Abstract
BACKGROUND
JC virus (JCV) has been implicated in the pathogenesis of colorectal cancer; however, its role in premalignant lesions is unknown. The hypothesis that JCV DNA sequences and T-antigen (T-Ag) expression may be present in adenomatous polyps of the colon was tested. Furthermore, an association between JCV and microsatellite instability (MSI) was also sought in these lesions.
METHODS
DNA was extracted from 74 paraffin-embedded adenomatous polyps. JCV gene sequences were amplified by polymerase chain reaction (PCR), and the specificity confirmed by DNA sequencing. Immunohistochemical staining was performed to localize T-Ag expression in the adenomas using a monoclonal antibody. For microsatellite instability analysis, 5 mononucleotide repeat markers (BAT-25, BAT-26, NR-21, NR-24, and NR-27) were coamplified in a pentaplex PCR and analyzed for deletion mutations.
RESULTS
JCV T-Ag sequences were found in 82% (61 of 74) of adenomas, and T-Ag protein was expressed in 16% (12 of 74) of these polyps. The T-Ag staining was localized exclusively in the nuclei of adenoma cells, but never in the cytoplasm or the adjacent nonneoplastic cells. The prevalence of MSI-H and non-MSI-H (MSI-L/MSS) in adenomatous polyps was 9.5% (7 of 74) and 90.5% (67 of 74), respectively. Among the 61 adenomas that harbored JCV sequences, 8% (5 of 61) were MSI-H, and similarly among 12 adenomatous polyps expressing T-Ag protein 8% (1 of 12) of the adenomatous polyps were MSI-H.
CONCLUSIONS
JCV T-Ag DNA sequences are frequently present in adenomatous polyps of the colon, and T-Ag is expressed specifically in the nuclei of these premalignant lesions. This study indicates that JCV T-Ag is present in the early stage of colonic carcinogenesis. Future studies will be required to determine the molecular mechanism of carcinogenesis in these JCV-infected lesions.
Keywords: JC virus, polyomaviruses, T-antigen, genomic instability, microsatellite instability, adenomatous polyp of the colon, carcinogenesis
JC virus (JCV), a member of the polyomaviridae family, ubiquitously infects humans, and as many as 70% to 80% of the adult population have JCV-specific antibodies.1,2 Epidemiological studies have demonstrated that JCV infection takes place during early childhood and usually remains subclinical. After primary infection, JCV infection can be found in the kidneys, B lymphocytes, and gut mucosa. However, under conditions of severe immunosuppression such as AIDS, cancer chemotherapy, or organ transplantation the virus may become reactivated and induce the fatal demyelinating disease, progressive multifocal leukoencephalopathy (PML).3 In addition to its essential role in PML, there is mounting evidence that JCV may be associated with several human cancers even in the absence of immunosuppression or PML. JCV genomic sequences and onco-genic T-antigen (T-Ag) expression have been reported in a variety of human malignancies, including brain tumors,4,5 colon cancer,6–9 gastric cancer,10 and esophageal cancer.11
JCV is a 5.13 kb closed, circular, supercoiled, double-stranded DNA virus. The viral genome consists of 3 functional regions: the ‘early’ and ‘late’ coding regions and a noncoding regulatory region. The early region encodes the large T-Ag and small t-antigens (t-Ag), whereas the late region encodes the viral capsid proteins (VP1, VP2, and VP3) and an agnoprotein that is involved in viral assembly. The early and late regions are separated by a bidirectional, noncoding regulatory region, also known as the transcriptional control region (TCR). The TCR contains the origin of replication (ori), and the promoter and enhancer elements that control viral replication.12,13 Although the precise mechanisms responsible for JCV-induced cellular transformation and tumor development are not completely understood, it is believed that T-Ag plays a critical role in malignant transformation by interacting with several cell regulatory proteins, including p5314 and pRb,15 and also by modulating several critical growth signaling pathways, such as the insulin-like growth factor-I receptor (IGF-IR)16 and Wnt signaling pathways.17
Colorectal cancer (CRC) develops through a stepwise progression of multiple genetic and epigenetic alterations, which manifest the transition from normal colonic mucosa to adenocarcinomas via adenomas (or adenomatous polyps) of the colorectum. In CRCs, 3 types of genomic instability, microsatellite instability (MSI), chromosomal instability (CIN), and the CpG island methylator phenotype (CIMP), are frequently observed and can account for almost all CRCs. In sporadic CRCs, CIN and CIMP are represented in almost equal frequencies, and the MSI cancers arise as a consequence of methylation of hMLH1, a key DNA mismatch repair (MMR) gene.18 MSI is characterized by accumulation of somatic alterations in the length of simple repeat sequences called microsatellites19–21 and is representative of tumors from hereditary nonpolyposis colorectal cancer (HNPCC). MSI may be present in greater than 95% of CRCs and in 50% to 80% of colorectal adenomas in HNPCC patients.22–26 Conversely, MSI in sporadic colorectal tumors may be present in 10% to 15% of sporadic CRCs,19–21,27–30 and in less than 10% of sporadic adenomas.28,30,31 The molecular mechanisms responsible for both CIN and CIMP are elusive and are a matter of intense investigation.
Recently, work from our laboratory reported that JCV T-Ag expression frequently associates with both CIN and CIMP CRCs and suggested that this viral oncogene may be involved in CRC through multiple mechanisms of genetic and epigenetic instability.32 These data are consistent with several previous studies where JCV sequences were frequently demonstrated in as much as 80% to 90% of CRCs,6,8,32,33 whereas T-Ag protein expression was found in a smaller subset of these neoplasms.8,32 Interestingly, in these studies it was clearly noted that T-Ag expression was present in a tumor-specific manner, and was never present in the corresponding nonneoplastic tissues.8,32 JCV T-Ag DNA sequences have also been found in adenomatous polyps of the colon, but no studies have investigated JCV T-Ag protein expression in these premalignant polyps.9,34 These studies provide a strong rationale for the potential role of T-Ag in colon tumorigenesis. However, to appreciate the role of JCV infection and CRC it would be meaningful to determine the timing of JCV activation in the normal-adenoma-carcinoma multistep process. To the best of our knowledge no previous studies have interrogated this important question, providing strength to the role of JCV in various gastrointestinal cancers. In addition, unlike CRC, the relationship between JCV T-Ag expression and genomic instability in precursor adenomas is also currently uncertain. Therefore, the objectives of our study were to test the hypothesis that JCV is present, that T-Ag is expressed in sporadic adenomatous polyps of the colon, and to evaluate associations between JCV T-Ag expression and specific forms of genomic instability in these lesions.
