SUMMARY
Heart diseases are the most common causes of morbidity and death in humans. Using cardiac-specific RNAi-silencing in Drosophila, we knocked-down 7061 evolutionarily conserved genes under conditions of stress. We present a first global road-map of pathways potentially playing conserved roles in the cardiovascular system. One critical pathway identified was the CCR4-Not complex implicated in transcriptional and post-transcriptional regulatory mechanisms. Silencing of the CCR4-Not components in adult Drosophila resulted in myofibrillar disarray and dilated cardiomyopathy. Heterozygous not3 knockout mice showed spontaneous impairment of cardiac contractility and increased susceptibility to heart failure. These heart defects were reversed via inhibition of HDACs suggesting a mechanistic link to epigenetic chromatin remodeling. In humans, we show that a common NOT3 SNP correlates with altered cardiac QT intervals, a known cause of lethal arrhythmias. Thus, our functional genome-wide screen in Drosophila can identify candidates that directly translate into conserved mammalian genes involved in heart function.
INTRODUCTION
Cardiovascular diseases are the most common cause of death in North America and Europe (Yusuf et al., 2001) killing more than 860,000 people annually in the United States (A.H.A., 2005; Lloyd-Jones et al., 2009). Moreover, 80 million people in the USA are estimated to suffer from cardiovascular diseases (A.H.A., 2005, Lloyd-Jones et al., 2009). Known or associated causes of cardiovascular disease include diabetes mellitus, inflammation, high cholesterol, hypertension, overweight and obesity, physical inactivity, or smoking (A.H.A., 2005; Lloyd-Jones et al., 2009). Although there have been great advances in the understanding of heart failure in recent decades (Mudd and Kass, 2008), there is still a gap in understanding the genetic causes and an unmet need for better therapies. In particular, the complex interplay of lifestyle, genetic susceptibilities, diseases, and aging have made it difficult to understand the underlying pathogenic principles (Yusuf et al., 2001). In addition to large scale genetic mapping and phenotyping in humans (Gordon et al., 1977; Morita et al., 2005; Nabel, 2003), a genetic dissection of the cardiovascular system in less complex model organisms would greatly facilitate the understanding of basic controls of cardiac physiology and mechanisms of disease.
Multiple proteins that control contraction in cardiomyocytes are highly conserved between species. For instance, the fly heart is capable of spontaneous rhythmic activity required for the circulation of hemolymph, and the same genes control heart rhythm in humans and flies (Ocorr et al., 2007a). In aging flies, the heartbeat becomes irregular with increased episodes of arrhythmias (Ocorr et al., 2007b), reminiscent of increased atrial fibrillation and heart failure in older humans (Lakatta and Levy, 2003). Moreover, genes involved in specification and differentiation of the heart are also conserved between Drosophila and mammals [reviewed in (Bodmer, 1995; Cripps and Olson, 2002; Ocorr et al., 2007a)]. Mutations in subunits of repolarizing voltage gated potassium channels IKr (eag, KCNH2) and IKs (kvLQT1, KCNQ1) perturb heart function in Drosophila and in vertebrates can cause long QT syndrome (Ocorr et al., 2007b; Sanguinetti and Tristani-Firouzi, 2006). Moreover, the sarco-endoplasmic reticulum Ca2+-ATPase (serca2a, ATP2A2) and the Ca2+-channel Cacophony control heart function also in Drosophila (Ray and Dowse, 2005; Sanyal et al., 2006). Thus, Drosophila has become a powerful genetic model system to identify conserved genes involved in heart function.
RESULTS
A Drosophila high throughput assay to identify candidate heart genes
To identify candidate genes for heart development and heart function (Fig. 1A), we used cardiac tissue-specific RNAi silencing of all genes that we identified as showing possible conservation between mammalian species and Drosophila melanogaster (Table S1A). TinCΔ4-Gal4 specifically drives expression in cardioblasts (Lo and Frasch, 2001) and has been previously used to study genes involved in heart function of the adult fly (Qian et al., 2008). Because RNAi-mediated downregulation of gene expression in many cases permits the circumvention of lethality commonly associated with classical mutations (Dietzl et al., 2007), cardiac-tissue specific TinCΔ4-Gal4 RNAi-mediated gene silencing therefore allowed us to assay the functional roles of the respective target genes in adult flies. Since elevated ambient temperature results in an increase in Drosophila heart rate (Paternostro et al., 2001; Ray and Dowse, 2005), we combined cardiac-tissue specific RNAi knockdown with an increased ambient temperature to reveal cardiac phenotypes under conditions of stress. Elevated temperature also enhances the activity of the UAS/Gal4 system, without affecting survival within the timeframe of the experiment (Fig. S1A).
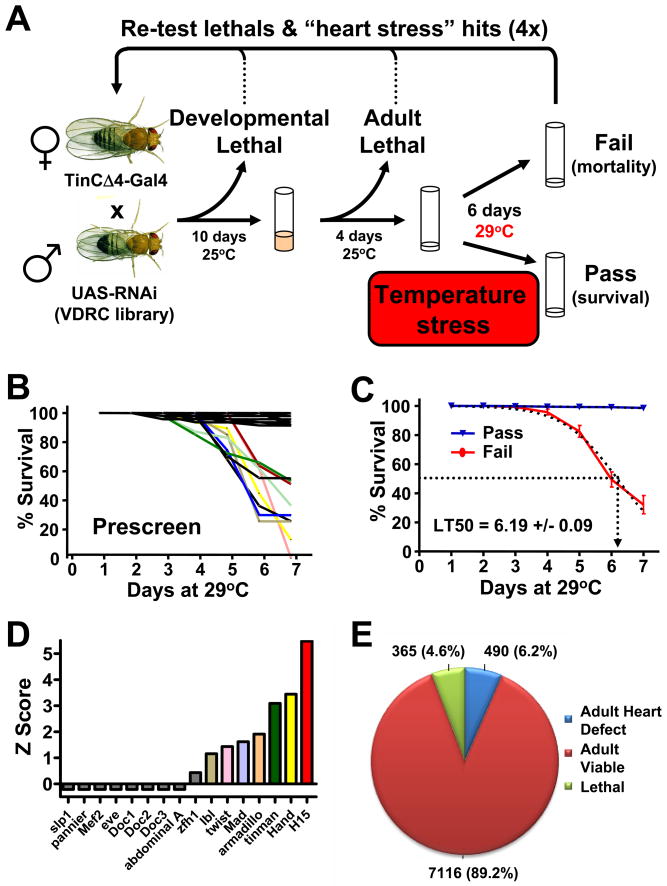
Figure 1. Genome wide screen for conserved heart genes.
(A) Schematic for screen setup. TinΔ4-Gal4, a cardiac tissue specific driver, was used to drive conserved UAS-RNAi hairpins in the developing heart. Developmental lethality and baseline adult viability was scored. Viable adult flies were then given a heart stress (continued exposure to 29°C) and survival was scored on day 6. Fly lines showing a potential developmental or heart function phenotype were then retested to confirm the candidate gene. (B) 80 randomly selected UAS-RNAi lines were crossed to TinCΔ4-Gal4 and evaluated for adult lethality following an increase in ambient temperate as cardiac stressor. Lines were either viable (black) or died starting around day 3. Data from individual lines are shown as % survival on the indicated days. (C) Mean responses from viable and failing (death after exposure to 29°C) flies revealed an average lethal time at which 50% of failing flies died (LT50) of 6.19 days. (D) Efficacy of TinCΔ4-Gal4 x UAS-RNAi lines to knock-down transcription factors known to play a role in heart formation. (E) Using this system a genome-wide screen was performed to search for conserved candidate genes for adult heart function under conditions of cardiac stress. 4.6% TinCΔ4-Gal4 x UAS-RNAi lines were developmental lethal. Among the 7971 viable lines, 490 transformant lines exhibited significantly increased death (Z-score >3, determined on day 6 after shifting the ambient temperature to 29°C). See also Figure S1 and Tables S1 and S2.
