Abstract
Virtually all animals and plants utilize small RNA molecules to control protein expression during different developmental stages and in response to viral infection. Structural and mechanistic studies have begun to illuminate three fundamental aspects of these pathways: small RNA biogenesis, formation of RNA-induced silencing complexes (RISCs) and targeting of complementary mRNAs. Here we review exciting recent progress in understanding how regulatory RNAs are produced and how they trigger specific destruction of mRNAs during RNA interference (RNAi).
Introduction
Small RNA molecules play central roles in regulating eukaryotic gene expression by means of RNA interference (RNAi) and related pathways [1],[2]. Fundamentally, RNA-mediated genetic control begins with the production of ~20-30-nucleotide RNAs whose sequences can base pair with segments of mRNA transcripts. Once generated, these microRNAs (miRNAs) or small interfering RNAs (siRNAs) assemble into large multi-protein effectors, called RNA-induced silencing complexes (RISCs), which bind to target transcripts and trigger their destruction.
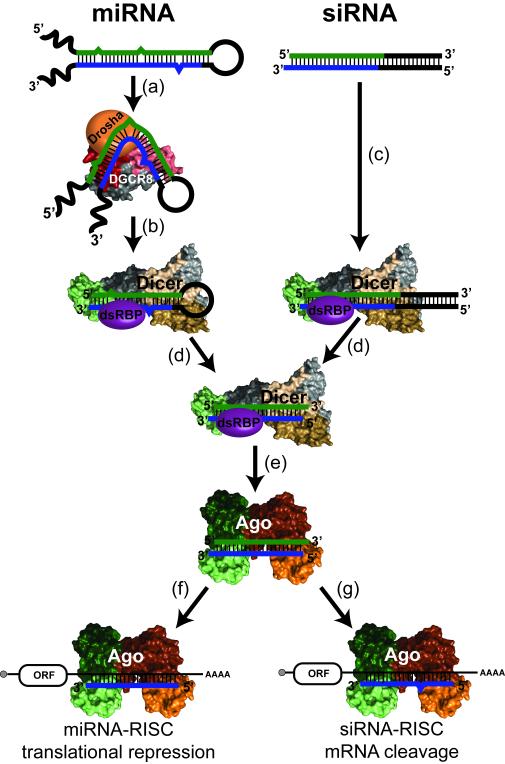
The production of miRNAs and siRNAs requires endonucleolytic processing of double-stranded precursors. Transcribed initially as long stem-loop structures in the nucleus, miRNAs are recognized by the protein DGCR8, which recruits the RNA to the RNase III-family endonuclease Drosha for initial double-strand cleavage (Fig. 1a). The resulting pre-miRNA is exported to the cytoplasm for cleavage into the mature miRNA duplex by a second RNase III-family endonuclease called Dicer (Fig. 1b). SiRNA production results from Dicer-mediated cleavage of long double-stranded RNAs that are either generated in the cell or introduced by viruses or transfection (Fig. 1c). In either case, the resulting duplex includes both the antisense guide strand and the sense, or passenger, strand (Fig. 1d).
Figure 1. Gene silencing by small RNAs.
Known structures of RNA silencing factors are represented with same coloring throughout the article, while unknown structures are shown as 3D shapes. RNA is depicted with thick lines where blue represents the guide strand sequence, green represents the passenger strand sequence, and black represents sequence that is cleaved off by RNase III enzymes. (a) Primary miRNA transcripts form hairpin structures that are recognized by the double-stranded RNA binding protein (dsRBP) DGCR8 and cleaved 11 bp away from the base of the stem by the RNase III enzyme Drosha. (b,c) Pre-miRNA hairpins and double stranded siRNA precursors are cleaved by Dicer, forming the mature miRNA duplex. Product length is determined by the distance between the 3′-overhang binding PAZ domain (green) and the RNase III catalytic center (beige, sand). In some organisms, dsRBPs associated with Dicer assist with substrate recognition. (d) The Dicer-dsRBP complex retains the mature product, handing it off to Argonaute (Ago), the effector of RISC. (e) After binding the duplex, the passenger strand (green) is removed while the guide strand (blue) remains bound to Ago. (f,g) The activated RISC can then go on to target mRNAs, acting through separate mechanisms depending on the type of guide RNA.