MATERIALS AND METHODS
Adenomatous Polyp Specimens
We obtained 74 adenomatous colonic polyps, removed by colonoscopic biopsy or polypectomy at Baylor University Medical Center (Dallas, Tex), and excluded any patients with a personal or family history of hereditary colorectal neoplastic syndromes. The patients included 27 females and 47 males with a mean age of 63.7 (range, 41–80) years. On the basis of histology, 60 polyps were tubular, 9 were tubulovillous, and 5 were serrated adenomas. Anatomically, 40 polyps were present in the right colon (from the ascending colon to the proximal splenic flexure) and 34 were located in the left colon (from the distal splenic flexure to the rectum). The mean sizes of polyps were 6.5 × 3.9 × 2.9 (range, 15 × 10 × 9 to 3 × 2 × 1) mm. The pathologic diagnosis was confirmed by experienced pathologists; the study was approved by the Baylor Institutional Review Board.
DNA Extraction
Paraffin-embedded tissue specimens were sectioned into 5-lm slices. Genomic DNA was extracted from the microdissected tissues using the QIAamp DNA mini kit (Qiagen, Valencia, Calif) according to the manufacturer’s instructions. Extreme caution was taken to perform all preparatory polymerase chain reaction (PCR) steps, including DNA extraction, in a separate room completely isolated from any post-PCR samples to prevent contamination.
Detection of JCV T-Antigen Sequences
PCR amplification for JCV T-Ag sequences was performed using gene-specific primers, which amplified a 154-bp NH2-terminal region of JCV T-Ag as described previously.32 Sequences for the sense and antisense primers were 5′-ATGTATTCCACCAGGA TTCCCATTCATC-3′ (nucleotide positions 4381–4408) and 5′ -AGTTCTTGGAGACACCCCCTACAG-3′ (nucleotide positions 4511–4534), respectively. Each PCR reaction consisted of a 25-µL reaction mixture containing 200 ng of genomic DNA, 1× HotStarTaq master mix (Qiagen), and 0.75 mmol/L of each primer (Invitrogen, Carlsbad, Calif). The PCR reactions were carried out in a PTC 200 DNA Engine System (MJ Research, Watertown, Mass). PCR conditions were initial activation of HotStar Taq polymerase at 94°C for 15 minutes, followed by 40 cycles of denaturation at 94°C, annealing at 58°C, and extension at 72°C for 30 seconds. The extension time for the final cycle was 10 minutes. The gene-specific PCR primers and the assay conditions have been optimized in our laboratory and were observed to be very robust and optimal for the amplification of genomic DNA from both frozen and formalin-fixed, paraffin-embedded tissue specimens. Along with each PCR amplification we included a positive template control (with the same amount of DNA from the SW480 cell line that was stably transfected with JCV) and multiple template blanks (with no DNA, but containing all other reagents). The PCR products were electrophoresed on an agarose gel in an isolated post-PCR room in the laboratory. A tissue specimen was judged positive for T-Ag if a 154-bp band could be seen on the gel. To further confirm the specificity of PCR results, all positive PCR products were subsequently confirmed through automated sequencing using an ABI PRISM BigDye Terminator v. 1.1 Cycle Sequencing Kit on an ABI PRISM 3100 Avant Genetic Analyzer (Applied Biosystems, Foster City, Calif). The sequencing data obtained were aligned with JCV sequences to ensure that the sequences were of JCV origin only, and did not match other polyomaviruses, BK virus or SV40.
Immunohistochemical Staining for JCV T-Antigen
Paraffin-embedded tissue was sectioned to 5-µm thickness and mounted onto positively charged slides. We used JCV-inoculated hamster brain tumor tissue (kindly provided by Kamel Khalili, Temple University, Philadelphia, Pa) as a positive control and normal colon tissue sections from our hospital as a negative control (by adding all reagents including primary antibodies). Tissue sections were placed in an oven at 60°C for 40 minutes to melt the paraffin. The sections were deparaffinized in 3 changes of xylene for 30 minutes each and then rehydrated through a graded series of alcohols. For nonenzymatic antigen retrieval, sections were immersed in 10 mM citrate buffer (pH 6.0) and autoclaved at 100°C for 15 minutes. After an initial cooling period of 20 minutes, sections were incubated in 10% normal goat serum for 1 hour at room temperature to block nonspecific binding of immunoglobulin. Incubation with the primary antibodies was done overnight at room temperature in a humidified chamber. The primary antibody used in this study was a mouse monoclonal antibody specific for JCV T-Ag (clone PAb 2003, 1:100 dilution; kindly provided by Richard Frisque, Pennsylvania State University, Hershey). PAb2003 is a JCV T-Ag-specific monoclonal antibody and has comparable specificity to the commercially available PAb416 (anti-SV40-T-Ag antibody).35 Endogenous peroxidase was quenched by incubating in 3% H2O2/phosphate-buffered saline (PBS) for 10 minutes, followed by incubation with Dako EnVision+ Dual Link System peroxidase-labeled polymer (Dako Cytomation, Carpinteria, Calif) for 30 minutes at room temperature. Staining was developed by reaction with diaminobenzidine chromogen for 5–10 minutes and then counterstained with hematoxylin for 1–2 minutes. Sections were dehydrated through a graded series of alcohols, cleared in xylene, and mounted with Cytoseal 60 (Richard-Allan Scientific, Kalamazoo, Mich). The brown chromagen complexes indicated T-Ag expression.
