders. Ann NY Acad Sci. 1998 Jun 30;850:251–69. doi: 10.1111/j.1749-6632.1998.tb10482.x. [DOI] [PubMed] [Google Scholar]
- 2.Weatherall DJ, Clegg JB. Inherited haemoglobin disorders: an increasing global health problem. Bull World Health Organ. 2001;79(8):704–12. [PMC free article] [PubMed] [Google Scholar]
- 3.Steinberg MH. Predicting clinical severity in sickle cell anaemia. Br J Haematol. 2005 May;129(4):465–81. doi: 10.1111/j.1365-2141.2005.05411.x. [DOI] [PubMed] [Google Scholar]
- 4.Kutlar A. Sickle cell disease: a multigenic perspective of a single gene disorder. Hemoglobin. 2007;31(2):209–24. doi: 10.1080/03630260701290233. [DOI] [PubMed] [Google Scholar]
- 5.Rother RP, Bell L, Hillmen P, Gladwin MT. The clinical sequelae of intravascular hemolysis and extracellular plasma hemoglobin: a novel mechanism of human disease. Jama. 2005 Apr 6;293(13):1653–62. doi: 10.1001/jama.293.13.1653. [DOI] [PubMed] [Google Scholar]
- 6.Steinberg MH. Modulation of fetal hemoglobin in sickle cell anemia. Hemoglobin. 2001 May;25(2):195–211. doi: 10.1081/hem-100104028. [DOI] [PubMed] [Google Scholar]
- 7.Ingram VM. Separation of the peptide chains of human globin. Nature. 1959 Jun 27;183:1795–98. doi: 10.1038/1831795a0. [DOI] [PubMed] [Google Scholar]
- 8.Bunn HF. Pathogenesis and treatment of sickle cell disease. N Engl J Med. 1997 Sep 11;337(11):762–9. doi: 10.1056/NEJM199709113371107. [DOI] [PubMed] [Google Scholar]
- 9.Steinberg MH. Management of sickle cell disease. N Engl J Med. 1999 Apr 1;340(13):1021–30. doi: 10.1056/NEJM199904013401307. [DOI] [PubMed] [Google Scholar]
- 10.Platt OS, Thorington BD, Brambilla DJ, et al. Pain in sickle cell disease. Rates and risk factors. N Engl J Med. 1991 Jul 4;325(1):11–6. doi: 10.1056/NEJM199107043250103. [DOI] [PubMed] [Google Scholar]
- 11.Williams TN. Human red blood cell polymorphisms and malaria. Curr Opin Microbiol. 2006 Aug;9(4):388–94. doi: 10.1016/j.mib.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Aidoo M, Terlouw DJ, Kolczak MS, et al. Protective effects of the sickle cell gene against malaria morbidity and mortality. Lancet. 2002 Apr 13;359(9314):1311–2. doi: 10.1016/S0140-6736(02)08273-9. [DOI] [PubMed] [Google Scholar]
- 13.Williams TN, Mwangi TW, Wambua S, et al. Negative epistasis between the malaria-protective effects of alpha+-thalassemia and the sickle cell trait. Nat Genet. 2005 Nov;37(11):1253–7. doi: 10.1038/ng1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Serjeant GR. Natural history and determinants of clinical severity of sickle cell disease. Curr Opin Hematol. 1995 Mar;2(2):103–8. doi: 10.1097/00062752-199502020-00001. [DOI] [PubMed] [Google Scholar]
- 15.Steinberg MH. Review: sickle cell disease: present and future treatment. Am J Med Sci. 1996 Oct;312(4):166–74. doi: 10.1097/00000441-199610000-00004. [DOI] [PubMed] [Google Scholar]
- 16.Noguchi CT, Dover GJ, Rodgers GP, et al. Alpha thalassemia changes erythrocyte heterogeneity in sickle cell disease. J Clin Invest. 1985 May;75(5):1632–7. doi: 10.1172/JCI111870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steinberg MH, West MS, Gallagher D, Mentzer W. Effects of glucose-6-phosphate dehydrogenase deficiency upon sickle cell anemia. Blood. 1988 Mar;71(3):748–52. [PubMed] [Google Scholar]
- 18.Kimmick G, Owen J. Rhabdomyolysis and hemolysis associated with sickle cell trait and glucose-6-phosphate dehydrogenase deficiency. South Med J. 1996 Nov;89(11):1097–8. doi: 10.1097/00007611-199611000-00015. [DOI] [PubMed] [Google Scholar]
- 19.Cohen-Solal M, Prehu C, Wajcman H, et al. A new sickle cell disease phenotype associating Hb S trait, severe pyruvate kinase deficiency (PK Conakry), and an alpha2 globin gene variant (Hb Conakry) Br J Haematol. 1998 Dec;103(4):950–6. doi: 10.1046/j.1365-2141.1998.01094.x. [DOI] [PubMed] [Google Scholar]
- 20.Warkentin TE, Barr RD, Ali MA, Mohandas N. Recurrent acute splenic sequestration crisis due to interacting genetic defects: hemoglobin SC disease and hereditary spherocytosis. Blood. 1990 Jan 1;75(1):266–70. [PubMed] [Google Scholar]
- 21.Yang YM, Donnell C, Wilborn W, et al. Splenic sequestration associated with sickle cell trait and hereditary spherocytosis. Am J Hematol. 1992 Jun;40(2):110–6. doi: 10.1002/ajh.2830400207. [DOI] [PubMed] [Google Scholar]
- 22.Adekile AD. Mild-phenotype sickle cell disease: molecular basis, clinical presentation and management recommendations. CURRENT PAEDIATRICS. 2005 Feb;15(1):57–61. [Google Scholar]
