Summary
MicroRNAs (miRNAs) regulate various biological processes, but evidence for miRNAs that control the differentiation program of specific neural cell types has been elusive. To determine the role of miRNAs in the formation of myelinating oligodendrocytes, we selectively deleted a miRNA-processing enzyme Dicer1 in oligodendrocyte lineage cells. Mice lacking Dicer1 display severe myelinating deficits despite an expansion of oligodendrocyte progenitor pool. To search for miRNAs responsible for the induction of oligodendrocyte maturation, we identified miR-219 and miR-338 as oligodendrocyte-specific miRNAs in spinal cord. Overexpression of these miRNAs is sufficient to promote oligodendrocyte differentiation. Additionally, blockage of these miRNA activities in oligodendrocyte precursor culture and knockdown of miR-219 in zebrafish inhibit oligodendrocyte maturation. miR-219 and miR-338 function in part by directly repressing negative regulators of oligodendrocyte differentiation, including transcription factors Sox6 and Hes5. These findings illustrate that miRNAs are important regulators of oligodendrocyte differentiation, providing new targets for myelin repair.
Keywords: Myelination, Dicer, differentiation inhibitors, oligodendrocytes, small non-coding RNAs, miR-219, miR-338, transcriptional regulation
Introduction
Oligodendrocytes, a fundamental and unique cell type in the CNS, synthesize multilamellar myelin membranes that ensheath axons. They play a critical role for development and function of the CNS. Failure of remyelination by oligodendrocytes disrupts saltatory nerve conduction, leading to nerve degeneration associated with acquired and inherited disorders such as multiple sclerosis (MS) and leukodystrophies (Berger et al., 2001; Trapp et al., 1998). At present, molecular mechanisms controlling oligodendrocyte differentiation and myelination are poorly understood.
Myelinating oligodendrocytes are derived from multipotent neural progenitor cells. Numerous signaling pathways such as Shh, Wnt, Bmp, and Notch, appear to regulate oligodendrocyte specification and differentiation (Miller, 2002; Rowitch, 2004). Such pathways often modulate transcriptional events via either DNA-binding transcription factors or chromatin-remodeling factors to dictate oligodendrocyte formation (Copray et al., 2009; Shen and Casaccia-Bonnefil, 2008). As a result, transcriptional regulation of oligodendrocyte lineage development has been a major focus in understanding mechanisms promoting differentiation of oligodendrocyte precursor cells (OPCs). A balance in the activity of transcriptional activators and repressors controls the formation of myelinating oligodendrocytes in a spatially and temporally specific manner (Emery et al., 2009; Rowitch, 2004; Wegner, 2008; Ye et al., 2009).
In addition to transcriptional regulation, posttranscriptional control by small noncoding RNAs has emerged as a central regulator of many biological and disease processes such as cell proliferation, differentiation, apoptosis, stress response, and tumorigenesis (reviewed in Bartel, 2004; Stefani and Slack, 2008). miRNAs are a class of small non-coding RNAs (∼22nt) that negatively regulate gene expression post-transcriptionally, either through translational inhibition or degradation of target mRNAs (Flynt and Lai, 2008; Hobert, 2004). miRNAs are initially transcribed as long primary transcripts (pri-miRNAs) and cleaved first by a nuclear RNase enzyme complex, Drosha/DGCR8. After exported into the cytoplasm mediated by exportin 5, they are processed by a RNase-III-type enzyme Dicer to form mature miRNAs, which are then incorporated into the RNA-induced silencing complex (RISC) to direct posttranscriptional repression (Bartel, 2004; Stefani and Slack, 2008). A short sequence complementarity between the “seed” sequence in 5′ miRNAs and the 3′ untranslated region (3′UTR) of mRNA targets is an important determinant of miRNA targeting, rendering the ability of a miRNA to potentially regulate multiple target genes simultaneously and shape transcriptional networks (Bartel, 2009).
Given the critical role of miRNAs in neurogenesis (Cheng et al., 2009; Kawase-Koga et al., 2009; Leucht et al., 2008; Visvanathan et al., 2007), we hypothesized that oligodendroglial lineage specification and differentiation might be guided at least in part by a posttranscriptional mechanism of gene regulation, namely by miRNAs. In this study, we deleted Dicer1, an enzyme essential for the biogenesis of most miRNAs, in the oligodendrocyte lineage cells and observed severe myelination deficits in the Dicer1 mutants. By miRNA profiling, we identified a cohort of miRNAs reduced in the Dicer1 mutants. Among them, miR-219 and miR-338 are specifically expressed in oligodendrocytes and their expression is significantly downregulated in myelin-deficient Dicer1 or Olig1 mutants. Through gain- and loss of function experiments in vitro and in vivo, we demonstrated that miR-219 and miR-338 positively regulate oligodendrocyte differentiation. Furthermore, we showed that miR-219 and miR-338 control oligodendrogenesis by directly targeting negative regulators of oligodendrocyte differentiation such as Hes5 and Sox6, while inhibiting genes involved in neuronal differentiation. Our present study provides the first insight to the regulation of oligodendrocyte differentiation and myelination by miRNAs. This study further suggests that repression of oligodendrocyte differentiation inhibitors by miRNAs is a critical mechanism for the generation of terminally differentiated oligodendrocytes.