Microsatellite Instability Analysis
Microsatellite analyses on all adenomatous polyp tissues were performed in a pentaplex PCR using 5 mononucleotide repeat markers, BAT-25, BAT-26, NR-21, NR-24, and NR-27, as described previously.36 These 5 markers have a comparable degree of sensitivity and specificity as the original panel of NCI markers, are quasi-monomorphic, do not require simultaneous amplification of matching normal DNA, and can be amplified in a single multiplex PCR-amplification reaction. Because of the these advantages the recent revised NCI guidelines support the use of these markers for MSI analyses.37 The anti-sense primer in each pair was labeled with a fluorescent dye, FAM for BAT-26 and NR-21, HEX for BAT-25 and NR-27, and NED for NR-24 (Table 1). Each PCR reaction consisted of a 25-µL reaction mixture containing 100 ng of genomic DNA, 1× HotStarTaq master mix (Qiagen) and 1 µM of each primer. The PCR reactions were carried out in a PTC 200 DNA Engine System (MJ Research). PCR conditions were an initial denaturation at 94°C for 15 minutes, followed by 40 cycles of denaturation at 94°C, annealing at 55°C, and extension at 72°C for 30 seconds. The extension time of the final cycle was 10 minutes. With each PCR we used a positive template control (with the same amount of DNA from microsatellite unstable colon cancer cell line HCT 116) and a template blank (with no DNA but containing all other reagents). Microsatellite instability was detected with the aid of an ABI PRISM 3100 Avant Genetic Analyzer, analyzed with GeneMapper software (v. 3.5; Applied Biosystems). We classified the MSI status of adenomatous polyps as MSI-high (MSI-H), MSI-low (MSI-L), or microsatellite stable (MSS). MSI-H was defined when a tumor specimen showed a definitive mutation of BAT-26 or mutations in ≥2 of the other 4 markers. MSI-L was defined as a tumor showing a mutation of any 1 of the markers other than BAT-26, and MSS when none of the markers was mutated.
TABLE 1.
Five Mononucleotide Repeat Markers Used in Pentaplex PCR for the Determination of MSI Status
| Name | Gene | GenBank Number |
Length and location of the repeat |
Primer sequences | Size bp |
Fluorescent marker |
|---|---|---|---|---|---|---|
| BAT-26 | hMSH2 | U04045 | 26 (T) | CTGCGGTAATCAAGTTTTTAG | 183 | FAM |
| Intron 5 | AACCATTCAACATTTTTAACCC | |||||
| BAT-25 | c-kit | X06182 | 25 (T) | TACCAGGTGGCAAAGGGCA | 153 | HEX |
| Intron 16 | TCTGCATTTTAACTATGGCTC | |||||
| NR-21 | SLC7A8 | XM_033393 | 21 (T) | GAGTCGCTGGCACAGTTCTA | 109 | FAM |
| 5’ UTR | CTGGTCACTCGCGTTTACAA | |||||
| NR-24 | Zinc finger 2 (ZNF-2) |
X60152 | 24 (T) | GCTGAATTTTACCTCCTGAC | 131 | NED |
| 3’ UTR | ATTGTGCCATTGCATTCCAA | |||||
| NR-27 | Inhibitor of apoptosis protein-1 |
AF070674 | 27 (A) | AACCATGCTTGCAAACCACT | 87 | HEX |
| 5’ UTR | CGATAATACTAGCAATGACC |
PCR indicates polymerase chain reaction; MSI, microsatellite instability; bp, basepair.
Statistical Analyses
Statistical analyses were performed using the Fisher exact test.
RESULTS
JCV T-Ag DNA Sequences Are Frequently Present in Adenomatous Polyps of the Colon
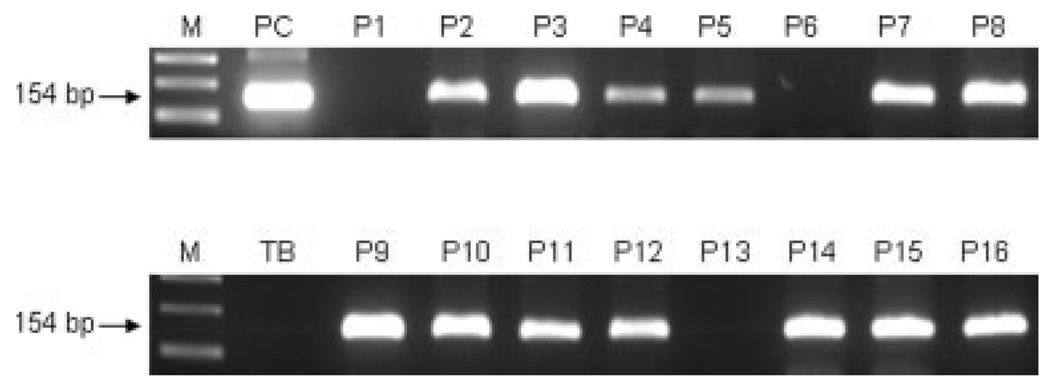
We designed PCR primers to specifically amplify JCV T-Ag sequences, which is possible because, although JCV T-Ag shares approximately 70% sequence homology with SV40 and BKV T-Ag at the amino acid level, the sequences are more variable at the nucleotide level. Figure 1 shows the results of PCR amplification for JCV T-Ag using these specific primers. We observed that 82.4% (61 of 74) of adenomatous polyps harbored JCV T-Ag sequences. Each T-Ag-positive PCR product was subsequently subjected to DNA sequencing and in each case the amplified sequences was confirmed to be JCV, and none matched other polyomaviruses, BK virus or SV40. We did not observe statistically significant associations between the presence of JCV T-Ag and any of the clinical-pathological data, including age, sex, polyp size, location, or pathologic features (Table 2).
FIGURE 1.