- 23.Sebastiani P, Nolan VG, Baldwin CT, et al. A network model to predict the risk of death in sickle cell disease. Blood. 2007 Oct 1;110(7):2727–35. doi: 10.1182/blood-2007-04-084921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinberg MH, Ballas SK, Brunson CY, Bookchin R. Sickle cell anemia in septuagenarians. Blood. 1995 Nov 15;86(10):3997–8. [PubMed] [Google Scholar]
- 25.Hebbel RP, Vercellotti GM. The endothelial biology of sickle cell disease. J Lab Clin Med. 1997 Mar;129(3):288–93. doi: 10.1016/s0022-2143(97)90176-1. [DOI] [PubMed] [Google Scholar]
- 26.Charache S, Terrin ML, Moore RD, et al. Effect of hydroxyurea on the frequency of painful crises in sickle cell anemia. Investigators of the Multicenter Study of Hydroxyurea in Sickle Cell Anemia. N Engl J Med. 1995 May 18;332(20):1317–22. doi: 10.1056/NEJM199505183322001. [DOI] [PubMed] [Google Scholar]
- 27.Dover GJ, Chang VT, Boyer SH, Serjeant GR, Antonarakis S, Higgs DR. The cellular basis for different fetal hemoglobin levels among sickle cell individuals with two, three, and four alpha-globin genes. Blood. 1987 Jan;69(1):341–4. [PubMed] [Google Scholar]
- 28.Ofori-Acquah SF, Lalloz MR, Serjeant G, Layton DM. Dominant influence of gamma-globin promoter polymorphisms on fetal haemoglobin expression in sickle cell disease. Cell Mol Biol (Noisy-le-grand) 2004 Feb;50(1):35–42. [PubMed] [Google Scholar]
- 29.Wyszynski DF, Baldwin CT, Cleves MA, et al. Polymorphisms near a chromosome 6q QTL area are associated with modulation of fetal hemoglobin levels in sickle cell anemia. Cell Mol Biol (Noisy-le-grand) 2004 Feb;50(1):23–33. [PubMed] [Google Scholar]
- 30.Rucknagel DL, Hanash SH, Sing CF, Winter WP, Whitten CF, Prasad AS. Age and sex effects on hemoglobin F in sickle cell anemia. In: Nienhuis GSAW, editor. Cellular and Molecular Regulation of Hemoglobin Switching. New York: Grune and Stratton; 1979. pp. 107–18. [Google Scholar]
- 31.Morris J, Dunn D, Beckford M, et al. The haematology of homozygous sickle cell disease after the age of 40 years. Br J Haematol. 1991 Mar;77(3):382–5. doi: 10.1111/j.1365-2141.1991.tb08588.x. [DOI] [PubMed] [Google Scholar]
- 32.Dover GJ, Boyer SH. The cellular distribution of fetal hemoglobin: normal adults and hemoglobinopathies. Tex Rep Biol Med. 1980;40:43–54. [PubMed] [Google Scholar]
- 33.Nagel RL, Labie D. DNA haplotypes and the beta s globin gene. Prog Clin Biol Res. 1989;316B:371–93. [PubMed] [Google Scholar]
- 34.Nagel RLS, MH . Genetics of the bS gene: origins, epidemiology, and epistasis. In: MH, Steinberg BGF, Higgs DR, Nagel RL, editors. Disorders of Hemoglobin: Genetics, Pathophysiology, and Clinical Management. Cambridge: Cambridge University Press; 2001. pp. 711–55. [Google Scholar]
- 35.Labie D, Pagnier J, Lapoumeroulie C, et al. Common haplotype dependency of high G gamma-globin gene expression and high Hb F levels in beta-thalassemia and sickle cell anemia patients. Proc Natl Acad Sci U S A. 1985 Apr;82(7):2111–4. doi: 10.1073/pnas.82.7.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagel RL, Fabry ME, Pagnier J, et al. Hematologically and genetically distinct forms of sickle cell anemia in Africa. The Senegal type and the Benin type. N Engl J Med. 1985 Apr 4;312(14):880–4. doi: 10.1056/NEJM198504043121403. [DOI] [PubMed] [Google Scholar]
- 37.Steinberg MH, Hsu H, Nagel RL, et al. Gender and haplotype effects upon hematological manifestations of adult sickle cell anemia. Am J Hematol. 1995 Mar;48(3):175–81. doi: 10.1002/ajh.2830480307. [DOI] [PubMed] [Google Scholar]
- 38.Bakanay SM, Dainer E, Clair B, et al. Mortality in sickle cell patients on hydroxyurea therapy. Blood. 2005 Jan 15;105(2):545–7. doi: 10.1182/blood-2004-01-0322. [DOI] [PubMed] [Google Scholar]
- 39.Dover GJ, Smith KD, Chang YC, et al. Fetal hemoglobin levels in sickle cell disease and normal individuals are partially controlled by an X-linked gene located at Xp22.2. Blood. 1992 Aug 1;80(3):816–24. [PubMed] [Google Scholar]
- 40.Thein SL, Sampietro M, Rohde K, et al. Detection of a major gene for heterocellular hereditary persistence of fetal hemoglobin after accounting for genetic modifiers. Am J Hum Genet. 1994 Feb;54(2):214–28. [PMC free article] [PubMed] [Google Scholar]
- 41.Chang YC, Smith KD, Moore RD, Serjeant GR, Dover GJ. An analysis of fetal hemoglobin variation in sickle cell disease: the relative contributions of the X-linked factor, beta-globin haplotypes, alpha-globin gene number, gender, and age. Blood. 1995 Feb 15;85(4):1111–7. [PubMed] [Google Scholar]