To evaluate the efficacy of this experimental set-up (Fig. 1A), we performed a pre-screen with 80 randomly selected genes that were targeted by TinCΔ4-Gal4_RNAi (Table S1B). Whereas ~10% of these TinCΔ4-Gal4 RNAi lines started to die at the increased ambient temperature, the vast majority survived for more than 7 days (Fig. 1B). From these pilot experiments we calculated an average time of 6.19 days at which 50% of flies among the susceptible lines had died (lethal time 50 (LT50) (Fig. 1C). Thus, our large scale genome-wide screen was carried out at 29°C and lethality was recorded for each line at day 6. As a control, TinCΔ4-Gal4/RNAi knockdown of known cardiogenic transcription factors resulted in viable lines at 25°C (not shown), but a shift to 29°C resulted in increased death of nearly half of the transcription factor RNAi lines tested, including Tinman, Hand, or H15 (neuromancer-1/Tbx20) (Fig. 1D). Cardiac knockdown of pannier/Gata4 and the Doc genes (Tbx5/6) did not cause premature lethality at 29°C, even though they are known to contribute to adult heart function (Qian and Bodmer, 2009; Qian et al., 2008). As negative controls we used RNAi lines targeting eve and zfh-1, which are not expressed in the myocardium targeted by TinCΔ4-Gal4 (Fig. 1D). Thus, we have set-up a model system that allows for efficient high-throughput screening and has the capacity to pick up known heart genes.
A genome-wide in vivo fly RNAi screen for conserved genes
In total we screened 8417 transgenic RNAi lines corresponding to 7061 conserved genes for potential developmental and adult heart functional defects (Table S1C). We only included 7971 lines representing 6751 genes that fit the previously defined criteria of specificity (Dietzl et al., 2007) for further analyses, i.e. only lines with an S19 score ≥ 0.8 and having 6 or less CAN repeats were considered specific (Table S1D). Progeny of each RNAi line crossed to TinCΔ4-Gal4 were first monitored for viability (reared at 25°C). Among these 7971 RNAi lines, 365 lines resulted in lethality (Fig. 1E and Table S1E) indicating that many of these genes function in heart development. Developmental lethality was further staged as lethal (embryonic lethal or we never observed any offspring), larval lethal, pupal lethal, or early adult (within 4 days after eclosion) lethal (Table S1F).
To identify candidate genes for adult heart function, we assayed 7804 adult TinCΔ4-Gal4_RNAi progeny (Dietzl et al., 2007) for survival after shifting the flies to 29°C (Fig. S1B–D). To categorize our hits from the screen, we used the Z score, which is a measure of the distance in standard deviations of a sample from the mean. All RNAi lines with a Z-score of 2 in the primary screen were tested on average 4.18 independent times (an average of 90 flies per genotype) using in some cases second RNAi transformants to control for transgenic insertion effects and second independent RNAi hairpins to target a different region of the gene (Table S2). After repeated screening we identified 498 genes that passed the more stringent Z-score of 3 (Fig. 1E and Table S2) indicating that these hits exhibit a death score of 3 standard deviations from the mean. Using gene ontology (GO) annotations, our candidate hits were classified according to their predicted biological processes (BP), molecular functions (MF), and cellular components (CC). Of the classified genes, those involved in signaling, ion transporter activity, metabolism and mitochondrial structure, development and morphogenesis, transcriptional regulation, or nucleic acid binding were highly represented among the entire data set (Fig. S2 and Table S4A). To remove any artificial bias in the gene list created by the ad-hoc Z score cut-off >3, we performed a Gene Set Analysis (GSA) to confirm enrichment of selected GO terms (Table S4B). In addition, 121 candidate heart genes had no annotated function by GO. Using panther (http://www.pantherdb.org/) we were able to functionally annotate 116 of these genes (Table S4C).
Given that the RNAi library screened is known to generate a level of false negative phenotypes due to inefficient targeting of genes to levels required to reveal phenotypes (Dietzl et al., 2007), and based on the assumption that our candidate heart hits perform some of their functions in protein complexes, we next identified first degree binding partners (Table S4D). Using this list of primary heart hits and their binding partners, we performed fly KEGG pathway analyses. Moreover, we included developmental lethal hits to generate a global interaction network. KEGG analyses showed enrichment of multiple pathways, such as mTOR signaling and PI3K/AKT, amino acid metabolism, JAK-STAT signaling, ErbB signaling, the Wnt, Notch, hedgehog, or TGFβ pathways, protein degradation, VEGF signaling, DNA repair, and Calcium homeostasis (Table S3 and Table S4E). Besides the identification of multiple known genes, our screen has also revealed hundreds of candidate genes and pathways that have not been previously associated with heart function.
A global view of heart function
To extend our Drosophila results to mammalian systems we used the power of data-mining and bioinformatics at a global systems level. Potential mouse and human orthologues of our candidate heart screen hits were evaluated for GO enrichment. The GO analyses of the human and mouse orthologues showed marked enrichment of genes involved in PIP3 and calcium signaling, ion transporter activity, metabolism, development, fatty acid metabolism, or muscle contraction (Table S4F). We next performed KEGG pathway as well as Broad Institute C2 gene set analysis on the mouse and human orthologues and their first degree binding partners. Based on the mammalian KEGG (Table S4E) and C2 (Table S4G) analyses, we found significant enrichment for gene sets involved in signaling, metabolism, ion channels, inflammation, aging, and transcription.
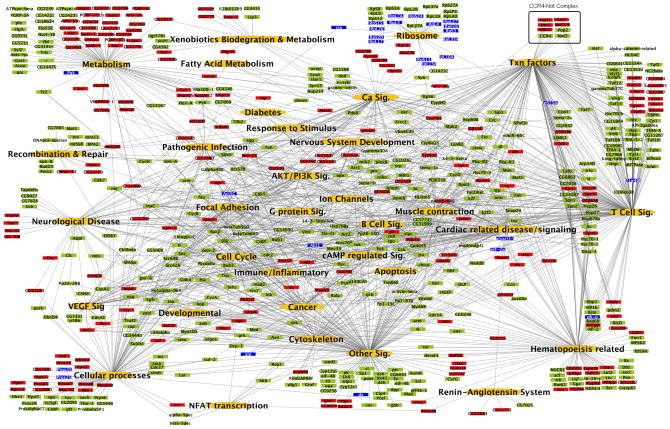
To generate a network map that includes our functional data in Drosophila, their human and mouse orthologues, and first degree binding partners, KEGG pathways from Drosophila, mouse and human were combined with relevant gene sets from the Broad Institute C2 annotations (Tables S4). A combined systems map and the interactions between the individual genes in the indicated nodes are shown in Fig. 2 and Table S4H. A systems map using only direct screening hits was also generated, yielding a comparable network map (Table S3). Importantly, using this network approach we identified multiple pathways known to play key roles in heart function and cardiovascular disease. For instance, we found significant enrichment in NFAT transcription, AKT activation and PI3K signaling, calcium signaling and muscle contraction, GPCR- and cAMP signaling, ion channels and proton-transporting ATPase complexes, and transcription. We also found associations with the renin-angiotensin system, a key pathway involved in cardiovascular function in humans (Fig. 2 and Table S4H). In support of our network approach, advanced data mining revealed that 171 of our primary fly hits and their first degree binding partners corresponded to mouse knock-outs with known cardiovascular phenotypes (Table S5). Thus, our genome-wide screen for candidate heart genes and in silico analyses provides a first attempt at a global road-map of essential molecular components and key pathways potentially involved in heart function and cardiac failure.
Figure 2. A global network of heart function.