Polymerase chain reaction (PCR) amplification of JC virus (JCV) T-antigen (T-Ag) DNA sequences in adenomatous polyps of the colon using specific primers that amplify a 154-bp NH2-terminal region of JCV T-Ag. Top: Results from 8 different polyps and a positive control. Bottom: Results from 8 different polyps and a template blank. M = 100 bp size markers; P1-P16 = 16 different adenomatous polyp samples; PC = positive control from SW480 cells that were stably transfected with JCV; TB = template blank lacking DNA, but containing all other reagents.
TABLE 2.
Clinicopathologic Characteristics and Their Relationship With JCV T-Antigen Expression and MSI Status
| JCV T-Ag DNA |
JCV T-Ag protein |
MSI status |
||||
|---|---|---|---|---|---|---|
| Positive |
Negative |
Positive |
Negative |
MSI-H |
Non-MSI-H |
|
| 61/74 (82%) | 13/74 (18%) | 12/74 (16%) | 62/74 (84%) | 7/74 (9.5%) | 67/74 (90.5%) | |
| Age | ||||||
| ≤60, n = 25 | 20 (80%) | 5 (20%) | 2 (8%) | 23 (92%) | 2 (8%) | 23 (92%) |
| >60, n = 49 | 41 (84%) | 8 (16%) | 10 (20%) | 39 (80%) | 5 (10%) | 44 (90%) |
| Sex | ||||||
| Men, n = 47 | 37 (79%) | 10 (21%) | 7 (15%) | 40 (85%) | 3 (6%) | 44 (94%) |
| Women, n = 27 | 24 (89%) | 3 (11%) | 5 (19%) | 22 (81%) | 4 (15%) | 23 (85%) |
| Polyp Size | ||||||
| ≥10mm, n = 16 | 15 (94%) | 1 (6%) | 2 (12.5%) | 14 (87.5%) | 1 (6%) | 15 (94%) |
| <10mm, n = 58 | 46 (79%) | 12 (21%) | 10 (17%) | 48 (83%) | 6 (10%) | 52 (90%) |
| Location | ||||||
| Right colon, n = 40 | 33 (82.5%) | 7 (17.5%) | 5 (12.5%) | 35 (87.5%) | 4 (10%) | 36 (90%) |
| Left colon, n = 34 | 28 (82%) | 6 (18%) | 7 (21%) | 27 (79%) | 3 (9%) | 31 (91%) |
| Histologic type | ||||||
| Tubular, n = 60 | 49 (82%) | 11 (18%) | 6 (10%) | 54 (90%) | 7 (12%) | 53 (88%) |
| Tubulovillous, n = 9 | 8 (89%) | 1 (11%) | 4 (44%) | 5 (56%) | 0 (0%) | 9 (100%) |
| Serrated, n = 5 | 4 (80%) | 1 (20%) | 2 (40%) | 3 (60%) | 0 (0%) | 5 (100%) |
| Grade of dysplasia | ||||||
| High grade, n = 3 | 3 (100%) | 0 (0%) | 1 (33%) | 2 (67%) | 0 (0%) | 3 (100%) |
| Low grade, n = 71 | 58 (82%) | 13 (18%) | 11 (15%) | 60 (85%) | 7 (10%) | 64 (90%) |
JCV indicates JC virus; MSI, microsatellite instability; T-Ag, T-antigen.
T-Ag Protein Expression Is Present in Adenomatous Polyps
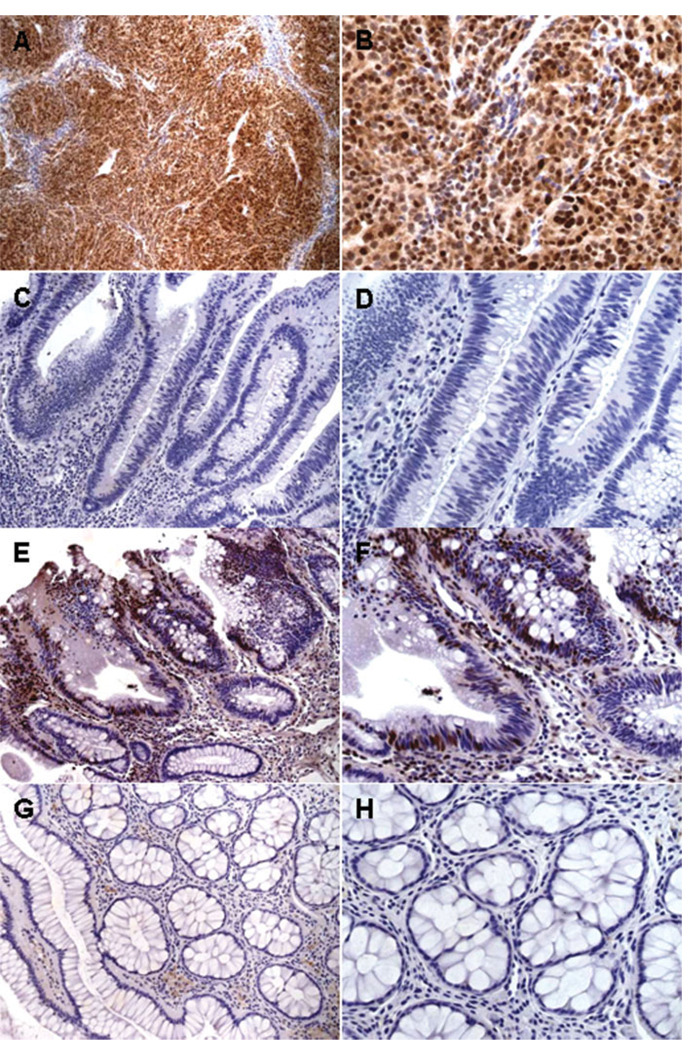
We performed immunohistochemical (IHC) staining to determine the prevalence of JCV T-Ag expression in the 74 adenomatous polyps. T-Ag expression data reported in this study were obtained using the JCV T-Ag-specific monoclonal antibody PAb2003, although PAb416 (the anti SV-40 T-Ag antibody that cross-reacts with JCV T-Ag) was also used for IHC and yielded concordant results (data not shown). Figure 2 shows IHC data, including a positive control (using JCV-inoculated hamster brain tumor tissue), positive and negative samples, and normal glandular epithelium adjacent to a positive sample. Overall, 16% (12 of 74) of the adenomatous polyps demonstrated nuclear T-Ag expression. As expected, all 12 T-Ag-positive adenomatous polyps harbored JCV T-Ag DNA sequences, but not all adenomatous polyps that carried T-Ag sequences expressed T-Ag protein, as shown in Table 3. As depicted in Figure 2A,B, T-Ag staining was exclusively present in the nuclei of almost all neurogenic tumor cells of hamster brain tumor tissue, but never showed positive staining in any the normal colon tissue controls. Similarly, as illustrated in Figure 2E–H, T-Ag expression was observed specifically in the nuclei of adenoma cells, was never cytoplasmic, and was not seen in any adjacent normal epithelial cells, which is consistent with the proposed role for this oncogenic protein in neoplastic transformation of normal colonic epithelial cells.