- 42.Garner C, Mitchell J, Hatzis T, Reittie J, Farrall M, Thein SL. Haplotype mapping of a major quantitative-trait locus for fetal hemoglobin production, on chromosome 6q23. Am J Hum Genet. 1998 Jun;62(6):1468–74. doi: 10.1086/301859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Close J, Game L, Clark B, Bergounioux J, Gerovassili A, Thein SL. Genome annotation of a 1.5 Mb region of human chromosome 6q23 encompassing a quantitative trait locus for fetal hemoglobin expression in adults. BMC Genomics. 2004 May 31;5(1):33. doi: 10.1186/1471-2164-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garner CP, Tatu T, Best S, Creary L, Thein SL. Evidence of genetic interaction between the beta-globin complex and chromosome 8q in the expression of fetal hemoglobin. Am J Hum Genet. 2002 Mar;70(3):793–99. doi: 10.1086/339248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garner C, Silver N, Best S, et al. Quantitative trait locus on chromosome 8q influences the switch from fetal to adult hemoglobin. Blood. 2004 Oct 1;104(7):2184–6. doi: 10.1182/blood-2004-02-0527. [DOI] [PubMed] [Google Scholar]
- 46.Thein SL, Menzel S, Peng X, et al. Intergenic variants of HBS1L-MYB are responsible for a major quantitative trait locus on chromosome 6q23 influencing fetal hemoglobin levels in adults. Proc Natl Acad Sci U S A. 2007 Jul 3;104(27):11346–51. doi: 10.1073/pnas.0611393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garner C, Tatu T, Reittie JE, et al. Genetic influences on F cells and other hematologic variables: a twin heritability study. Blood. 2000 Jan 1;95(1):342–6. [PubMed] [Google Scholar]
- 48.Ma Q, Wyszynski DF, Farrell JJ, et al. Fetal hemoglobin in sickle cell anemia: genetic determinants of response to hydroxyurea. Pharmacogenomics J. 2007 Dec;7(6):386–94. doi: 10.1038/sj.tpj.6500433. [DOI] [PubMed] [Google Scholar]
- 49.Gladwin MT, Schechter AN, Shelhamer JH, Ognibene FP. The acute chest syndrome in sickle cell disease. Possible role of nitric oxide in its pathophysiology and treatment. Am J Respir Crit Care Med. 1999 May;159(5 Pt 1):1368–76. doi: 10.1164/ajrccm.159.5.9810094. [DOI] [PubMed] [Google Scholar]
- 50.Powars D, Wilson B, Imbus C, Pegelow C, Allen J. The natural history of stroke in sickle cell disease. Am J Med. 1978 Sep;65(3):461–71. doi: 10.1016/0002-9343(78)90772-6. [DOI] [PubMed] [Google Scholar]
- 51.Ohene-Frempong K, Weiner SJ, Sleeper LA, et al. Cerebrovascular accidents in sickle cell disease: rates and risk factors. Blood. 1998 Jan 1;91(1):288–94. [PubMed] [Google Scholar]
- 52.Hassan A, Markus HS. Genetics and ischaemic stroke. Brain. 2000 Sep;123(Pt 9):1784–812. doi: 10.1093/brain/123.9.1784. [DOI] [PubMed] [Google Scholar]
- 53.Laine M, Salmelin R, Helenius P, Marttila R. Brain activation during reading in deep dyslexia: an MEG study. J Cogn Neurosci. 2000 Jul;12(4):622–34. doi: 10.1162/089892900562381. [DOI] [PubMed] [Google Scholar]
- 54.Hademenos GJ, Alberts MJ, Awad I, et al. Advances in the genetics of cerebrovascular disease and stroke. Neurology. 2001 Apr 24;56(8):997–1008. doi: 10.1212/wnl.56.8.997. [DOI] [PubMed] [Google Scholar]
- 55.Matarin M, Brown WM, Scholz S, et al. A genome-wide genotyping study in patients with ischaemic stroke: initial analysis and data release. Lancet Neurol. 2007 May;6(5):414–20. doi: 10.1016/S1474-4422(07)70081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams RJ, Kutlar A, McKie V, et al. Alpha thalassemia and stroke risk in sickle cell anemia. Am J Hematol. 1994 Apr;45(4):279–82. doi: 10.1002/ajh.2830450402. [DOI] [PubMed] [Google Scholar]
- 57.Platt OS, Brambilla DJ, Rosse WF, et al. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N Engl J Med. 1994 Jun 9;330(23):1639–44. doi: 10.1056/NEJM199406093302303. [DOI] [PubMed] [Google Scholar]
- 58.Adams RJ, McKie VC, Hsu L, et al. Prevention of a first stroke by transfusions in children with sickle cell anemia and abnormal results on transcranial Doppler ultrasonography. N Engl J Med. 1998 Jul 2;339(1):5–11. doi: 10.1056/NEJM199807023390102. [DOI] [PubMed] [Google Scholar]
- 59.Driscoll MC, Hurlet A, Styles L, et al. Stroke risk in siblings with sickle cell anemia. Blood. 2003 Mar 15;101(6):2401–4. doi: 10.1182/blood.V101.6.2401. [DOI] [PubMed] [Google Scholar]