The systems network includes data from the significantly enriched Drosophila KEGG and mouse and human KEGG and C2 data sets. Pathways and gene sets from the same biological processes were grouped into common functional categories. Green nodes represent statistically enriched functional categories of pathways; red nodes represent direct primary fly RNAi hits; light red nodes represent their first degree binding partners; and blue nodes indicate genes that were scored as developmentally lethal in our Drosophila heart screen. Lines indicate associations of the genes to the appropriate functional category. All KEGG pathways and selected C2 gene sets have been represented in the systems map. See also Tables S3–S5.
RNAi silencing of not3 and UBC4 result in dilated cardiomyopathy in Drosophila
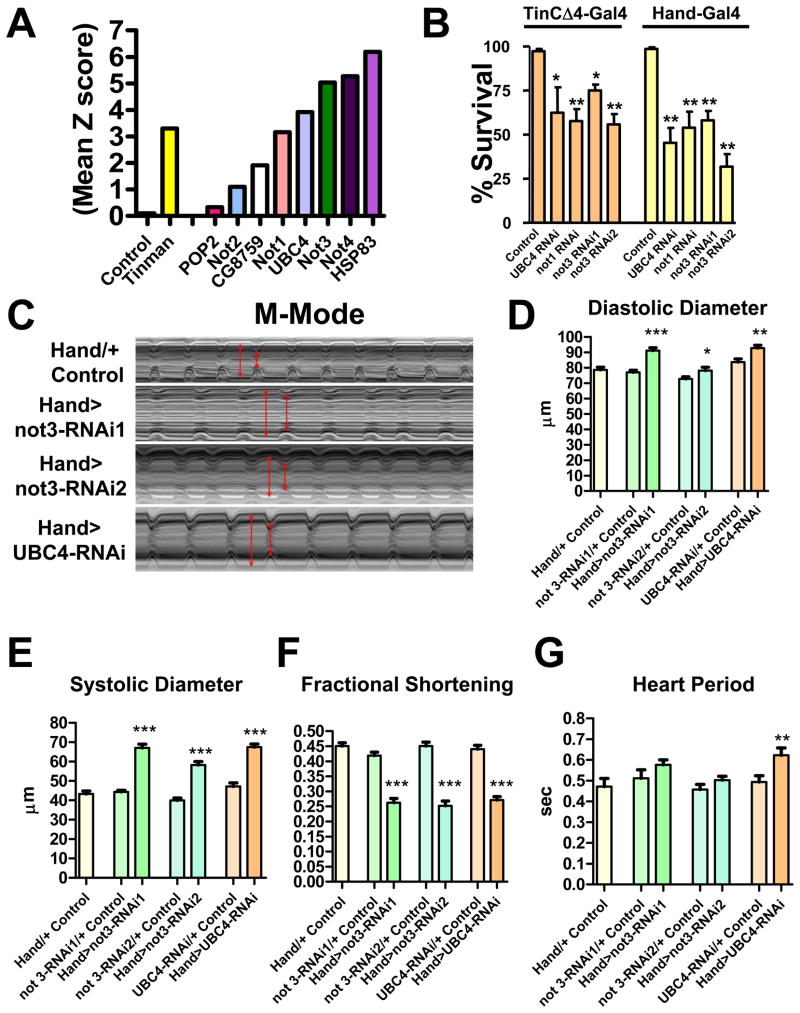
One of the novel pathways we found in our global network analyses was the CCR4-Not complex (Fig. 2 and Table S3). Intriguingly, among the 8 members of this complex assayed, we hit the subunits not1, not3 (not3/5 in fly), not4, UBC4, and Hsp83 (Fig. 3A). In addition, the subunits not2 and CG8759 were “weak” hits (Fig. 3A). The CCR4-Not complex was first identified in yeast (Denis, 1984) and is highly conserved in evolution (Albert et al., 2000). Components of the CCR4-Not complex have not yet been associated with cardiovascular function. We therefore re-tested components of this pathway using TinCΔ4-Gal4-driven knockdown in the heart, which confirmed the phenotype (Fig. 3B). Moreover, use of a second heart driver, Hand-Gal4, which is expressed with high specificity in myocardial and pericardial cells throughout development and in the adult fly heart (Han and Olson, 2005), showed that silencing of not1, not3, and UBC4 resulted in early death when adult flies were shifted to 29°C (Fig. 3B).
Figure 3. The CCR4-Not complex is a central regulator of adult heart function and loss of not3 results in dilated cardiomyopathy in Drosophila.
(A) Mean Z scores for TinCΔ4-Gal4 x UAS-RNAi lines targeting the indicated members of the fly CCR4-Not complex. A negative control (w1118 [isogenic to the RNAi library] X TinΔ4-Gal4) and the positive control Tinman RNAi line are shown. (B) Not1, not3, and UBC4 are essential for proper adult heart function in both Tinman and Hand-expressing cells. Data are shown as mean +/− SEM for at least 3 replicates. RNAi1 and RNAi2 indicate different transgenic hairpins targeting not3. * p<0.05, ** p<0.01 by ANOVA. (C) 1 week old adult flies with Hand-Gal4 driving not3 or UBC4 cardiac specific knock-down exhibit impaired heart function. M-modes provide traces of the heart contractions to document the movements in a 1 pixel-wide region of the heart tube over time. HandG4/+ control are the progeny of Hand-Gal4 crossed to w1118. Hand>Not3-RNAi are the progeny of Hand-Gal4 crossed to either UAS-not3-RNAi (−1 or −2) or UAS-UBC4-RNAi lines. Fly heart analysis was generated using a MatLab based image analysis program (Fink et al., 2009; Ocorr et al., 2007b). M-modes of the RNAi knockdown hearts reveal dilated diastolic and systolic diameters (double-headed red arrows) and reduced shortening properties (difference between diameters) when compared to M-modes of control hearts. Each trace represents a 5 second recording. (D–G) Not3 or UBC4 heart specific knock-down perturbs several indices of cardiac performance. Progeny of Hand-Gal4 crossed to two different UAS-not3-RNAi lines or an UAS-UBC4-RNAi line (experimental), and w1118 crossed to UAS-RNAi or Hand-Gal4 driver (controls) were used for these experiments as in C. not3 and UBC4 knockdown led to significantly wider (D) diastolic and (E) systolic diameters, and as a result significantly depressed (F) fractional shortening in all experimental lines relative to controls. (G) not3 knockdown trended toward a slight lengthening in the heart period (time between consecutive diastolic intervals) while UBC4 knockdown led to a significant increase in heart period. Mean values ± SEM are shown for each group (n = 29–40). Unpaired t-tests were performed between each Hand-Gal4>UAS-RNAi and each corresponding UAS-RNAi/+ control (progeny of w1118 crossed to UAS-RNAi line). Additionally, one-way ANOVAs with Bonferroni multiple comparison tests revealed no significant differences between the HandG4/+ control and all UAS-RNAi/+ control lines, for any cardiac parameter measured. * p<0.05, ** p<0.01, *** p<0.001. See also Figure S3 and Video S1.
Since not3 RNAi lines gave a strong phenotype with two different UAS-RNAi lines (Fig. 3B), we focused on the CCR4-Not component not3. Cardiac specific knockdown of not3 using two different RNAi lines (Hand>not3-RNAi: progeny from Hand-Gal4 crossed to UAS-not3-RNAi) significantly increased both diastolic and systolic diameters and resulted in a marked reduction in systolic fractional shortening relative to control flies (Fig. 3C–F and supplemental video 1). Hearts with cardiac not3 knockdown also showed slight increases in heart periods (Fig. 3G), however this was not statistically significant. Fluorescent microscopy revealed that not3 RNAi lines exhibit marked myofibrillar disarray, especially in the conical chamber (Fig. 4A–D). Heart-restricted not3-RNAi-mediated knockdown was confirmed by qRT-PCR (Fig. S3). In addition, we observed transcriptional downregulation of the Sarcoplasmic/endoplasmic reticulum calcium ATPase (Serca2a, ATP2A2), myosin heavy chain (mhc, MYH7), and the potassium channel KCNQ (kcnq1, KCNQ1) (Fig. S3) involved in heart rhythm control. Cardiac-specific knockdown of not3 increased the number of flies exhibiting contractile irregularities (Fig. S3H,I), a finding similar to what is seen in response to cardiac-specific knockdown of the KCNQ K+ channel (Fig. S3I) and what has been reported for KCNQ mutant flies (Ocorr et al, 2007). Of note, a not3 P-element mutant was developmentally lethal, exhibiting a late stage defect in embryonic heart tube organization, which could be rescued by P-element excision (Fig. S3C).