FIGURE 2.
Immunohistochemical detection of JC virus (JCV) T-antigen (T-Ag). (A,B) Examples of positive controls using JCV-inoculated hamster brain tumor tissue. T-Ag staining (shown in brown) was exclusively nuclear in all neurogenic tumor cells. (C,D) Examples of negative samples. No nuclear T-Ag staining was evident in any nuclei of the adenoma cells or lymphocytes. (E,F) Examples of T-Ag-expressing adenomatous cells and nonstained stromal lymphocytes. (G,H) The corresponding adjacent normal glandular portion of a positive sample. There was no evidence of immunoreactivity for T-Ag in this portion. Original magnification: ×100 (A), ×200 (C,E,G), ×400 (B,D,F,H).
TABLE 3.
Association of MSI Status With Expression of JCV T-Ag DNA Sequences and T-Ag Protein
| JCV T-Ag DNA |
JCV T-Ag Protein |
MSI Status |
|||||
|---|---|---|---|---|---|---|---|
| Positive, n = 61 | Negative, n = 13 | Positive, n = 12 | Negative, n = 62 | MSI-H, n = 7 | Non-MSI-H, n = 67 | ||
| JCV T-Ag DNA | Positive, n = 61 | — | — | 12 (20%) | 49 (80%) | 5 (8%) | 56 (92%) |
| Negative, n = 13 | — | — | 0 (0%) | 13 (100%) | 2 (15%) | 11 (85%) | |
| JCV T-Ag protein | Positive, n = 12 | 12 (100%) | 0 (0%) | — | — | 1 (8%) | 11 (92%) |
| Negative, n = 62 | 49 (79%) | 13 (21%) | — | — | 6 (10%) | 56 (90%) | |
| MSI status | MSI-H, n = 7 | 5 (71%) | 2 (29%) | 1 (14%) | 6 (86%) | — | — |
| non-MSI-H, n = 67 | 56 (84%) | 11 (16%) | 11 (16%) | 56 (84%) | — | — | |
JCV indicates JC virus; MSI, microsatellite instability; T-Ag, T-antigen.
T-Ag Expression May Be Present in Either MSI-H or Non-MSI-H Adenomas
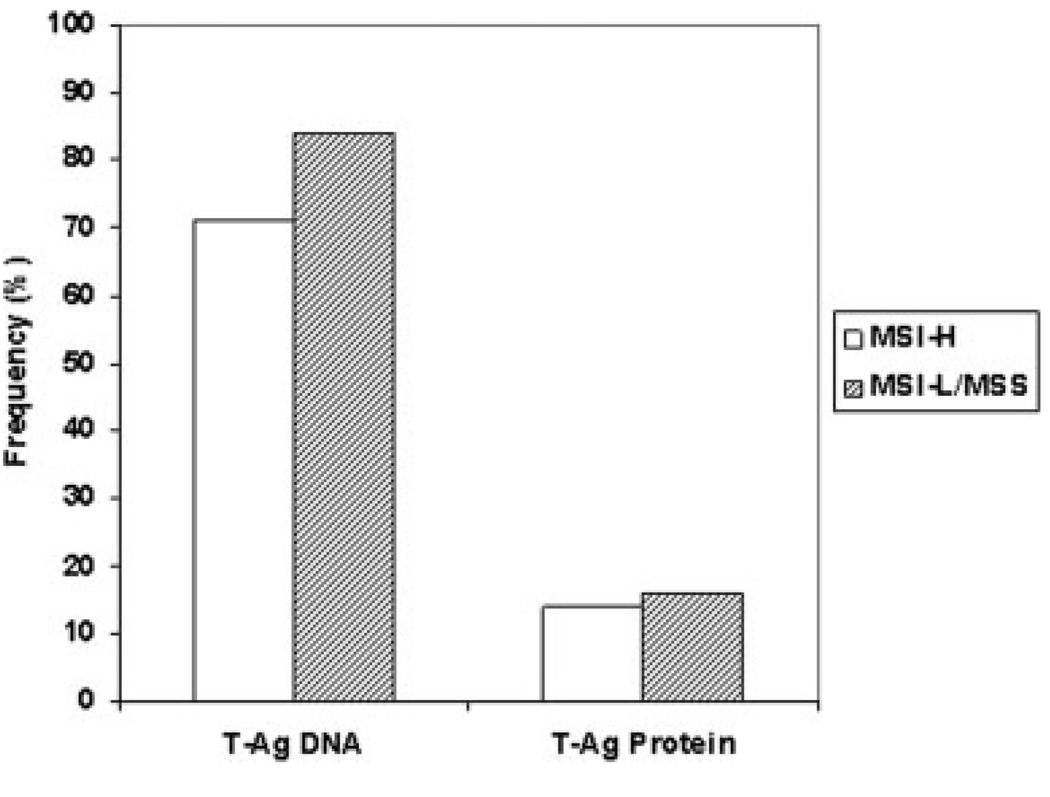
We performed pentaplex PCR for the detection of MSI in the 74 adenomatous polyps to determine whether there was any relationship between the presence of JCV T-Ag DNA sequences or T-Ag protein expression with this form of genomic instability in these lesions. We grouped MSI-L and MSS polyps together for comparative purposes because both have similar clinical, pathologic, and mutational features in colorectal cancers.38 Of the 74 adenomatous polyps studied, 9.5% (7 of 74) were MSI-H, and the remainder were MSI-L or MSS. MSI data were in agreement with IHC analyses for the loss of DNA MMR protein expression (data not shown). We observed that JCV TAg DNA sequences and protein expression were present in similar frequencies in MSI-H (5 of 7, 71% harbored DNA sequences; 1 of 7, 14% showed protein expression) and MSI-L/MSS (56 of 67, 84% harbored DNA sequences: 11 of 67, 16% showed protein expression) adenomatous polyps, as demonstrated in Table 3 and Figure 3. We also observed that among the 61 adenomas that harbored JCV sequences, 8% (5 of 61) were MSI-H, and similarly among 12 adenomatous polyps expressing T-Ag protein 8% (1 of 12) of the adenomatous polyps were MSI-H.