Two features of miRNAs and siRNAs ensure their efficient incorporation into the RISC complex: a specific length of the RNA duplex, and characteristic 5′ and 3′ ends carrying a phosphate group and a dinucleotide overhang, respectively. Structural studies of a prokaryotic RNase III and a eukaryotic Dicer enzyme have provided insights into how these critical features of siRNAs and miRNAs are generated [3-7].
MiRNAs and siRNAs subsequently associate with members of the Argonaute (Ago) family of proteins, which function as the core components of RISCs (Fig. 1e). A key aspect of this process is the loading of a guide strand into Ago, and the dissociation and/or destruction of the passenger strand. RISCs use the small RNAs as guides for sequence-specific silencing of complementary target RNA molecules by repressing their translation (Fig. 1f) or inducing their degradation (Fig. 1g). A specialized nuclear Argonaute-containing complex, known as the RNA-induced transcriptional silencing (RITS) complex, mediates transcriptional gene silencing by inducing heterochromatin formation in certain organisms [8]. Proteins belonging to the Argonaute family are also involved in the biogenesis of a third class of small RNAs, the Piwi-interacting RNAs (piRNAs) [9].
Key to the activities outlined above is the ability to specifically recognize, process and traffic small RNAs and their target mRNAs. Here we highlight recent structural and mechanistic insights into small RNA biogenesis, assembly with RISC and target recognition.
RNA manipulation by dsRBPs during biogenesis
Small RNA precursors contain distinctive characteristics that contribute to their recognition by the biogenesis machinery. In some cases, domains within partner proteins are uniquely tailored to recognize these features. For example, the PAZ domain found within Dicer and Ago specifically binds the 3′ overhangs of Drosha and Dicer products. In contrast, several silencing factors utilize more generic dsRNA binding domains (dsRBDs) to recognize the double stranded elements within precursor RNAs. Both Drosha and Dicer associate with dsRBD-containing proteins (dsRBPs), which are critical for the specific binding and cleavage of their substrates. These RNA-binding cofactors generally contain more than one dsRBD, providing a surprising diversity of lengths of RNAs bound by various Dicer-associated dsRBPs. For example, the Caenorhabditis elegans dsRBP RDE-4 binds RNA cooperatively, affording it an increased affinity for the longer siRNA precursors targeted by its associated Dicer [10]. In contrast, human TRBP does not display cooperative dsRNA binding, and instead binds RNAs of various lengths with similar affinities [10]. TRBP plays an important role in transferring mature siRNA duplexes to RISC [11-13], so must be able to bind short RNAs in addition to longer substrates.
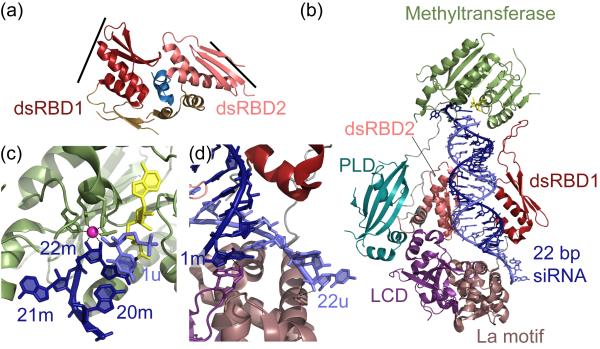
While dsRBD-RNA interactions have been a popular target of structural studies, structures of multiple dsRBD-containing proteins are rare. The recent crystal structure of the dsRBD region of human DGCR8, the RNA-binding partner of Drosha, has provided important insights into the mechanism of substrate recognition and product length determination by a protein containing two dsRBDs [14]. Individual dsRBDs adopt αβββα folds, forming a compact α/β sandwich that contacts both the major and minor grooves on one face of the RNA [15]. In the crystal structure of DGCR8, the two dsRBDs form a pseudo two-fold symmetry based mainly on mutual interactions with a C-terminal α-helix (Fig. 2a). The α-helices and loop that contact RNA lie on opposite sides of the molecule, suggesting that a single RNA helix bound by DGCR8 would be substantially kinked by interactions with the two dsRBDs (Fig. 1a, 2a). Intriguingly, the approximate length of an RNA that could bind in this conformation is 33 base pairs, similar to the length of a typical pri-miRNA hairpin, suggesting a mechanism by which DGCR8 identifies the correct substrates for Drosha.