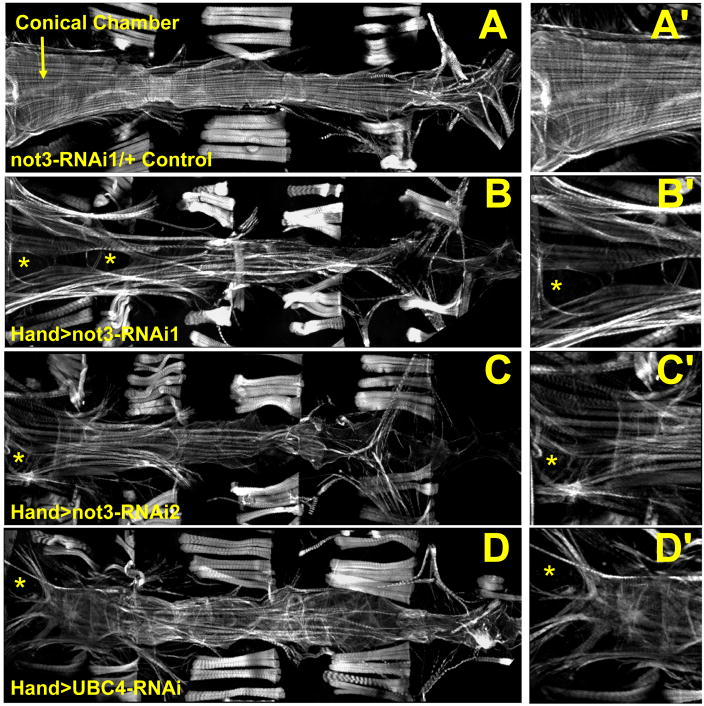
Figure 4. not3 and UBC4 cardiac specific RNAi-knockdown substantially perturb myofibrillar organization and content.
(A) Alexa584-phalloidin staining of control Drosophila cardiac tubes reveals typical spiraling myofibrillar arrangements within the cardiomyocytes. The fibers, especially those in the conical chamber, located anteriorly, are densely packed with f-actin. (B–D) Relative to control hearts, not3 or UBC4 RNAi knockdown severely disrupts myofibrillar organization and leads to an apparent loss of myofilaments as noted by large gaps in f-actin staining (*) as well as by a lack of myosin heavy chain transcripts (Fig. S4F). (A′-D′) Enlarged images of the conical chambers from A–D, respectively, which illustrate the high degree of myofibrillar disarray and large gaps in f-actin staining within the cardiomyocytes of not3 and UBC4 RNAi knockdown hearts. Original images taken at 10X magnification with a Zeiss Imager Z1 fluorescent microscope.
The CCR4-Not complex component UBC4 was also a major hit identified by our heart screen. Moreover, UBC4 expression was reduced following not3 knockdown (Fig. S3B). Hand-Gal4>UAS-UBC4-RNAi flies also exhibited significantly longer heart periods and showed dramatically altered diastolic and systolic diameters and reduced fractional shortening relative to control hearts (Fig. 3C–G). Fluorescent imaging again revealed severe myofibrillar disarray (Fig. 4D). that was strikingly similar to that observed in not3 knock-down hearts. Further, we observed similar structural and functional phenotypes in not1 cardiac-specific knock-down flies (A.C., G.N., J.P. and R.B., unpublished observation). Thus, knock-down of different components of the CCR4-Not complex result in abnormal heart structure and severely impaired cardiac function indicative of dilated cardiomyopathy.
Functional assessment of additional Drosophila heart hits
To extend our confirmations beyond the CCR4-Not complex, we assayed heart function in adult flies with heart-specific knock-down of four additional candidates identified in our heart screen (Fig. S3). One candidate heart gene tested was CG1216 (Mrityu), a mesoderm-expressed BTB-POZ domain containing protein (Rusconi and Challa, 2007). Cardiac knock-down of CG1216 resulted in a significant increase in systolic diameter. Another candidate heart gene is CG8933 (extradenticle), a PBX-family transcription factor. Cardiac knock-down of CG8933 resulted in increased systolic diameter and reduced fractional shortening. Cardiac knockdown of CG33261 (Trithorax-like) resulted in significantly altered diastolic and systolic diameters as well as impaired fractional shortening. Finally, knock-down of CG7371, a Vps52-domain containing protein predicted to participate in Golgi trafficking, resulted in a marked increase in heart period and affected the diastolic diameter. These data further demonstrate that our screen indeed has the capacity to identify novel factors involved in and required for normal adult heart function.
Generation of not3 knock-out mice
We next tested whether our data on Drosophila can be directly translated into a mammalian species. The mouse and human not3 proteins (official gene name cnot3) share 60% identity with the Drosophila not3 orthologue. Expression of human and mouse not3 mRNA transcripts can be found in the majority of tissues analysed. Although not3 is evolutionarily conserved from yeast to mammals, essentially nothing is known about the in vivo role of mammalian not3. We therefore generated not3 knock-out mice.
We disrupted the not3 gene in murine embryonic stem (ES) cells using a targeting vector in which nucleotides encompassing exons 2 through 9 are deleted (Fig. 5A and Fig. S4A). Both not3+/− male and not3+/− female mice are viable and exhibit normal fertility. We never obtained viable not3−/− newborn mice indicating that loss of not3 results in embryonic lethality. We staged embryonic development but failed to recover not3 null embryos from placental implantations (Fig. S4B,C). We therefore assayed early embryogenesis and observed that not3−/− blastocysts can develop. These mutant blastocysts have a normal appearance (Fig. S6D), occur at Mendelian frequencies (Fig. S4E,F), and express key markers of early embryonic differentiation at normal levels (Fig. S4G). not3 mRNA transcripts and not3 protein were undetectable in not3−/− blastocysts by RT-PCR and immunostaining (Fig. S4F,G). In not3+/+ and not3−/− epiblast cultures (Fig. S4H), trophoblast cells started to spread and supported the outgrowths of the inner cell mass (ICM). While the ICM of not3+/+ blastocysts continued to grow, not3−/− ICM cells exhibited a severe outgrowth defect. Thus, complete loss of mouse not3 results in early embryonic death at the implantation stage.