FIGURE 3.
Prevalence of T-antigen (T-Ag) DNA sequences and protein expression stratified by microsatellite instability (MSI) status in adenomatous polyps. T-Ag DNA sequences were present in comparable frequencies in polyps that are MSI-H (71%) or MSI-L/MSS (84%). By immunohistochemistry, T-Ag protein expression was detected in 14% and 16% of MSI-H and MSI-L/ MSS polyps. There were no statistically significant associations between T-Ag and MSI status.
In addition, we did not find any significant differences between MSI status and any of the clinicopathologic factors (Table 2). However, all 7 MSI-H polyps were tubular adenomas with low-grade dysplasia, limiting the power of this analysis.
DISCUSSION
In light of the compelling evidence that JCV has been implicated in CRC, and the suggestion that the T-Ag may be involved in CRC through multiple mechanisms of genetic and epigenetic instability,32 we were interested in extending our studies to adenomatous polyps of the colon. We report here that JCV T-Ag sequences have been detected in 82% of adenomatous polyps, similar to that observed in CRC,6,8,32,33 but at a higher frequency than has been reported previously.9,34 In addition, we detected JCV T-Ag protein expression in 16% of adenomatous polyps, which is about one-third the frequency seen in CRC.8,32
JCV T-Ag is a multifunctional oncogenic protein that can bind and break DNA, has helicase, α-polymerase, and ATPase activities,39–42 interacts with the tumor suppressor proteins p53, pRb, p130, and p107,14,15 and deregulates the IGF-IR and Wnt signaling pathways.16,17 In addition to its established role in causing PML in immunocompromised patients, attention has been drawn to the potential oncogenic role of T-Ag in gastrointestinal malignancies such colon cancer,6–9 stomach cancer,10 and esophageal cancer.11 Previous work from our laboratory and others has consistently shown the presence of T-Ag DNA sequences in the DNA extracted from both normal colonic epithelium and CRC tissues,6,8,43 but the exclusive expression of T-Ag protein in neoplastic cells using IHC.8,32 T-Ag expression in CRCs, and its absence in normal colonic epithelium, clearly underscored the need to investigate the presence and expression of T-Ag in adenomatous polyps to better understand the role of JCV in gastrointestinal carcinogenesis.
In a prior report, JCV T-Ag DNA sequences were detected in just 1 of 21 colorectal adenomas (4.8%) by nested PCR, but T-Ag protein expression was not investigated.34 In a quantitative (real time) PCR study, JCV T-Ag DNA sequences were detected in 15 of 25 adenomas (60%), but no T-Ag expression studies were performed. Interestingly, similar quantitative viral loads were found in DNA extracted from CRCs and colorectal adenomas (9000–20,000 viral copies per µg of extracted DNA), but the viral load was 100-fold lower in the adjacent colonic epithelium (50–450 viral copies per µg of DNA).9
Our data confirming the frequent presence of JCV sequences in sporadic adenomas of the colon is consistent with the hypothesis that JCV infection is ubiquitous, and that the gastrointestinal tract is a reservoir for this polyomavirus.43 However, the present study is the first demonstration of JCV sequences and simultaneous protein expression of T-Ag in colonic adenomas, because the prior 2 studies only investigated the presence of T-Ag DNA sequences.9,34 The frequency of T-Ag expression in colonic adenomas was less than what is found in CRCs (16% vs 40%–50%) and needs further exploration in future studies using larger sample sizes. However, it is possible that the timing of T-Ag protein expression in adenomas may be important. Perhaps T-Ag expression occurs principally in late-stage adenomas, or in early CRCs. Our studies examined each lesion at a single timepoint in its natural history.
From a technical standpoint, we were stringent with our criteria to score a polyp positive for T-Ag staining. Moreover, it is possible that the frequency of T-Ag expression would be higher if we had studied more advanced polyps, because a majority of the polyps in our study were obtained from biopsies rather than polypectomies, and provided us with a relatively small area to examine for T-Ag expression. Our finding that T-Ag is expressed in adenomas is relevant because the presence of JCV T-Ag DNA sequences alone does not prove an active biological role for this polyomavirus. Furthermore, T-Ag DNA sequences are frequently present even in normal, healthy individuals.6,8,43 Therefore, the demonstration of T-Ag expression in a subset of adenomatous polyps, its presence exclusively in the nuclei of adenoma cells, and its absence from normal cells provides additional evidence that the JCV may play an important role in the early stages of colorectal tumorigenesis.