- 60.Hoppe C, Klitz W, Noble J, Vigil L, Vichinsky E, Styles L. Distinct HLA associations by stroke subtype in children with sickle cell anemia. Blood. 2003 Apr 1;101(7):2865–9. doi: 10.1182/blood-2002-09-2791. [DOI] [PubMed] [Google Scholar]
- 61.Hoppe C, Klitz W, Cheng S, et al. Gene interactions and stroke risk in children with sickle cell anemia. Blood. 2004 Mar 15;103(6):2391–6. doi: 10.1182/blood-2003-09-3015. [DOI] [PubMed] [Google Scholar]
- 62.Styles LA, Aarsman AJ, Vichinsky EP, Kuypers FA. Secretory phospholipase A(2) predicts impending acute chest syndrome in sickle cell disease. Blood. 2000 Nov 1;96(9):3276–8. [PubMed] [Google Scholar]
- 63.Hoppe C, Klitz W, D’Harlingue K, et al. Confirmation of an association between the TNF(-308) promoter polymorphism and stroke risk in children with sickle cell anemia. Stroke. 2007 Aug;38(8):2241–6. doi: 10.1161/STROKEAHA.107.483115. [DOI] [PubMed] [Google Scholar]
- 64.Taylor JGt, Tang DC, Savage SA, et al. Variants in the VCAM1 gene and risk for symptomatic stroke in sickle cell disease. Blood. 2002 Dec 15;100(13):4303–9. doi: 10.1182/blood-2001-12-0306. [DOI] [PubMed] [Google Scholar]
- 65.Kutlar A BD, Clair B, Haghighat A, et al. ASH. 11. Vol. 110. Atlanta GA USA: Blood; Candidate Gene Polymorphisms and Their Association with TCD Velocities in Children with Sickle Cell Disease; p. 2007. [Google Scholar]
- 66.Sebastiani P, Ramoni MF, Nolan V, Baldwin CT, Steinberg MH. Genetic dissection and prognostic modeling of overt stroke in sickle cell anemia. Nat Genet. 2005 Apr;37(4):435–40. doi: 10.1038/ng1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zee RY, Cook NR, Cheng S, et al. Polymorphism in the P-selectin and interleukin-4 genes as determinants of stroke: a population-based, prospective genetic analysis. Hum Mol Genet. 2004 Feb 15;13(4):389–96. doi: 10.1093/hmg/ddh039. [DOI] [PubMed] [Google Scholar]
- 68.Lauritzen SLS, NA Graphical models for genetic analysis. Statist Sci. 2004;18:489–514. [Google Scholar]
- 69.Romana M, Diara JP, Doumbo L, et al. Angiotensinogen gene associated polymorphisms and risk of stroke in sickle cell anemia: Additional data supporting an association. Am J Hematol. 2004 Jul;76(3):310–11. doi: 10.1002/ajh.20078. [DOI] [PubMed] [Google Scholar]
- 70.Steinberg MH, Voskaridou E, Kutlar A, et al. Concordant fetal hemoglobin response to hydroxyurea in siblings with sickle cell disease. Am J Hematol. 2003 Feb;72(2):121–6. doi: 10.1002/ajh.10264. [DOI] [PubMed] [Google Scholar]
- 71.Kahn MJ, Scher C, Rozans M, Michaels RK, Leissinger C, Krause J. Factor V Leiden is not responsible for stroke in patients with sickling disorders and is uncommon in African Americans with sickle cell disease. Am J Hematol. 1997 Jan;54(1):12–5. doi: 10.1002/(sici)1096-8652(199701)54:1<12::aid-ajh2>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 72.Andrade FL, Annichino-Bizzacchi JM, Saad ST, Costa FF, Arruda VR. Prothrombin mutant, factor V Leiden, and thermolabile variant of methylenetetrahydrofolate reductase among patients with sickle cell disease in Brazil. Am J Hematol. 1998 Sep;59(1):46–50. doi: 10.1002/(sici)1096-8652(199809)59:1<46::aid-ajh9>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 73.Liesner R, Mackie I, Cookson J, et al. Prothrombotic changes in children with sickle cell disease: relationships to cerebrovascular disease and transfusion. Br J Haematol. 1998 Dec;103(4):1037–44. doi: 10.1046/j.1365-2141.1998.01121.x. [DOI] [PubMed] [Google Scholar]
- 74.Setty BN, Stuart MJ, Dampier C, Brodecki D, Allen JL. Hypoxaemia in sickle cell disease: biomarker modulation and relevance to pathophysiology. Lancet. 2003 Nov 1;362(9394):1450–5. doi: 10.1016/S0140-6736(03)14689-2. [DOI] [PubMed] [Google Scholar]
- 75.Milner PF, Kraus AP, Sebes JI, et al. Sickle cell disease as a cause of osteonecrosis of the femoral head. N Engl J Med. 1991 Nov 21;325(21):1476–81. doi: 10.1056/NEJM199111213252104. [DOI] [PubMed] [Google Scholar]
- 76.Kutlar A, Kutlar F, Turker I, Tural C. The methylene tetrahydrofolate reductase (C677T) mutation as a potential risk factor for avascular necrosis in sickle cell disease. Hemoglobin. 2001 May;25(2):213–7. doi: 10.1081/hem-100104029. [DOI] [PubMed] [Google Scholar]
- 77.Moreira Neto F, Lourenco DM, Noguti MA, et al. The clinical impact of MTHFR polymorphism on the vascular complications of sickle cell disease. Braz J Med Biol Res. 2006 Oct;39(10):1291–5. doi: 10.1590/s0100-879x2006001000004. [DOI] [PubMed] [Google Scholar]