Figure 2. Structures of two dsRBPs involved in small RNA biogenesis.
(a) Crystal structure of the RNA binding core (residues 493-720) of human DGCR8 (PDB: 2YT4). The two dsRBDs pack against a C-terminal α-helix (blue), forming a rigid RNA-binding platform that causes a sharp kink in the dsRNA target. The RNA binding surfaces on the dsRBDs are indicated with lines. (b) Crystal structure of full length HEN1 from Arabidopsis thaliana bound to a 22 base pair siRNA duplex (PDB: 3HTX). The two dsRBPs bind a 16 base pair stretch of the RNA simultaneously on two sides of the helix. The length of the substrate is specified by interactions with the methyltransferase domain and La motif. PLD: PPIase-like domain. LCD: La motif-containing domain. (c) Close up of methyltransferase-RNA interaction. The 2′ and 3′ hydroxyls of the modified nucleotide (22m) interact with a Mg2+ ion (magenta) that is coordinated by 4 conserved residues in the protein. The base of 22m stacks with the AdoHcy cofactor (yellow) ribose and the final base pair of the RNA helix (20m-1u). The penultimate nucleotide, 21m, flips out of the helix to allow this interaction. (d) Close up of La motif-RNA interaction. The 3′ nucleotide of the unmodified strand (22u) interacts with a tyrosine residue in the La motif, but this interaction is not essential for substrate binding. The 5′ nucleotide of the modified strand (1m) stacks with a conserved tryptophan and hydrogen bonds with an arginine.
Although the structure of DGCR8 provides preliminary insight into how multiple dsRBDs within a single protein may cooperatively bind an RNA target, several questions still remain. Biochemical evidence suggests that DGCR8 binds to the base of pri-miRNA hairpins, an interaction that is facilitated by association with flanking single-stranded sequence on either side of the RNA stem. Cleavage occurs ~11 base-pairs into the hairpin [15], suggesting that DGCR8 positions Drosha at an appropriate distance from the base of the stem (Fig. 1a). This mechanism for generating products of a specific length is reminiscent of Dicer, in which the distance between the 3′-overhang-binding PAZ domain and the catalytic center of the enzyme determines the length of the product (Fig. 1b, c) [5]. However, for the Drosha-DGCR8 complex, the structural basis for product length definition remains unclear. Structures of DGCR8 bound to RNA or Drosha would greatly increase our understanding of how the dsRBP recognizes the ssRNA junction at the base of the hairpin, while concurrently positioning Drosha for cleavage.
In plants, an additional processing step occurs following Dicer cleavage, in which the 2′-hydroxyl on the 3′ end of the siRNA duplex is methylated by the methyltransferase HEN1. In Arabidopsis thaliana, HEN1 displays a preference for 21-24 nucleotide duplexes with 2 nucleotide overhangs, suggesting that RNA recognition may proceed through mechanisms similar to those observed for other components of the biogenesis machinery [16]. Indeed, the recent crystal structure of HEN1 elegantly condenses several of the themes of small RNA recognition discussed above into a single molecule (Fig. 2b) [17]. In the structure, HEN1 is bound to a 22-nucleotide RNA duplex, an interaction that is mainly mediated by two dsRBDs that simultaneously bind the same stretch of RNA on opposite faces (Fig. 2b). This arrangement is in sharp contrast to the dsRBDs of DGCR8, where the rigid orientation of the domains restricts the protein from wrapping around the RNA, allowing the protein to bind RNAs of a specific length along only one side of the duplex. The HEN1 dsRBDs span only 16-base pairs of the helix, requiring that the final few base pairs on either end interact with other domains to determine substrate length.
One domain that restricts substrate length is the catalytic methyltransferase domain, which caps the RNA and interacts with an elongated loop within the first dsRBD. The 3′ nucleotide in the active site is mainly oriented through coordination of the 2′ and 3′ hydroxyls by a Mg2+ ion that is bound by four conserved residues, as well as stacking interactions with the ribose group of the cofactor adenosyl-L-homocysteine (AdoHcy) (Fig. 2c). The penultimate nucleotide of the modified strand flips out of the A-form helix, making specific contacts with two residues distant from the active site. This conformation allows the 3′ nucleotide to stack on the final base pair of the helix, likely providing stability for substrates with 2 nucleotide overhangs (Fig. 2c).