Approximately 15% of sporadic CRCs demonstrate MSI,19–21,27–30 but the frequency of MSI is lower in sporadic colorectal adenomas.28,30,31 Kim et al.30 reported MSI in just 3 of 40 (7.5%) colorectal adenomas using BAT-26 as a solitary marker, whereas Ishiguro et al.31 reported that 10% of adenomas were MSI-H using multiple MSI markers including the 5 National Cancer Institute panel of markers. In this study we observed MSI-H in 9.5% of adenomas. More important, we were interested in the relationship between JCV T-Ag expression and genomic instability. However, we did not observe any significant association between T-Ag expression and MSI, suggesting that JCV T-Ag may involve other mechanisms of genomic instability such as CIN or CIMP, which could not be addressed in this study. Also, most of our polyps (60 of 74) were tubular adenomas, most had no more than low-grade dysplasia (71 of 74), and none were villous adenomas. Thus, events occurring in the later stages of adenoma development could have been missed in this study.
In conclusion, our results demonstrate that JCV T-Ag DNA sequences are frequently present in adenomatous polyps of the colon, and T-Ag expression can be found in these premalignant lesions. T-Ag was exclusively expressed in the nuclei of adenoma cells, but not in the corresponding normal epithelial cells. Therefore, this study suggests that JCV T-Ag is associated with the premalignant stage of colonic carcinogenesis. No specific relation was found between the presence of JCV DNA, the expression of T-Ag, and MSI. The evidence for the role of JCV in colorectal carcinogenesis continues to mount.
Acknowledgments
Supported by grant R-01 CA98572 from the NCI, NIH, and the Baylor Research Institute.
REFERENCES
- 1.Taguchi F, Kajioka J, Miyamura T. Prevalence rate and age of acquisition of antibodies against JC virus and BK virus in human sera. Microbiol Immunol. 1982;26:1057–1064. doi: 10.1111/j.1348-0421.1982.tb00254.x. [DOI] [PubMed] [Google Scholar]
- 2.Stolt A, Sasnauskas K, Koskela P, Lehtinen M, Dillner J. Seroepidemiology of the human polyomaviruses. J Gen Virol. 2003;84:1499–1504. doi: 10.1099/vir.0.18842-0. [DOI] [PubMed] [Google Scholar]
- 3.Gordon J, Khalili K. The human polyomavirus, JCV, and neurological diseases [review] Int J Mol Med. 1998;1:647–655. doi: 10.3892/ijmm.1.4.647. [DOI] [PubMed] [Google Scholar]
- 4.Del Valle L, Gordon J, Assimakopoulou M, et al. Detection of JC virus DNA sequences and expression of the viral regulatory protein T-antigen in tumors of the central nervous system. Cancer Res. 2001;61:4287–4293. [PubMed] [Google Scholar]
- 5.Del Valle L, Gordon J, Enam S, et al. Expression of human neurotropic polyomavirus JCV late gene product agnoprotein in human medulloblastoma. J Natl Cancer Inst. 2002;94:267–273. doi: 10.1093/jnci/94.4.267. [DOI] [PubMed] [Google Scholar]
- 6.Laghi L, Randolph AE, Chauhan DP, et al. JC virus DNA is present in the mucosa of the human colon and in colorectal cancers. Proc Natl Acad Sci U S A. 1999;96:7484–7489. doi: 10.1073/pnas.96.13.7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ricciardiello L, Chang DK, Laghi L, Goel A, Chang CL, Boland CR. Mad-1 is the exclusive JC virus strain present in the human colon, and its transcriptional control region has a deleted 98-base-pair sequence in colon cancer tissues. J Virol. 2001;75:1996–2001. doi: 10.1128/JVI.75.4.1996-2001.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enam S, Del Valle L, Lara C, et al. Association of human polyomavirus JCV with colon cancer: evidence for interaction of viral T-antigen and beta-catenin. Cancer Res. 2002;62:7093–7101. [PubMed] [Google Scholar]
- 9.Theodoropoulos G, Panoussopoulos D, Papaconstantinou I, et al. Assessment of JC polyoma virus in colon neoplasms. Dis Colon Rectum. 2005;48:86–91. doi: 10.1007/s10350-004-0737-2. [DOI] [PubMed] [Google Scholar]
- 10.Shin SK, Li MS, Fuerst F, et al. Oncogenic T-antigen of JC virus is present frequently in human gastric cancers. Cancer. 2006;107:481–488. doi: 10.1002/cncr.22028. [DOI] [PubMed] [Google Scholar]
- 11.Del Valle L, White MK, Enam S, et al. Detection of JC virus DNA sequences and expression of viral T antigen and agnoprotein in esophageal carcinoma. Cancer. 2005;103:516–527. doi: 10.1002/cncr.20806. [DOI] [PubMed] [Google Scholar]
- 12.Frisque RJ, Bream GL, Cannella MT. Human polyomavirus JC virus genome. J Virol. 1984;51:458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boland CR, Luciani MG, Gasche C, Goel A. Infection, inflammation, and gastrointestinal cancer. Gut. 2005;54:1321–1331. doi: 10.1136/gut.2004.060079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bollag B, Chuke WF, Frisque RJ. Hybrid genomes of the polyomaviruses JC virus, BK virus, and simian virus 40: identification of sequences important for efficient transformation. J Virol. 1989;63:863–872. doi: 10.1128/jvi.63.2.863-872.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyson N, Bernards R, Friend SH, et al. Large T antigens of many polyomaviruses are able to form complexes with the retinoblastoma protein. J Virol. 1990;64:1353–1356. doi: 10.1128/jvi.64.3.1353-1356.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Valle L, Wang JY, Lassak A, et al. Insulin-like growth factor I receptor signaling system in JC virus T antigeninduced primitive neuroectodermal tumors—medulloblastomas. J Neurovirol. 2002;8 suppl 2:138–147. doi: 10.1080/13550280290101111. [DOI] [PubMed] [Google Scholar]
- 17.Gan DD, Reiss K, Carrill T, et al. Involvement of Wnt signaling pathway in murine medulloblastoma induced by human neurotropic JC virus. Oncogene. 2001;20:4864–4870. doi: 10.1038/sj.onc.1204670. [DOI] [PubMed] [Google Scholar]