- 78.Adekile AD, Kutlar F, Haider MZ, Kutlar A. Frequency of the 677 C−>T mutation of the methylenetetrahydrofolate reductase gene among Kuwaiti sickle cell disease patients. Am J Hematol. 2001 Apr;66(4):263–6. doi: 10.1002/ajh.1055. [DOI] [PubMed] [Google Scholar]
- 79.De Castro L RH, Howe JG, Smith BR. Thrombophilic genotypes do not adversely affect the course of sickle cell disease (SCD) Blood. 1998;92(Suppl):161a. [Google Scholar]
- 80.Zimmerman SA, Ware RE. Inherited DNA mutations contributing to thrombotic complications in patients with sickle cell disease. Am J Hematol. 1998 Dec;59(4):267–72. doi: 10.1002/(sici)1096-8652(199812)59:4<267::aid-ajh1>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 81.Baldwin C, Nolan VG, Wyszynski DF, et al. Association of klotho, bone morphogenic protein 6, and annexin A2 polymorphisms with sickle cell osteonecrosis. Blood. 2005 Jul 1;106(1):372–5. doi: 10.1182/blood-2005-02-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sharan K, Surrey S, Ballas S, et al. Association of T-786C eNOS gene polymorphism with increased susceptibility to acute chest syndrome in females with sickle cell disease. Br J Haematol. 2004 Jan;124(2):240–3. doi: 10.1046/j.1365-2141.2003.04762.x. [DOI] [PubMed] [Google Scholar]
- 83.Grasemann H, Ratjen F. Gender-specific disease modification by NOS3. Br J Haematol. 2004 Jul;126(1):160. doi: 10.1111/j.1365-2141.2004.05001.x. author reply 161. [DOI] [PubMed] [Google Scholar]
- 84.Sullivan KJ, Kissoon N, Duckworth LJ, et al. Low exhaled nitric oxide and a polymorphism in the NOS I gene is associated with acute chest syndrome. Am J Respir Crit Care Med. 2001 Dec 15;164(12):2186–90. doi: 10.1164/ajrccm.164.12.2012090. [DOI] [PubMed] [Google Scholar]
- 85.Chaar V, Tarer V, Etienne-Julan M, Diara JP, Elion J, Romana M. ET-1 and ecNOS gene polymorphisms andsusceptibility to acute chest syndrome and painful vaso-occlusive crises in children with sickle cell anemia. Haematologica. 2006 Sep;91(9):1277–8. [PubMed] [Google Scholar]
- 86.Styles LA, Schalkwijk CG, Aarsman AJ, Vichinsky EP, Lubin BH, Kuypers FA. Phospholipase A2 levels in acute chest syndrome of sickle cell disease. Blood. 1996 Mar 15;87(6):2573–8. [PubMed] [Google Scholar]
- 87.Nolan VG, Baldwin C, Ma Q, et al. Association of single nucleotide polymorphisms in klotho with priapism in sickle cell anaemia. Br J Haematol. 2005 Jan;128(2):266–72. doi: 10.1111/j.1365-2141.2004.05295.x. [DOI] [PubMed] [Google Scholar]
- 88.Elliott L, Ashley-Koch AE, De Castro L, et al. Genetic polymorphisms associated with priapism in sickle cell disease. Br J Haematol. 2007 May;137(3):262–7. doi: 10.1111/j.1365-2141.2007.06560.x. [DOI] [PubMed] [Google Scholar]
- 89.Koshy M, Entsuah R, Koranda A, et al. Leg ulcers in patients with sickle cell disease. Blood. 1989 Sep;74(4):1403–8. [PubMed] [Google Scholar]
- 90.Clare A, FitzHenley M, Harris J, Hambleton I, Serjeant GR. Chronic leg ulceration in homozygous sickle cell disease: the role of venous incompetence. Br J Haematol. 2002 Nov;119(2):567–571. doi: 10.1046/j.1365-2141.2002.03833.x. [DOI] [PubMed] [Google Scholar]
- 91.Akinyanju O, Akinsete I. Leg ulceration in sickle cell disease in Nigeria. Trop Geogr Med. 1979 Mar;31(1):87–91. [PubMed] [Google Scholar]
- 92.Knox-Macaulay HH. Sickle cell disease in Sierra Leone: a clinical and haematological analysis in older children and adults. Ann Trop Med Parasitol. 1983 Aug;77(4):411–9. doi: 10.1080/00034983.1983.11811730. [DOI] [PubMed] [Google Scholar]
- 93.Durosinmi MA, Gevao SM, Esan GJ. Chronic leg ulcers in sickle cell disease: experience in Ibadan, Nigeria. Afr J Med Med Sci. 1991 Mar;20(1):11–4. [PubMed] [Google Scholar]
- 94.Nolan VG, Ma Q, Cohen HT, et al. Estimated glomerular filtration rate in sickle cell anemia is associated with polymorphisms of bone morphogenetic protein receptor 1B. Am J Hematol. 2007 Mar;82(3):179–84. doi: 10.1002/ajh.20800. [DOI] [PubMed] [Google Scholar]
- 95.Lammert F, Matern S. The genetic background of cholesterol gallstone formation: an inventory of human lithogenic genes. Curr Drug Targets Immune Endocr Metabol Disord. 2005 Jun;5(2):163–70. doi: 10.2174/1568008054064841. [DOI] [PubMed] [Google Scholar]
- 96.Lammert F, Sauerbruch T. Mechanisms of disease: the genetic epidemiology of gallbladder stones. Nat Clin Pract Gastroenterol Hepatol. 2005 Sep;2(9):423–33. doi: 10.1038/ncpgasthep0257. [DOI] [PubMed] [Google Scholar]