On the other end of the RNA, a La motif completes the substrate length determination. La protein, from which the La motif originates, recognizes the 3′ ends of nascent RNA Polymerase III transcripts, forming specific contacts between its N-terminal domain (NTD) and the terminal 3 nucleotides of the transcript [18]. However, the RNA interactions observed for the HEN1 La motif differs significantly from the RNA-NTD interaction. The HEN1 La motif replaces the 3′ binding pocket of the NTD with two conserved loops that form a platform on which the 5′ end of the modified strand stacks (Fig. 2d), an interaction that is essential for substrate binding and methylation. This variation in specific RNA contacts between the La-NTD and the HEN1 La motif presents an intriguing example of how a single RNA binding motif can be modified slightly to provide starkly different binding environments. The 2-nucleotide overhang of the unmodified strand is also bound by the La motif (Fig. 2d); however, this contact is not essential, consistent with biochemical evidence that the length of the unmodified overhang is not important for HEN1 activity [16]. Overall the HEN1 structure is an excellent example of how RNA silencing factors utilize a large variety of motifs to specify substrate length, and provides an interesting counterpoint to the molecular ruler observed for Dicer product length determination [5].
RISC Loading
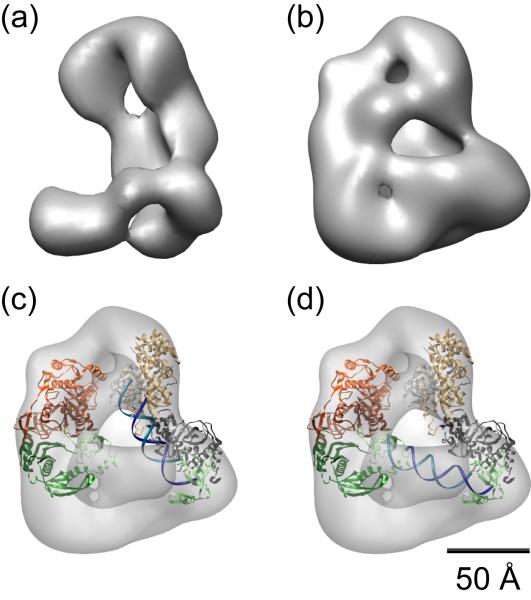
Following Dicer cleavage, mature miRNA or siRNA duplexes must be transferred from Dicer to Argonaute to form the active RISC. This process leads to the selection of the guide strand and removal of the passenger strand. In humans, the minimal components required for loading Argonaute 2 (Ago2) are Dicer complexed with its dsRBP cofactor, TRBP, and a mature siRNA duplex [13]. The human RISC loading complex (RLC) and its components have recently been analyzed by negative-stain electron microscopy (Fig. 3a, b), providing a structural framework for testing how siRNA duplexes may be passed from Dicer-TRBP to Ago2 during RISC loading [19, 20].
Figure 3. Cryo-EM reconstructions of the human RISC loading complex.
Volumes representing (a) Dicer alone and (b) Dicer plus Ago2 reflect the relative locations of the two larger RLC factors. TRBP volume is not detectable in (b) due to flexibility of its interaction with Dicer. (c, d) Model of the locations of Dicer and Ago, using structures of Thermus theromphilus Ago (PDB: 3DLH, left) and Giardia intestinalis Dicer (PDB: 2QVW, right). The large volume below Dicer likely corresponds to the helicase domain, which is not present in G. intestinalis Dicer. (c) A 22 bp siRNA modeled between the PAZ domain (green) and RNase III active sites (beige) of Dicer mimics binding of the siRNA duplex during initial stages of RISC loading. The space between Ago and Dicer in the reconstruction is sufficient to accommodate the duplex RNA. (d) Alternatively, the 22 bp RNA can be modeled to stretch between the two PAZ domains of Dicer and Ago, each of which could bind the 2 nucleotide overhangs on either end of the duplex. The distance between the PAZ domains of Ago and Dicer is approximately the same length as the 22 bp duplex, suggesting a possible intermediate in the hand-off of RNA during RISC loading.