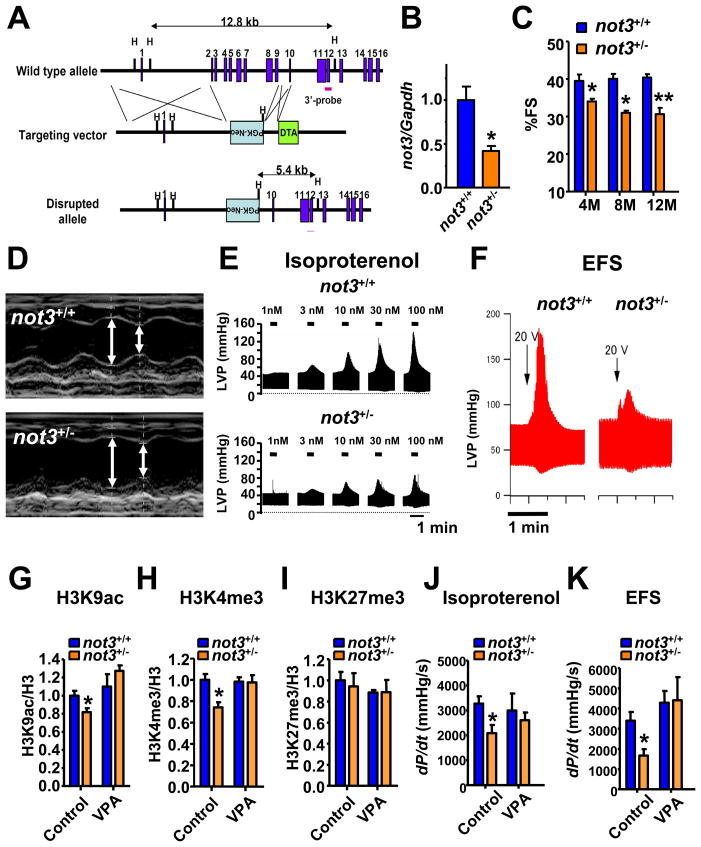
Figure 5. not3+/− mice exhibit reduced heart contractility, ex vivo function, and histone modifications that can be rescued by treatment with HDAC inhibitors.
(A) Gene targeting strategy. Exons 2 to 9 of the not3 gene (official symbol cnot3) were replaced with a PGK-Neo cassette by homologous recombination in A9 ES cells. The wild type allele, the targeting vector, and mutant allele, and the PGK-Neo and DTH selection cassettes are shown. Blue boxes indicate exons. (B) Real-time PCR analyses for not3 mRNA expression in 3 month old wild-type and not3+/− hearts. Values were normalized to gapdh mRNA expression. n = 6 mice per group. (C) not3+/− mice display a significant reduction in % fractional shortening at 4 months of age, which became more pronounced with age. n = 6–8 mice per group. Fractional shortening was determined by echocardiography. (D) Representative M-mode echocardiography for wild-type and not3+/− mice at 8 months of age. (E) Left ventricular pressure (LVP) measurements in isolated ex vivo not3+/− and not3+/+ hearts under isoproterenol perfusion. not3+/− hearts from 4 months old mice showed impaired contractile responses to different doses of isoproterenol perfusion in the retrograde Langendorff mode as compared to age matched controls. (F) Impaired contractile response of ex vivo not3+/− hearts to electrical field stimulation (EFS) compared with littermate not3+/+ hearts. Representative data for left ventricular pressure (LVP) at 20V stimulation are shown. (G) H3K9 acetylation (H3K9ac), (H) H3K4 trimethylation (H3K4me3) and (I) H3K27 trimethylation (H3K27me3) levels were analyzed by Western blot for acid-extracted histones from whole heart ventricles of wild type and not3+/− mice treated with vehicle or VPA (0.71% w/v in drinking water for 1 week) Band intensities were normalized to total H3 levels. (J and K) Treatment (1 week) with the HDAC inhibitor VPA rescue impaired ex vivo heart contractility of not3+/− hearts to isoproterenol (100nM) perfusion or 25V EFS. All values are mean +/− SEM. *; P < 0.05, **; P < 0.01. n = 5–12 per group. See also Figure S4 and S5.
not3 haploinsufficiency results in impaired heart function
We speculated that similar to RNAi-mediated down-regulation of not3 in Drosophila, not3 haploinsufficiency might also reveal a role in mammalian heart function. In not3 heterozygote mice, not3 expression is indeed downregulated in the heart (Fig. 5B). We failed to observe overt structural changes in the hearts of not3+/− mice. However, both male and female not3 haploinsufficient mice exhibited a reduction in cardiac contractility as determined by decreased left ventricle fractional shortening and increased left ventricular diameter in systole (Fig. 5C,D).
To address whether the defects in cardiac function are intrinsic to the heart per se or the observed impairment of contractility was secondary due to haploinsufficiency of not3 in other tissues, we subjected explanted hearts from wild-type and not3+/− littermate mice to Langendorff perfusion, assessing ex vivo heart function (Joza et al., 2005). When isoproterenol was used to activate β-adrenergic receptors, not3+/− hearts exhibited severe contractile abnormalities as defined by impaired generation of left ventricular pressure (LVP) (Fig. 5E and Fig. S5A). Hemodynamic measurements confirmed that all functional heart parameters such as dP/dTmax or dP/dTmin, indicative of generated contractile pressure, were markedly reduced in not3+/− hearts (not shown). Moreover, when explanted hearts were electrically stimulated, not3+/− hearts exhibited a striking defect in contractility (Fig. 5F). Thus, downregulation of not3 expression in not3 haploinsufficient mice results in an intrinsic impairment in heart function.
Yeast strains mutant for components of the CCR4-Not complex, including not3, display reduced acetylation levels of lysine residues on histone tails (e.g. H3K9) (Peng et al., 2008) and/or reduced trimethylation of H3K4 (Laribee et al., 2007). H3K9 acetylation and H3K4 trimethylation are indicative of transcriptionally active states of chromatin. Moreover, promoter regions of not3 target genes were shown to recruit trimethylated H3K4 in mouse ES cells (Hu et al., 2009) suggesting that not3 may regulate chromatin modifications. Our gene expression and bioinformatic analyses of mouse not3 knockout cells revealed that histone deacetylases (HDACs) and mRNA metabolisms are localized central in gene networks (unpublished). We therefore assessed the state of histone modifications in hearts from not3+/− mice. Histone extracts of whole hearts from not3 haploinsufficient mice showed a slight but significant reduction in active histone marks such as acetylation of H3K9 and trimethylation of H3K4 (Fig. 5G, 5H and Fig S5). H3K27 trimethylation was not changed (Fig. 5I and Fig S5). Treatment of not3+/− hearts with the HDAC inhibitor VPA restored the reduced acetylation of H3K9 and H3K4 trimethylation to that of wild type levels (Fig. 5G–I and Fig. S5). Most importantly, administration of HDAC inhibitors rescued the impairment in heart function in not3+/− mice; i.e. ex vivo heart functions of VPA treated mice were similar to control mice in response to both isoproterenol (Fig. 5J) and electrical stimulation (Fig. 5K). These data were confirmed using TSA, a second HDAC inhibitor (Fig. S5H,I). Taken together, not3+/− mice exhibit a spontaneous and intrinsic defect in cardiac function which can be rescued with HDAC inhibitors.
not3+/− mice develop severe cardiomyopathy in response to cardiac stress
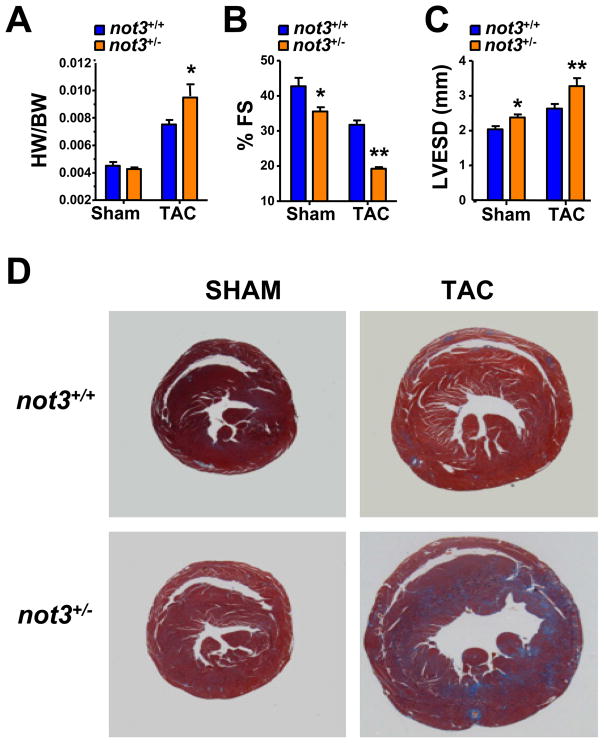
We next exposed control and not3+/− littermates to chronic pressure overload by surgical constriction of the aorta (transverse aortic constriction, TAC). Three weeks after TAC, heart weight/body weight ratios (HW/BW) increased in both not3+/+ and not3+/− mice, albeit this increase was significantly larger in the not3+/− mice (Fig. 6A). Cardiac hypertrophy was also seen by histology (Fig. 6D and S6A). Aortic banding of not3+/− mice resulted in severe heart failure characterized by decreased fractional shortening (Fig. 6B) and a dilation of the left ventrical as determined by echocardiography (Fig. 6C). In addition, not3+/− mice develop severe cardiac fibrosis following TAC, as shown by Masson-trichrome staining of hearts 3 weeks after TAC (Fig. 6D and Fig. S6B). Thus, not3+/− mice develop severe symptoms of heart failure in response to cardiac stress.