- 97.Lammert F, Wang DQ. New insights into the genetic regulation of intestinal cholesterol absorption. Gastroenterology. 2005 Aug;129(2):718–34. doi: 10.1016/j.gastro.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 98.Bosma PJ, Chowdhury JR, Bakker C, et al. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert’s syndrome. N Engl J Med. 1995 Nov 2;333(18):1171–5. doi: 10.1056/NEJM199511023331802. [DOI] [PubMed] [Google Scholar]
- 99.Monaghan G, Ryan M, Seddon R, Hume R, Burchell B. Genetic variation in bilirubin UPD-glucuronosyltransferase gene promoter and Gilbert’s syndrome. Lancet. 1996 Mar 2;347(9001):578–81. doi: 10.1016/s0140-6736(96)91273-8. [DOI] [PubMed] [Google Scholar]
- 100.Beutler E, Gelbart T, Demina A. Racial variability in the UDP-glucuronosyltransferase 1 (UGT1A1) promoter: a balanced polymorphism for regulation of bilirubin metabolism? Proc Natl Acad Sci U S A. 1998 Jul 7;95(14):8170–4. doi: 10.1073/pnas.95.14.8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Passon RG, Howard TA, Zimmerman SA, Schultz WH, Ware RE. Influence of bilirubin uridine diphosphate-glucuronosyltransferase 1A promoter polymorphisms on serum bilirubin levels and cholelithiasis in children with sickle cell anemia. J Pediatr Hematol Oncol. 2001 Oct;23(7):448–51. doi: 10.1097/00043426-200110000-00011. [DOI] [PubMed] [Google Scholar]
- 102.Fertrin KY, Melo MB, Assis AM, Saad ST, Costa FF. UDP-glucuronosyltransferase 1 gene promoter polymorphism is associated with increased serum bilirubin levels and cholecystectomy in patients with sickle cell anemia. Clin Genet. 2003 Aug;64(2):160–2. doi: 10.1034/j.1399-0004.2003.00113.x. [DOI] [PubMed] [Google Scholar]
- 103.Haverfield EV, McKenzie CA, Forrester T, et al. UGT1A1 variation and gallstone formation in sickle cell disease. Blood. 2005 Feb 1;105(3):968–72. doi: 10.1182/blood-2004-02-0521. [DOI] [PubMed] [Google Scholar]
- 104.Heeney MM, Howard TA, Zimmerman SA, Ware RE. UGT1A promoter polymorphisms influence bilirubin response to hydroxyurea therapy in sickle cell anemia. J Lab Clin Med. 2003 Apr;141(4):279–82. doi: 10.1067/mlc.2003.28. [DOI] [PubMed] [Google Scholar]
- 105.Kinney TR, Helms RW, O’Branski EE, et al. Safety of hydroxyurea in children with sickle cell anemia: results of the HUG-KIDS study, a phase I/II trial. Pediatric Hydroxyurea Group. Blood. 1999 Sep 1;94(5):1550–4. [PubMed] [Google Scholar]
- 106.Chaar V, Keclard L, Diara JP, et al. Association of UGT1A1 polymorphism with prevalence and age at onset of cholelithiasis in sickle cell anemia. Haematologica. 2005 Feb;90(2):188–99. [PubMed] [Google Scholar]
- 107.Kalotychou V, Antonatou K, Tzanetea R, Terpos E, Loukopoulos D, Rombos Y. Analysis of the A(TA)(n)TAA configuration in the promoter region of the UGT1 A1 gene in Greek patients with thalassemia intermedia and sickle cell disease. Blood Cells Mol Dis. 2003 Jul-Aug;31(1):38–42. doi: 10.1016/s1079-9796(03)00118-9. [DOI] [PubMed] [Google Scholar]
- 108.Adewoye AH, Nolan VG, Ma Q, et al. Association of polymorphisms of IGF1R and genes in the transforming growth factor- beta/bone morphogenetic protein pathway with bacteremia in sickle cell anemia. Clin Infect Dis. 2006 Sep 1;43(5):593–8. doi: 10.1086/506356. [DOI] [PubMed] [Google Scholar]
- 109.Tamouza R, Neonato MG, Busson M, et al. Infectious complications in sickle cell disease are influenced by HLA class II alleles. Hum Immunol. 2002 Mar;63(3):194–99. doi: 10.1016/s0198-8859(01)00378-0. [DOI] [PubMed] [Google Scholar]
- 110.Tamouza R, Busson M, Fortier C, et al. HLA-E*0101 allele in homozygous state favors severe bacterial infections in sickle cell anemia. Hum Immunol. 2007 Oct;68(10):849–53. doi: 10.1016/j.humimm.2007.08.260. [DOI] [PubMed] [Google Scholar]
- 111.Costa RN, Conran N, Albuquerque DM, Soares PH, Saad ST, Costa FF. Association of the G-463A myeloperoxidase polymorphism with infection in sickle cell anemia. Haematologica. 2005 Jul;90(7):977–9. [PubMed] [Google Scholar]
- 112.Thomas AN, Pattison C, Serjeant GR. Causes of death in sickle-cell disease in Jamaica. Br Med J (Clin Res Ed) 1982 Aug 28 4;Sep 28 4;285(6342):633–5. doi: 10.1136/bmj.285.6342.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McKie KT, Hanevold CD, Hernandez C, Waller JL, Ortiz L, McKie KM. Prevalence, prevention, and treatment of microalbuminuria and proteinuria in children with sickle cell disease. J Pediatr Hematol Oncol. 2007 Mar;29(3):140–4. doi: 10.1097/MPH.0b013e3180335081. [DOI] [PubMed] [Google Scholar]