The EM reconstruction of human Dicer reveals an L-shaped molecule containing a central cleft within the long axis (Fig. 3a) [19, 20]. The short arm of the L corresponds to the N-terminal helicase (DE×H/D) domain, based on analysis of a deletion mutant missing the ~80 kD helicase region [19]. The crystal structure of Giardia intestinalis Dicer, which lacks the helicase domain, can be modeled into the Dicer reconstruction in two different orientations, with the RNase III domains positioned toward either the base or the top of the L [5, 20]. However, given the location of the N-terminal helicase domain at the base of the L, the latter orientation seems more likely, as the C-terminal RNase III domains are distant from the N-terminus in the G. intestinalis Dicer structure (Fig. 3c).
EM reconstructions of various combinations of RLC components were used to dissect the remaining make-up of the full RLC reconstruction [19]. These analyses suggest that TRBP interacts flexibly with the helicase domain of Dicer, while Ago2 stretches between TRBP and the long arm of Dicer, forming a closed complex in some particles (Fig. 3b). The Ago2-Dicer interaction is thought to occur between the catalytic PIWI domain of Ago2 and the RNase IIIa domain of Dicer, based on previous biochemical studies [21]. This information, coupled with the positioning of G. intestinalis Dicer, suggests the location of Ago2 within the EM reconstruction of Dicer plus Ago2, allowing for the modeling of the Thermus thermophilus Ago crystal structure [22] into this reconstruction (Fig. 3b, c).
In order to determine how an siRNA duplex could be accommodated within the RLC, a 22 base-pair siRNA was modeled along the putative RNA-binding face of Dicer, revealing that the dsRNA fits perfectly within the gap between Ago2 and Dicer (Fig. 3c). In an alternative RNA-binding model, the 22 base-pair duplex can also be stretched between the PAZ domains of Ago2 and Dicer, suggesting that the two domains could bind both ends of the siRNA simultaneously (Fig. 3d). This observation suggests an intriguing possibility for an intermediate RLC conformation during RNA hand-off. Although TRBP is known to be important for recruiting siRNAs to Ago2 [11], its exact role in this process is unknown. The location and flexibility of TRBP suggested by the EM model may allow the dsRBP access to the PAZ domain of Ago2. TRBP could therefore bridge the gap between Dicer and Ago2, retaining the siRNA within the RLC once Dicer releases it, and positioning the correct end of the siRNA for binding by the Ago2 PAZ domain. The process of orienting the siRNA during transfer to Ago2 is particularly important for strand selection, which is based primarily on thermodynamic asymmetry of the small RNA duplex. TRBP may have an additional proofreading role for strand selectivity during RISC loading by sensing the relative thermodynamic stability of either end of the duplex.
The final step of RISC activation, passenger strand removal, has recently been the subject of several biochemical studies. In Drosophila melanogaster, passenger strand removal proceeds through different mechanisms depending on which Argonaute homolog is present in RISC. Upon transfer of duplex siRNAs to RISC, Drosophila Ago2 cleaves the passenger strand [23-25] and the endonuclease C3PO removes the cleavage products, activating the Ago2-guide strand complex [26]. In contrast, Ago1, which binds miRNAs and has relatively weak cleavage activity compared to Ago2 [27], unwinds the miRNA duplex through a slicer-independent mechanism [28]. Human Ago2-containing RISC likely follows the slicer-dependent mechanism for passenger strand removal [29], although human C3PO has not been confirmed as a RISC activator. Future biochemical and structural studies of the minimal human RLC associated with its other binding partners may shed more light on the activation of Ago2-RISC.