Figure 6. Not3+/− mice exhibit severe heart failure in response to pressure overload.
(A) Heart weight to body weight ratios (HW/BW) in not3+/− and not3+/+ littermate mice 3 weeks after transverse aortic constriction (TAC). Animals receiving sham surgery are shown as controls. (B) and (C) Echocardiography of male not3+/− and wild type littermates 3 weeks after TAC. not3+/− mice with TAC show (B) decreased % fractional shortening (%FS) and (C) increased left ventricular diameter in systolic phase (LVESD) compared with not3+/+ mice that received TAC. (D) Representative sections of not3+/+ and not3+/− hearts analyzed 3 weeks after sham or TAC surgery. Masson-trichrome staining are shown to visualize collagen deposits indicative of fibrotic changes. Note the severe cardiac hypertrophy and ventricular dilation in not3+/− mice following TAC.
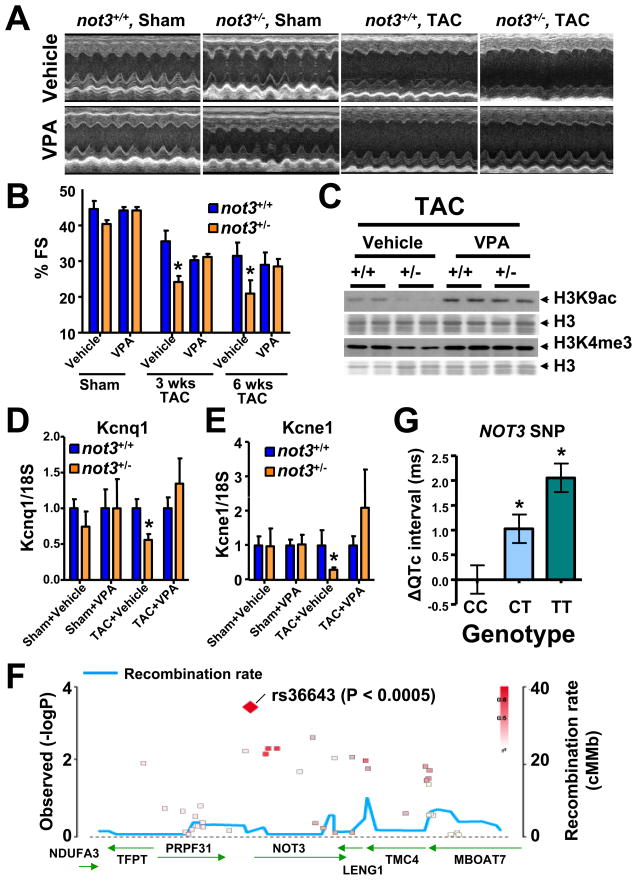
We next assessed whether HDAC inhibitors can also rescue stress-induced heart failure. HDAC inhibitor treatment could indeed block the augmented loss of cardiac function observed in not3+/− mice following TAC (Fig. 7A,B). In vivo treatment of not3+/− mice with HDAC inhibitors also blocked the exaggerated induction of heart failure markers such as ANF (Fig. S6C) and βMyhc (Fig. S6D). Moreover, treatment with an HDAC inhibitor restored the observed histone alterations in not3+/− mice to that of wild type littermates (Fig. 7C and Fig. S6E,F). Thus, not3 haploinsufficiency results in exaggerated heart failure which can be rescued by HDAC inhibition in vivo.
Figure 7. Not3 is a conserved regulator of heart function.
(A and B) Rescue of severe heart failure in TAC not3+/− hearts by the HDAC inhibitor VPA. One day after TAC or sham surgery the mice received treatment with vehicle or VPA (0.71% w/v in drinking water) for 6 weeks. (A) Representative M-mode echocardiography and (B) % FS in not3+/− and not3+/+ littermate mice 6 weeks after TAC or sham surgery with or without VPA treatment. (C) Reduced H3K9 acetylation (H3K9ac) and H3K4 trimethylation (H3K4me3) levels were rescued by VPA treatment. Acid-extracted histones from the hearts 6 weeks after TAC surgery were immunoblotted with antibodies for H3K9ac and H3K4me3. H3 is shown as a loading control. (D and E) Real time PCR analyses for the QT interval-associated potassium channel genes Kcnq1 and Kcne1. Total RNA was isolated from hearts 6 weeks after TAC or sham surgery with or without VPA treatment, and Kcnq1 and Kcne1 mRNA levels were measured and normalized to 18S mRNA. Data are shown as fold changes compared to not3+/+ mice for each treatment group. Values are mean +/− SEM. *; P < 0.05, **; P < 0.01. n = 5–10 per group. (F) Regional visualization of the association signal between common variants in the NOT3 region and the adjusted QT interval (QTc). SNP rs36643 in the 5′ region of NOT3 (−969bp from the transcription start and −924 from the TATA box) showed a significant regional association (p=0.000366). (G) Association between the T allele of SNP rs36643 and a prolongation of QTc. * P< 0.0005 from linear regression with inverse variance weighting using an additive genetic model. Data is derived from a meta-analysis of genome-wide association scans in several populations (Pfeufer et al., 2009). See also Figure S6.
A common genomic variation in the NOT3 promoter correlates with cardiac repolarization duration in humans
Using an in silico search to identify not3 target genes, we found that not3 has been shown to bind to the Kcnq1 promoter in ES cells (Hu et al., 2009). Kcnq1 encodes the α-subunit of the repolarizing voltage gated potassium channel IKs, mutations in which are the most common cause of long-QT syndrome (LQT1) in humans (Wang et al., 1996). Abnormalities of cardiac repolarization, measured as alterations in QT interval, predispose to sudden cardiac death in humans (Moss and Kass, 2005). Indeed, while sham-operated not3+/− mice exhibit a subtle reduction in cardiac Kcnq1 expression, this decrease was pronounced following TAC (Fig. 7D). Reduced Kcnq1 expression was rescued following HDAC inhibitor treatment. Also for Kcne1, the β-subunit of IKs, we observed a TAC-inducible and HDAC-sensitive defect in expression in not3+/− hearts (Fig. 7E). In fly not3-RNAi hearts, KCNQ expression is also reduced (Fig. S3D), and these flies exhibit cardiac contractile irregularities (Fig. S3H,I).