- 114.Thompson J, Reid M, Hambleton I, Serjeant GR. Albuminuria and renal function in homozygous sickle cell disease: observations from a cohort study. Arch Intern Med. 2007 Apr 9;167(7):701–8. doi: 10.1001/archinte.167.7.701. [DOI] [PubMed] [Google Scholar]
- 115.Ashley-Koch AE, Elliott L, Kail ME, et al. Identification of genetic polymorphisms associated with risk for pulmonary hypertension in sickle cell disease. Blood. 2008 Jun 15;111(12):5721–6. doi: 10.1182/blood-2007-02-074849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yildiz P, Oflaz H, Cine N, Erginel-Unaltuna N, Erzengin F, Yilmaz V. Gene polymorphisms of endothelial nitric oxide synthase enzyme associated with pulmonary hypertension in patients with COPD. Respir Med. 2003 Dec;97(12):1282–8. doi: 10.1016/j.rmed.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 117.Yildiz P, Oflaz H, Cine N, et al. Endothelial dysfunction in patients with asthma: the role of polymorphisms of ACE and endothelial NOS genes. J Asthma. 2004 Apr;41(2):159–66. doi: 10.1081/jas-120026073. [DOI] [PubMed] [Google Scholar]
- 118.Hines PC, Zen Q, Burney SN, et al. Novel epinephrine and cyclic AMP-mediated activation of BCAM/Lu-dependent sickle (SS) RBC adhesion. Blood. 2003 Apr 15;101(8):3281–7. doi: 10.1182/blood-2001-12-0289. [DOI] [PubMed] [Google Scholar]
- 119.Zennadi R, Hines PC, De Castro LM, Cartron JP, Parise LV, Telen MJ. Epinephrine acts through erythroid signaling pathways to activate sickle cell adhesion to endothelium via LW-alphavbeta3 interactions. Blood. 2004 Dec 1;104(12):3774–81. doi: 10.1182/blood-2004-01-0042. [DOI] [PubMed] [Google Scholar]
- 120.Zennadi R, Moeller BJ, Whalen EJ, et al. Epinephrine-induced activation of LW-mediated sickle cell adhesion and vaso-occlusion in vivo. Blood. 2007 Oct 1;110(7):2708–17. doi: 10.1182/blood-2006-11-056101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Eyler CE, Jackson T, Elliott LE, et al. beta(2)-Adrenergic receptor and adenylate cyclase gene polymorphisms affect sickle red cell adhesion. Br J Haematol. 2008 Apr;141(1):105–8. doi: 10.1111/j.1365-2141.2008.07008.x. [DOI] [PubMed] [Google Scholar]
- 122.Steinberg MH, Adewoye AH. Modifier genes and sickle cell anemia. Curr Opin Hematol. 2006 May;13(3):131–6. doi: 10.1097/01.moh.0000219656.50291.73. [DOI] [PubMed] [Google Scholar]
- 123.Steinberg MH, Nagel RL, Lawrence C, et al. Beta-globin gene haplotype in Hb SC disease. Am J Hematol. 1996 Jul;52(3):189–91. doi: 10.1002/(SICI)1096-8652(199607)52:3<189::AID-AJH9>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 124.Wierenga KJ, Hambleton IR, Lewis NA. Survival estimates for patients with homozygous sickle-cell disease in Jamaica: a clinic-based population study. Lancet. 2001 Mar 3;357(9257):680–3. doi: 10.1016/s0140-6736(00)04132-5. [DOI] [PubMed] [Google Scholar]
- 125.Pagnier J, Mears JG, Dunda-Belkhodja O, et al. Evidence for the multicentric origin of the sickle cell hemoglobin gene in Africa. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1771–3. doi: 10.1073/pnas.81.6.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nagel RL, Fabry ME. The many pathophysiologies of sickle cell anemia. Am J Hematol. 1985 Oct;20(2):195–9. doi: 10.1002/ajh.2830200216. [DOI] [PubMed] [Google Scholar]
- 127.Kurnit DM. Evolution of sickle variant gene. Lancet. 1979 Jan;1(8107):104. doi: 10.1016/s0140-6736(79)90093-x. [DOI] [PubMed] [Google Scholar]
- 128.Angastiniotis M, Modell B, Englezos P, Boulyjenkov V. Prevention and control of haemoglobinopathies. Bull World Health Organ. 1995;73(3):375–86. [PMC free article] [PubMed] [Google Scholar]
- 129.Ofori-Acquah SF OFK. Beyond National Borders: A global perspective on advances in sickle cell disease research and management, and new challenges in the genome era. In: Pace B, editor. Renaissance of Sickle Cell Disease Research in the Genome Era. London: Imperial College Press; 2007. pp. 333–45. [Google Scholar]