Mechanisms of RNA silencing
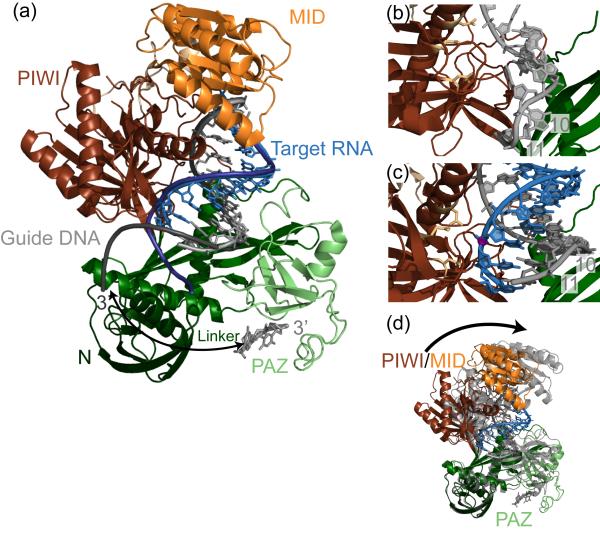
Following RISC loading, the Ago-guide strand complex binds its target mRNA, which occurs through base pairing of the guide strand with sequences within the 3′-untranslated region (UTR) of the mRNA. Full guide-target strand complementarity is required for target strand cleavage through the endonucleolytic “slicer” activity of Ago [30], a common mechanism for RNA silencing by siRNAs. Ago family proteins contain two distinctive domains: the PAZ domain, which binds the 3′-end of the guide strand, and the RNase H-like PIWI domain, which houses the slicing active site (Fig. 4a). A subset of Ago proteins also contain a MID domain, which forms a binding pocket for the 5′-phosphate of the guide strand through extensive hydrogen bonding with conserved residues and a Mg2+ ion. Several structures of prokaryotic Ago homologs have revealed the arrangement of these domains [31-38], and a recent set of structures of Thermus thermophilus Ago bound to several combinations of guide and target strands have provided insight into the mechanisms of substrate selection and slicing catalysis [22, 39, 40].
Figure 4. Structures of full length Argonaute from Thermus thermophilus bound to guide DNA and target RNA.
(a) Crystal structure of a catalytically inactive Ago bound to a 22 nt guide DNA (light gray) and 12 nt target RNA (light blue) (PDB: 3HO1). The 3′ end of the guide strand is buried in the PAZ binding pocket. Nucleation with a longer 19 nt target RNA (dark blue ribbon, PDB: 3HK2) causes the release of the guide DNA (dark gray ribbon) from the PAZ domain, as indicated by an arrow. (b) Close up of the active site of Ago with only the guide strand bound. The guide is slightly elongated, and kinked between nucleotides 10-11, which lie opposite site of cleavage. (c) This kinking is relieved in the active site in which the guide strand is bound to a target RNA. The catalytic aspartate residues (beige) are in close proximity to the cleavage site phosphate (purple). (d) The overall conformational change within Ago upon target binding. In the binary Ago-guide complex (transparent gray, PDB: 3DLH), the PIWI/MID lobe is closer to the PAZ lobe. Upon target binding, the PIWI/MID lobe pivots away from the PAZ, allowing for target-guide zippering with minimal movement of the guide DNA.
In the structure of T. thermophilus Ago bound to a 21-nucleotide guide strand DNA [40], the guide strand stretches between the PAZ and MID/PIWI domains, with nucleotides 2-10 adopting a stacked, partially helical conformation that is facilitated by an extensive hydrogen-bonding network between the phosphate backbone of nucleotides 2-6 and residues in the MID and PIWI domains. Nucleotides 12-17 are mostly disordered in the structure, suggesting that they do not strongly contact the protein, while 18-21 are stacked and anchored by backbone interactions with residues in the PAZ binding pocket. This arrangement presents the seed sequence, located between nucleotide 2-8, with its Watson-Crick face exposed to solvent, consistent with the importance of this region in target recognition [41]. Surprisingly, the guide strand is distorted between nucleotides 10 and 11, where the DNA forms interactions with two highly conserved arginine residues (Fig. 4b). This conformation suggests that the guide-Argonaute complex must undergo a significant structural change upon binding of the target strand, resulting in an uninterrupted helix at the cleavage site.