Recently two consortia have published genome-wide association studies (GWAS) for QT interval, QTSCD (Pfeufer et al., 2009) and QTGEN (Newton-Cheh et al, 2009). One of the 12 identified genomic regions contains the NOT1 gene which we also found as a hit in our Drosophila screen (Fig. 3A and B). Due to the stringent requirements to achieve a genome-wide significance threshold of p<5×10−8 (Dudbridge and Gusnanto, 2008), many genuinely associated alleles will be missed because of both a failure to exceed this statistical threshold and the absence of functional confirmatory data for genes within loci of interest. We therefore evaluated whether common variants in and near the human NOT3 locus are associated with alterations in QT interval. Intriguingly, SNP rs36643 (chromosome 19: 59,3 Mb), located in the promoter region ~969 base pairs upstream from the NOT3 transcriptional start site (924 bases upstream of the TATA box), significantly associates with QT interval in the QTSCD dataset (Fig. 7F). Patients carrying the common T allele (MAF = 0.65) showed a dose-dependent increase in QT intervals (ES= +1.03±0.29 ms QT interval per copy of T-allele, p=3.66E−04) (Fig. 7G). Of note, similar to adult kcnq1 mutant mice (Nerbonne, 2004), we did not observe an increased QT interval in not3 heterozygous mice (except for one mouse with arrhythmia, K.K., M.M., H.Y. and K.F. unpublished). Thus, our genome-wide screening data for death in flies can be used to identify candidate variants in humans that predispose individuals to heart disease, i.e. in the case of Not3 to arrhythmia and sudden death.
DISCUSSION
Here we present the first in vivo RNAi adult heart screen in Drosophila assaying conserved genes. Using functional imaging, we were able to observe cardiac defects in all flies with heart-specific knockdown of candidate genes evaluated to date. Our experimental approach to screen for conserved heart genes in Drosophila in concert with advanced bioinformatics has the potency to reveal human and mouse genes involved in heart function and heart disease. Moreover, we uncovered a plethora of additional genes, a large proportion of which had completely unknown functions until now. Future experiments are required to test whether our novel candidate genes indeed control cardiac development, regulate adult heart function, and/or influence the outcome of heart failure in response to cardiac stress.
One pathway we identified was the CCR4-Not complex. Functional heart analyses in Drosophila confirmed that RNAi-mediated silencing of the CCR4-Not components not3 and UBC4 resulted in a severe impairment of cardiac function that resembles dilated cardiomyopathy in experimental mouse models and human patients. To provide a first proof-of-principle that our fly hits can indeed have similar functions in the more complex mammalian heart, we generated knock-out mice for a component of the CCR4-Not complex. not3 haploinsufficient mice develop spontaneous impairment of heart function and severe heart failure following aortic banding. Mechanistically, not3 downregulation results in a defect in active histone marks and cardiac defects observed in not3+/− mice could be rescued by treatment with HDAC inhibitors. Besides regulating transcriptionally active states of chromatin (Hu et al., 2009; Jayne et al., 2006; Laribee et al., 2007; Peng et al., 2008), the CCR4-Not complex has also been implicated in RNA deadenylation (Tucker et al., 2001) and microRNA-mediated mRNA degradation (Behm-Ansmant et al., 2006). Thus, we cannot exclude that CCR4-Not components affect additional mechanisms regulating heart function. Importantly, our work on not3 in flies and mice has also allowed us to identify a SNP in the human NOT3 promoter that is associated with prolonged QT intervals and sudden cardiac death in humans. Thus, large scale screens in Drosophila can be directly translated to mammalian species, and in combination with other genome-wide approaches, can reveal novel regulators of heart function and heart failure.
EXPERIMENTAL PROCEDURES
Detailed experimental procedures are provided in the Supplemental Data.
Fly stocks
All RNAi transgenic fly lines were obtained from the VDRC RNAi stocks (Dietzl et al., 2007). The cardiac-tissue specific TinCΔ4 12a-Gal4 was a kind gift from Manfred Frasch, (Lo and Frasch, 2001) and Hand-Gal4 was a gift from Eric Olsen (Han and Olson, 2005).
Screening system
Transgenic RNAi males were crossed to TinCΔ4 virgin females. Viable lines were then incubated at 29°C for 6 days to expose flies to temperature stress (Paternostro et al., 2001). Initially a Z score cut-off of 2 (Mean control-test)/SD was used to select RNAi lines for re-testing.
Drosophila cardiac function, morphology and gene expression
UAS-RNAi fly lines obtained from the Vienna Drosophila RNAi Center were crossed to Hand-Gal4 (II) driver-flies and to w1118 wild type control flies. Flies were assessed for heart morphology and physiology using imaging (Ocorr et al., 2007b). M-modes were generated and cardiac parameters including heart periods, diastolic and systolic diameters and fractional shortening were recorded for each group using a MatLab-based image analysis program (Fink et al., 2009). Fluorescent imaging of Drosophila heart tubes was performed as described (Alayari et al., 2009).
Bioinformatics analysis
For a detailed description of full bioinformatics analysis, please see supplemental experimental procedures.
Phenotyping of not3 knockout mice
A targeting vector was constructed to replace exons 2 and 9 of the murine not3 gene. Fractional shortening (FS) was calculated as follows: FS = [(LVEDD − LVESD)/LVEDD] × 100. For ex vivo heart studies, hearts were assayed using a Langendorff apparatus. The heart was paced electrically at 400 beats/min (bpm) and the electrical field stimulation (EFS) was applied in conjunction with the pacing stimulation. Isoproterenol was perfused for 30 seconds using the indicated doses. For HDAC inhibition, wild type and not3+/− mice were treated with vehicle, Trichostatin A (TSA) or Valproic acid (VPA) for 1 week. Acid-extracted histones were prepared, resolved, and transferred to nitrocellulose membranes for Western blotting. Transverse aorta constriction (TAC) was performed as described (Kuba et al., 2007). For heart histology, hearts were arrested, fixed, embedded in paraffin and stained with hematoxylin and eosin (H&E) or Masson-Trichrome.
Human QT interval association
Human QT interval association signals over the NOT3 region were obtained from data generated by the QTSCD Consortium (Pfeufer et al., 2009).
Supplementary Material
Acknowledgments
We thank all members of our laboratories and the VDRC, the BERC (Akita University), and Vincent Chen for helpful discussions and excellent technical support. We thank Eric Olson for the Hand-Gal4 driver and Manfred Frasch for TinCΔ4-Gal4 driver stocks. We thank the members of the QTSCD consortium for valuable support and discussion. JMP is supported by IMBA, the Austrian Ministry of Science, an ERC Advanced Investigator grant, and EuGeneHeart. KK is supported by Kaken (21659198) from Japanese Ministry of Science, MTT Program, and Japan Heart Foundation. GGN was supported by a Marie Curie International Incoming Fellowship. AC was supported by a postdoctoral fellowship and KO by a Scientist Development Grant from the American Heart Association. RB was supported by grants from NHLBI of NIH. AP is supported by grants 01GR0803 and 01EZ0874 from the German federal ministry of research (BMBF), FSID-261/2008 from the The UK Foundation for the Study of Infant Deaths (FSID) and YGEIA/1104/17 and ERYEX/0406/06 from the Cyprus Cardiovascular disease Educational and Research Trust (CCDERT). YI is supported by Global COE program.