- 130.Barbujani G, Magagni A, Minch E, Cavalli-Sforza LL. An apportionment of human DNA diversity. Proc Natl Acad Sci U S A. 1997 Apr 29;94(9):4516–9. doi: 10.1073/pnas.94.9.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rosenberg NA, Pritchard JK, Weber JL, et al. Genetic structure of human populations. Science. 2002 Dec 20;298(5602):2381–5. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 132.Hibbert JM, Creary MS, Gee BE, Buchanan ID, Quarshie A, Hsu LL. Erythropoiesis and myocardial energy requirements contribute to the hypermetabolism of childhood sickle cell anemia. J Pediatr Gastroenterol Nutr. 2006 Nov;43(5):680–7. doi: 10.1097/01.mpg.0000228120.44606.d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Archer DR, Stiles JK, Newman GW, et al. C-reactive protein and interleukin-6 are decreased in transgenic sickle cell mice fed a high protein diet. J Nutr. 2008 Jun;138(6):1148–52. doi: 10.1093/jn/138.6.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sinha S, Mishra SK, Sharma S, et al. Polymorphisms of TNF-enhancer and gene for FcgammaRIIa correlate with the severity of falciparum malaria in the ethnically diverse Indian population. Malar J. 2008;7:13. doi: 10.1186/1475-2875-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Timmann C, Evans JA, Konig IR, et al. Genome-wide linkage analysis of malaria infection intensity and mild disease. PLoS Genet. 2007 Mar 23;3(3):e48. doi: 10.1371/journal.pgen.0030048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Marquet S, Doumbo O, Cabantous S, et al. A functional promoter variant in IL12B predisposes to cerebral malaria. Hum Mol Genet. 2008 Jul 15;17(14):2190–5. doi: 10.1093/hmg/ddn118. [DOI] [PubMed] [Google Scholar]
- 137.Soares-Souza GB, Tarazona-Santos E, Chanock SJ. Characterization of a candidate locus for malaria susceptibility in human populations: a (TA)n microsatellite in the promoter region of the CYBB gene, the gp91 phox subunit of the NADPH oxidase. Int J Immunogenet. 2008 Apr;35(2):107–9. doi: 10.1111/j.1744-313X.2008.00750.x. [DOI] [PubMed] [Google Scholar]
- 138.Cox SE, Doherty C, Atkinson SH, et al. Haplotype association between haptoglobin (Hp2) and Hp promoter SNP (A-61C) may explain previous controversy of haptoglobin and malaria protection. PLoS ONE. 2007;2(4):e362. doi: 10.1371/journal.pone.0000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Magi R, Pfeufer A, Nelis M, Montpetit A, Metspalu A, Remm M. Evaluating the performance of commercial whole-genome marker sets for capturing common genetic variation. BMC Genomics. 2007;8:159. doi: 10.1186/1471-2164-8-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Weatherall MW, Higgs DR, Weiss H, Weatherall DJ, Serjeant GR. Phenotype/genotype relationships in sickle cell disease: a pilot twin study. Clin Lab Haematol. 2005 Dec;27(6):384–90. doi: 10.1111/j.1365-2257.2005.00731.x. [DOI] [PubMed] [Google Scholar]
- 141.Nagel RL, Fabry ME. Sickle cell anemia as a multigenetic disease: new insights into the mechanism of painful crisis. Prog Clin Biol Res. 1984;165:93–102. [PubMed] [Google Scholar]
- 142.Lettre G, Sankaran VG, Bezerra MA, et al. DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc Natl Acad Sci U S A. 2008 Aug 19;105(33):11869–74. doi: 10.1073/pnas.0804799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sebastiani P, Wang L, Nolan VG, et al. Fetal hemoglobin in sickle cell anemia: Bayesian modeling of genetic associations. Am J Hematol. 2008 Mar;83(3):189–95. doi: 10.1002/ajh.21048. [DOI] [PubMed] [Google Scholar]
- 144.Uda M, Galanello R, Sanna S, et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci U S A. 2008 Feb 5;105(5):1620–5. doi: 10.1073/pnas.0711566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hoppe B, Heymann GA, Schoenemann C, Nagy M, Kiesewetter H, Salama A. Description of a novel HLA-B allele, B*5613, identified during HLA-typing using sequence-specific oligonucleotide hybridization and sequence-specific amplification. Tissue Antigens. 2004 Nov;64(5):616–8. doi: 10.1111/j.1399-0039.2004.00306.x. [DOI] [PubMed] [Google Scholar]
- 146.Styles LA, Hoppe C, Klitz W, Vichinsky E, Lubin B, Trachtenberg E. Evidence for HLA-related susceptibility for stroke in children with sickle cell disease. Blood. 2000 Jun 1;95(11):3562–7. [PubMed] [Google Scholar]
- 147.Tang DC, Prauner R, Liu W, et al. Polymorphisms within the angiotensinogen gene (GT-repeat) and the risk of stroke in pediatric patients with sickle cell disease: a case-control study. Am J Hematol. 2001 Nov;68(3):164–9. doi: 10.1002/ajh.1173. [DOI] [PubMed] [Google Scholar]
- 148.Nolan VG, Adewoye A, Baldwin C, et al. Sickle cell leg ulcers: associations with haemolysis and SNPs in Klotho, TEK and genes of the TGF-beta/BMP pathway. Br J Haematol. 2006 Jun;133(5):570–8. doi: 10.1111/j.1365-2141.2006.06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]