- 18.Goel A, Nagasaka T, Arnold CN, et al. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132:127–138. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 19.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 20.Aaltonen LA, Peltomaki P, Leach FS, et al. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 21.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 22.Aaltonen LA, Peltomaki P, Mecklin JP, et al. Replication errors in benign and malignant tumors from hereditary nonpolyposis colorectal cancer patients. Cancer Res. 1994;54:1645–1648. [PubMed] [Google Scholar]
- 23.Iino H, Simms L, Young J, et al. DNA microsatellite instability and mismatch repair protein loss in adenomas presenting in hereditary nonpolyposis colorectal cancer. Gut. 2000;47:37–42. doi: 10.1136/gut.47.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rijcken FE, Hollema H, Kleibeuker JH. Proximal adenomas in hereditary nonpolyposis colorectal cancer are prone to rapid malignant transformation. Gut. 2002;50:382–386. doi: 10.1136/gut.50.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Jong AE, Morreau H, van Puijenbroek M, et al. The role of mismatch repair gene defects in the development of adenomas in patients with HNPCC. Gastroenterology. 2004;126:42–48. doi: 10.1053/j.gastro.2003.10.043. [DOI] [PubMed] [Google Scholar]
- 26.Velayos FS, Allen BA, Conrad PG, et al. Low rate of microsatellite instability in young patients with adenomas: reassessing the Bethesda guidelines. Am J Gastroenterol. 2005;100:1143–1149. doi: 10.1111/j.1572-0241.2005.40862.x. [DOI] [PubMed] [Google Scholar]
- 27.Gryfe R, Kim H, Hsieh ET, et al. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 28.Brueckl WM, Jung A, Wein A, et al. Microsatellite instability in colorectal adenomas: relevance and clinical importance. Int J Colorectal Dis. 2000;15:189–196. doi: 10.1007/s003840000241. [DOI] [PubMed] [Google Scholar]
- 29.Benatti P, Gafa R, Barana D, et al. Microsatellite instability and colorectal cancer prognosis. Clin Cancer Res. 2005;11:8332–8340. doi: 10.1158/1078-0432.CCR-05-1030. [DOI] [PubMed] [Google Scholar]
- 30.Kim HC, Roh SA, Ga IH, Kim JS, Yu CS, Kim JC. CpG island methylation as an early event during adenoma progression in carcinogenesis of sporadic colorectal cancer. J Gastroenterol Hepatol. 2005;20:1920–1926. doi: 10.1111/j.1440-1746.2005.03943.x. [DOI] [PubMed] [Google Scholar]
- 31.Ishiguro K, Yoshida T, Yagishita H, Numata Y, Okayasu T. Epithelial and stromal genetic instability contributes to genesis of colorectal adenomas. Gut. 2006;55:695–702. doi: 10.1136/gut.2005.079459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goel A, Li MS, Nagasaka T, et al. Association of JC virus T-antigen expression with the methylator phenotype in sporadic colorectal cancers. Gastroenterology. 2006;130:1950–1961. doi: 10.1053/j.gastro.2006.02.061. [DOI] [PubMed] [Google Scholar]
- 33.Casini B, Borgese L, Del Nonno F, et al. Presence and incidence of DNA sequences of human polyomaviruses BKV and JCV in colorectal tumor tissues. Anticancer Res. 2005;25:1079–1085. [PubMed] [Google Scholar]
- 34.Hori R, Murai Y, Tsuneyama K, et al. Detection of JC virus DNA sequences in colorectal cancers in Japan. Virchows Arch. 2005;447:723–730. doi: 10.1007/s00428-005-0014-3. [DOI] [PubMed] [Google Scholar]
- 35.Munoz-Marmol AM, Mola G, Fernandez-Vasalo A, Vela E, Mate JL, Ariza A. JC virus early protein detection by immunohistochemistry in progressive multifocal leukoencephalopathy: a comparative study with in situ hybridization and polymerase chain reaction. J Neuropathol Exp Neurol. 2004;63:1124–1130. doi: 10.1093/jnen/63.11.1124. [DOI] [PubMed] [Google Scholar]
- 36.Suraweera N, Duval A, Reperant M, et al. Evaluation of tumor microsatellite instability using 5 quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 2002;123:1804–1811. doi: 10.1053/gast.2002.37070. [DOI] [PubMed] [Google Scholar]
- 37.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young J, Simms LA, Biden KG, et al. Features of colorectal cancers with high-level microsatellite instability occurring in familial and sporadic settings: parallel pathways of tumorigenesis. Am J Pathol. 2001;159:2107–2116. doi: 10.1016/S0002-9440(10)63062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pipas JM. Common and unique features of T antigens encoded by the polyomavirus group. J Virol. 1992;66:3979–3985. doi: 10.1128/jvi.66.7.3979-3985.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sullivan CS, Cantalupo P, Pipas JM. The molecular chaperone activity of simian virus 40 large T antigen is required to disrupt Rb-E2F family complexes by an ATP-dependent mechanism. Mol Cell Biol. 2000;20:6233–6243. doi: 10.1128/mcb.20.17.6233-6243.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bollag B, Prins C, Snyder EL, Frisque RJ. Purified JC virus T and T′ proteins differentially interact with the retinoblastoma family of tumor suppressor proteins. Virology. 2000;274:165–178. doi: 10.1006/viro.2000.0451. [DOI] [PubMed] [Google Scholar]
- 42.Khalili K, Del Valle L, Otte J, Weaver M, Gordon J. Human neurotropic polyomavirus, JCV, and its role in carcinogenesis. Oncogene. 2003;22:5181–5191. doi: 10.1038/sj.onc.1206559. [DOI] [PubMed] [Google Scholar]
- 43.Ricciardiello L, Laghi L, Ramamirtham P, et al. JC virus DNA sequences are frequently present in the human upper and lower gastrointestinal tract. Gastroenterology. 2000;119:1228–1235. doi: 10.1053/gast.2000.19269. [DOI] [PubMed] [Google Scholar]