Recent structural studies of a catalytically inactive Argonaute complexed with a fully complementary guide DNA strand and target RNAs of various length (Fig. 4a) offer the first detailed illustration of the slicer active site with bound substrate, while also providing insight into the conformational rearrangements that occur upon target binding [39]. The RNase H-like active site of T. thermophilus Argonaute contains three aspartate residues that contribute to catalysis. In the catalytic mutant structure of Argonaute, the scissile phosphate at the point of cleavage (10-11 step) in the target strand is adjacent to the three aspartate residues (Fig. 4c). Intermolecular hydrogen-bonds between the protein and the phosphate backbone of the target strand near the cleavage site stabilize the active site, but the target strand is mainly positioned through interactions with the guide strand. As observed for the binary guide-protein structure, the seed sequence of the guide strand is firmly held by the protein through a hydrogen-bonding network. Crystals grown in the presence of increasing amounts of Mg2+ contain one or two metal ions coordinated between the Asp residues and the cleavage site, underscoring the importance of divalent metal ions for slicing activity.
As predicted from the binary Argonaute-guide strand structure, a conformational change between nucleotides 10 and 11 of the guide strand is observed upon target binding, with the kinked arrangement giving way to a more canonical A-form helical geometry (Fig. 4b, c). This conformational change is accompanied by a shift in the overall structure of Argonaute, with the PAZ domain pivoting away from the MID/PIWI region to form a more “open” conformation (Fig. 4d). The opening of the protein structure allows the guide and target strand to zip up, as illustrated by structures of the ternary complex with target strands of increasing size. Nucleation of the guide-target helix leads to the eventual release of the guide 3′ end, as further movement of the PAZ domain becomes physically impossible (Fig. 4a); however, the N-terminal domain of the protein blocks base pairing beyond nucleotide 16 of the guide strand, indicating that the entire guide-target duplex never forms in the context of RISC. Taken together, these observations reveal a flexibility of Ago that allows it to facilitate target binding and form a catalytically competent active site.
Conclusions
Our understanding of the molecular mechanisms of RNA silencing has expanded tremendously over the last several years, due in large part to structural insights of RNAi-related proteins from lower organisms. High-resolution structures of silencing factors from plants and animals remain elusive and represent the next major step for structural biology of RNAi.
Acknowledgements
We thank D. Patel for communicating results in advance of publication and members of the Doudna laboratory for helpful discussions. Research in the Doudna laboratory is supported by the Howard Hughes Medical Institute and the National Institutes of Health. D.G.S. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 3.Blaszczyk J, Gan J, Tropea JE, Court DL, Waugh DS, Ji X. Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure. 2004;12:457–466. doi: 10.1016/j.str.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Gan J, Tropea JE, Austin BP, Court DL, Waugh DS, Ji X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. doi: 10.1016/j.cell.2005.11.034.* * Detailed structural and mechanistic analysis of RNase III activity.
- 5.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638.** ** The only full-length Dicer structure currently available reveals the relative orientation of PAZ and RNase III domains, and the structural basis for Dicer product length.
- 6.Du Z, Lee JK, Tjhen R, Stroud RM, James TL. Structural and biochemical insights into the dicing mechanism of mouse Dicer: a conserved lysine is critical for dsRNA cleavage. Proc Natl Acad Sci U S A. 2008;105:2391–2396. doi: 10.1073/pnas.0711506105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takeshita D, Zenno S, Lee WC, Nagata K, Saigo K, Tanokura M. Homodimeric structure and double-stranded RNA cleavage activity of the C-terminal RNase III domain of human dicer. J Mol Biol. 2007;374:106–120. doi: 10.1016/j.jmb.2007.08.069.* *Structural studies of eukaryotic Dicer truncations.
- 8.Ekwall K. The RITS complex-A direct link between small RNA and heterochromatin. Mol Cell. 2004;13:304–305. doi: 10.1016/s1097-2765(04)00057-7. [DOI] [PubMed] [Google Scholar]
- 9.Aravin AA, Hannon GJ. Small RNA silencing pathways in germ and stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:283–290. doi: 10.1101/sqb.2008.73.058. [DOI] [PubMed] [Google Scholar]
- 10.Parker GS, Maity TS, Bass BL. dsRNA binding properties of RDE-4 and TRBP reflect their distinct roles in RNAi. J Mol Biol. 2008;384:967–979. doi: 10.1016/j.jmb.2008.10.002.* *Biochemical investigation of dsRBPs reveals how cooperativity of RNA binding can lead to recognition of dsRNA length.
- 11.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868.* *Original characterization of TRBP activity in small RNA processing and RISC loading.
- 12.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci U S A. 2008;105:512–517. doi: 10.1073/pnas.0710869105.* * Identification of the minimal human RISC loading complex.