Footnotes
Supplemental data includes 6 figures, 5 tables, 1 video, and supplemental experimental procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- A.H.A. 2005 http://www.americanheart.org/
- Alayari NN, Vogler G, Taghli-Lamallem O, Ocorr K, Bodmer R, Cammarato A. Fluorescent Labeling of Drosophila Heart Structures. J Vis Exp. 2009 doi: 10.3791/1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert TK, Lemaire M, van Berkum NL, Gentz R, Collart MA, Timmers HT. Isolation and characterization of human orthologs of yeast CCR4-NOT complex subunits. Nucleic acids research. 2000;28:809–817. doi: 10.1093/nar/28.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & development. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer R. Heart development in Drosophila and its relationship to vertebrates Trends in cardiovascular medicine. 1995;5:21–28. doi: 10.1016/1050-1738(94)00032-Q. [DOI] [PubMed] [Google Scholar]
- Cripps RM, Olson EN. Control of cardiac development by an evolutionarily conserved transcriptional network. Developmental biology. 2002;246:14–28. doi: 10.1006/dbio.2002.0666. [DOI] [PubMed] [Google Scholar]
- Denis CL. Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics. 1984;108:833–844. doi: 10.1093/genetics/108.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genetic epidemiology. 2008;32:227–234. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink M, Callol-Massot C, Chu A, Ruiz-Lozano P, Izpisua Belmonte JC, Giles W, Bodmer R, Ocorr K. A new method for detection and quantification of heartbeat parameters in Drosophila, zebrafish, and embryonic mouse hearts. BioTechniques. 2009;46:101–113. doi: 10.2144/000113078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon T, Castelli WP, Hjortland MC, Kannel WB, Dawber TR. Predicting coronary heart disease in middle-aged and older persons. The Framington study. Jama. 1977;238:497–499. [PubMed] [Google Scholar]
- Han Z, Olson EN. Hand is a direct target of Tinman and GATA factors during Drosophila cardiogenesis and hematopoiesis. Development (Cambridge, England) 2005;132:3525–3536. doi: 10.1242/dev.01899. [DOI] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes & development. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayne S, Zwartjes CG, van Schaik FM, Timmers HT. Involvement of the SMRT/NCoR-HDAC3 complex in transcriptional repression by the CNOT2 subunit of the human Ccr4-Not complex. The Biochemical journal. 2006;398:461–467. doi: 10.1042/BJ20060406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joza N, Oudit GY, Brown D, Benit P, Kassiri Z, Vahsen N, Benoit L, Patel MM, Nowikovsky K, Vassault A, et al. Muscle-specific loss of apoptosis-inducing factor leads to mitochondrial dysfunction, skeletal muscle atrophy, and dilated cardiomyopathy. Molecular and cellular biology. 2005;25:10261–10272. doi: 10.1128/MCB.25.23.10261-10272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K, Zhang L, Imai Y, Arab S, Chen M, Maekawa Y, Leschnik M, Leibbrandt A, Markovic M, Schwaighofer J, et al. Impaired heart contractility in Apelin gene-deficient mice associated with aging and pressure overload. Circulation research. 2007;101:e32–42. doi: 10.1161/CIRCRESAHA.107.158659. [DOI] [PubMed] [Google Scholar]
- Lakatta EG, Levy D. Arterial and cardiac aging: major shareholders in cardiovascular disease enterprises: Part II: the aging heart in health: links to heart disease. Circulation. 2003;107:346–354. doi: 10.1161/01.cir.0000048893.62841.f7. [DOI] [PubMed] [Google Scholar]
- Laribee RN, Shibata Y, Mersman DP, Collins SR, Kemmeren P, Roguev A, Weissman JS, Briggs SD, Krogan NJ, Strahl BD. CCR4/NOT complex associates with the proteasome and regulates histone methylation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:5836–5841. doi: 10.1073/pnas.0607996104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Jones D, Adams R, Carnethon M, De Simone G, Ferguson TB, Flegal K, Ford E, Furie K, Go A, Greenlund K, et al. Heart disease and stroke statistics--2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2009;119:e21–181. doi: 10.1161/CIRCULATIONAHA.108.191261. [DOI] [PubMed] [Google Scholar]
- Lo PC, Frasch M. A role for the COUP-TF-related gene seven-up in the diversification of cardioblast identities in the dorsal vessel of Drosophila. Mechanisms of development. 2001;104:49–60. doi: 10.1016/s0925-4773(01)00361-6. [DOI] [PubMed] [Google Scholar]
- Morita H, Seidman J, Seidman CE. Genetic causes of human heart failure. The Journal of clinical investigation. 2005;115:518–526. doi: 10.1172/JCI200524351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss AJ, Kass RS. Long QT syndrome: from channels to cardiac arrhythmias. The Journal of clinical investigation. 2005;115:2018–2024. doi: 10.1172/JCI25537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd JO, Kass DA. Tackling heart failure in the twenty-first century. Nature. 2008;451:919–928. doi: 10.1038/nature06798. [DOI] [PubMed] [Google Scholar]
- Nabel EG. Cardiovascular disease. The New England journal of medicine. 2003;349:60–72. doi: 10.1056/NEJMra035098. [DOI] [PubMed] [Google Scholar]
- Nerbonne JM. Studying cardiac arrhythmias in the mouse--a reasonable model for probing mechanisms? Trends in cardiovascular medicine. 2004;14:83–93. doi: 10.1016/j.tcm.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Ocorr K, Perrin L, Lim HY, Qian L, Wu X, Bodmer R. Genetic control of heart function and aging in Drosophila. Trends in cardiovascular medicine. 2007a;17:177–182. doi: 10.1016/j.tcm.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocorr K, Reeves NL, Wessells RJ, Fink M, Chen HS, Akasaka T, Yasuda S, Metzger JM, Giles W, Posakony JW, et al. KCNQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. Proceedings of the National Academy of Sciences of the United States of America. 2007b;104:3943–3948. doi: 10.1073/pnas.0609278104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paternostro G, Vignola C, Bartsch DU, Omens JH, McCulloch AD, Reed JC. Age-associated cardiac dysfunction in Drosophila melanogaster. Circulation research. 2001;88:1053–1058. doi: 10.1161/hh1001.090857. [DOI] [PubMed] [Google Scholar]
- Peng W, Togawa C, Zhang K, Kurdistani SK. Regulators of cellular levels of histone acetylation in Saccharomyces cerevisiae. Genetics. 2008;179:277–289. doi: 10.1534/genetics.107.085068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeufer A, Sanna S, Arking DE, Muller M, Gateva V, Fuchsberger C, Ehret GB, Orru M, Pattaro C, Kottgen A, et al. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nature genetics. 2009;41:407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Bodmer R. Partial loss of GATA factor Pannier impairs adult heart function in Drosophila. Human molecular genetics. 2009;18:3153–3163. doi: 10.1093/hmg/ddp254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Mohapatra B, Akasaka T, Liu J, Ocorr K, Towbin JA, Bodmer R. Transcription factor neuromancer/TBX20 is required for cardiac function in Drosophila with implications for human heart disease. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19833–19838. doi: 10.1073/pnas.0808705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray VM, Dowse HB. Mutations in and deletions of the Ca2+ channel-encoding gene cacophony, which affect courtship song in Drosophila, have novel effects on heartbeating. Journal of neurogenetics. 2005;19:39–56. doi: 10.1080/01677060590953066. [DOI] [PubMed] [Google Scholar]
- Rusconi JC, Challa U. Drosophila Mrityu encodes a BTB/POZ domain-containing protein and is expressed dynamically during development. The International journal of developmental biology. 2007;51:259–263. doi: 10.1387/ijdb.062233jr. [DOI] [PubMed] [Google Scholar]
- Sanguinetti MC, Tristani-Firouzi M. hERG potassium channels and cardiac arrhythmia. Nature. 2006;440:463–469. doi: 10.1038/nature04710. [DOI] [PubMed] [Google Scholar]
- Sanyal S, Jennings T, Dowse H, Ramaswami M. Conditional mutations in SERCA, the Sarco-endoplasmic reticulum Ca2+-ATPase, alter heart rate and rhythmicity in Drosophila. Journal of comparative physiology. 2006;176:253–263. doi: 10.1007/s00360-005-0046-7. [DOI] [PubMed] [Google Scholar]
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- Wang Q, Curran ME, Splawski I, Burn TC, Millholland JM, VanRaay TJ, Shen J, Timothy KW, Vincent GM, de Jager T, et al. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nature genetics. 1996;12:17–23. doi: 10.1038/ng0196-17. [DOI] [PubMed] [Google Scholar]
- Yusuf S, Reddy S, Ounpuu S, Anand S. Global burden of cardiovascular diseases: part I: general considerations, the epidemiologic transition, risk factors, and impact of urbanization. Circulation. 2001;104:2746–2753. doi: 10.1161/hc4601.099487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.