- 14.Sohn SY, Bae WJ, Kim JJ, Yeom KH, Kim VN, Cho Y. Crystal structure of human DGCR8 core. Nat Struct Mol Biol. 2007;14:847–853. doi: 10.1038/nsmb1294.** **Structure reveals the orientation of dsRBDs in DGCR8 and the mechanism by which it recognizes dsRNA length.
- 15.Tian B, Bevilacqua PC, Diegelman-Parente A, Mathews MB. The double-stranded-RNA-binding motif: interference and much more. Nat Rev Mol Cell Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- 16.Yang Z, Ebright YW, Yu B, Chen X. HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 2006;34:667–675. doi: 10.1093/nar/gkj474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Ji L, Huang Q, Vassylyev DG, Chen X, Ma J-B. Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature. 2009;461:823–827. doi: 10.1038/nature08433.** **Structure of HEN1 bound to a 22 base pair siRNA duplex reveals the mode of substrate length recognition by the methyltransferase. The dsRBDs, La motif, and methyltransferase domain combine to restrict binding of RNAs over a certain length.
- 18.Curry S, Conte MR. A terminal affair: 3′-end recognition by the human La protein. Trends Biochem Sci. 2006;31:303–305. doi: 10.1016/j.tibs.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Wang HW, Noland C, Siridechadilok B, Taylor DW, Ma E, Felderer K, Doudna JA, Nogales E. Structural insights into RNA processing by the human RISC-loading complex. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1673.** **Cryo-EM reconstructions of various combinations of RLC components reveals the relative orientation of Ago2, Dicer, and TRBP. Modelling of RNA into the reconstruction provide insight into how dsRNA may be transferred from Dicer to Ago2.
- 20.Lau PW, Potter CS, Carragher B, MacRae IJ. Structure of the human Dicer-TRBP complex by electron microscopy. Structure. 2009;17:1326–1332. doi: 10.1016/j.str.2009.08.013.** **Cryo-EM reconstructions of human Dicer and TRBP. Two models for Dicer orientation are presented, including possible locations of dsRNA within the complex.
- 21.Tahbaz N, Kolb FA, Zhang H, Jaronczyk K, Filipowicz W, Hobman TC. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep. 2004;5:189–194. doi: 10.1038/sj.embor.7400070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666.** **Structure of Ago bound to guide strand alone. Compared with structures of ternary complex, this structure reveals the conformational changes which must occur in the guide strand and the protein as a whole in order to accommodate target binding.
- 23.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 24.Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Ye X, Jiang F, Liang C, Chen D, Peng J, Kinch LN, Grishin NV, Liu Q. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science. 2009;325:750–753. doi: 10.1126/science.1176325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawamata T, Seitz H, Tomari Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol. 2009;16:953–960. doi: 10.1038/nsmb.1630. [DOI] [PubMed] [Google Scholar]
- 29.Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nat Chem Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 31.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parker JS, Roe SM, Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. Embo J. 2004;23:4727–4737. doi: 10.1038/sj.emboj.7600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rashid UJ, Paterok D, Koglin A, Gohlke H, Piehler J, Chen JC. Structure of Aquifex aeolicus argonaute highlights conformational flexibility of the PAZ domain as a potential regulator of RNA-induced silencing complex function. J Biol Chem. 2007;282:13824–13832. doi: 10.1074/jbc.M608619200. [DOI] [PubMed] [Google Scholar]
- 36.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 37.Yuan YR, Pei Y, Chen HY, Tuschl T, Patel DJ. A potential protein-RNA recognition event along the RISC-loading pathway from the structure of A. aeolicus Argonaute with externally bound siRNA. Structure. 2006;14:1557–1565. doi: 10.1016/j.str.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011.* *Early structures of Argonaute proteins from various organisms provided important insights into the overall conformation of PAZ, PIWI, and MID domains.
- 39.Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009 doi: 10.1038/nature08434. In Press.** **Most complete structures of Ago bound to guide-target strand duplex to date. The use of catalytically inactive mutants allowed for complete complementary between guide and target strands, providing the first structure of the Ago active site with substrate in catalytically competent orientation.
- 40.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315.* *Structure of WT Ago bound to guide-target duplex with mismatches at cleavage site.
- 41.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]