Results
Deletion of Dicer1 in the Oligodendrocyte Lineage Impairs Myelination
To assess the role of Dicer1 in oligodendrocyte development in vivo, we selectively deleted Dicer1 in the oligodendrocyte lineage by crossing Dicer1lox/lox mice with an Olig1-Cre line. Olig1-promoter activity has been shown to direct Cre expression in the oligodendrocyte lineage cells including OPCs and mature myelinating oligodendrocytes in the CNS (Lu et al., 2002; Ye et al., 2009). The resulting Dicer1lox/lox;Olig1-Cre+/- mice (referred to as Dicer1CKO) were born at Mendelian ratio but developed severe tremor and ataxia, reminiscent of myelin-deficient mice, and died around postnatal week 3.
Immunohistochemistry and in situ hybridization analysis of Dicer1CKO mice revealed that expression of mature oligodendrocyte markers, myelin basic protein (MBP), CC1 and Plp1 encoding proteolipid protein, was essentially diminished in the spinal cord and brain (Figure 1A, B). We did not detect a substantial alteration in expression of an astrocyte marker GFAP (Figure 1B) or a neuronal marker NeuN (data not shown) in the mutant cortex.
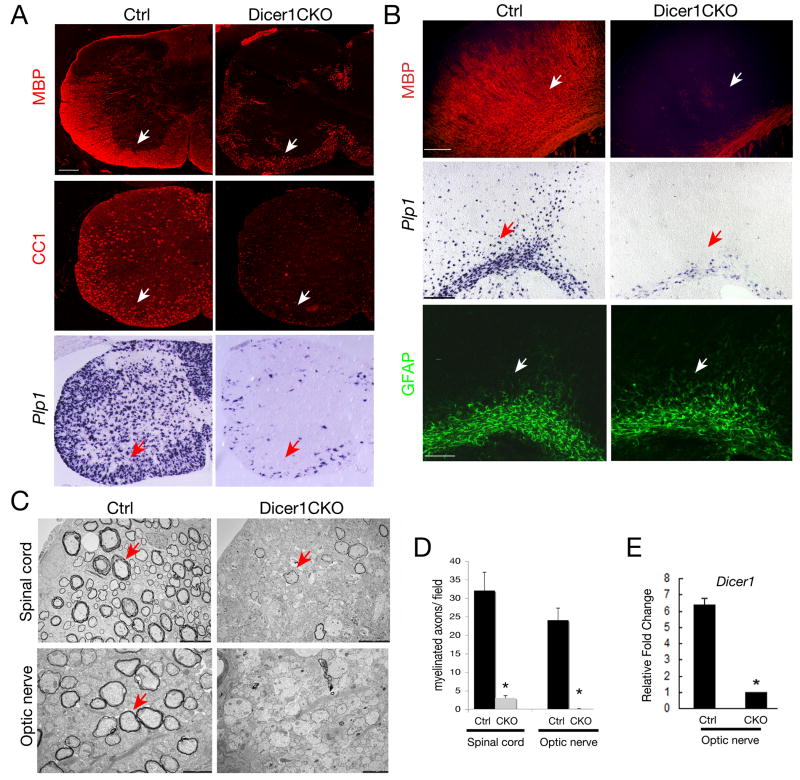
Figure 1. Dicer1 is Required for Oligodendrocyte Myelination.
A-B) Immunocytochemistry using antibodies to mature oligodendrocyte makers (MBP and CC1), an astrocyte marker GFAP and in situ hybridization using probes to Plp1 on frozen sections of P14 spinal cord and brains from control (Ctrl, Olig1Cre+/-;Dicer1lox/+) and Dicer1CKO (Olig1Cre+/-;Dicer1lox/lox) mice. Expression of MBP, CC1 and Plp1 is evident in white matter (arrows) of Ctrl but not Dicer1CKO mice.
C) Electron micrographs of spinal cord and optic nerves from Ctrl and Dicer1CKO mice at P14. Multilamellar myelin sheaths are apparent around many axons in Ctrl mice and axons in Dicer1CKO mice are essentially unmyelinated (red arrow).
D) Histogram depicts quantification of myelinated axons in the optic nerve and spinal cord of Ctrl and Dicer1CKO mice per defined area (100 μm2), respectively. Error bars indicated mean± S.D. * P < 0.01, two-tailed paired Student's t test.
E) Histogram shows the qRT-PCR analysis of Dicer1 expression from RNAs isolated from intact optic nerves of Dicer1lox/+;Olig1Cre (Ctrl) and Dicer1CKO mice (n=3) at p14 with primers against the floxed exon at the Dicer1 locus. GAPDH was used as internal control. *P<0.01.
Scale bars in A and B: 200 μm; in C, 5 μm for upper panel and 2 μm for lower panel.
Myelination deficits were further confirmed by electron microscopy in the spinal cord and the optic nerve of Dicer1 mutants. The number of myelinated axons at P14 was reduced significantly in the spinal cord and essentially absent in the optic nerve (Figure 1C-D). The few myelinated axons observed in the Dicer1CKO spinal cord were characterized by thinner myelin sheaths. Severe myelination deficiency in Dicer1 mutants suggests that maturation of miRNAs processed by Dicer1 is required for oligodendrocyte differentiation and myelination. The oligodendrocyte-specific deletion of the floxed Dicer1 allele by Olig1-Cre was verified by quantitative real-time RT-PCR (qRT-PCR) using primers specific to the floxed exon 23 and RNAs isolated from intact optic nerves, which are enriched with oligodendrocytes (Figure 1E).
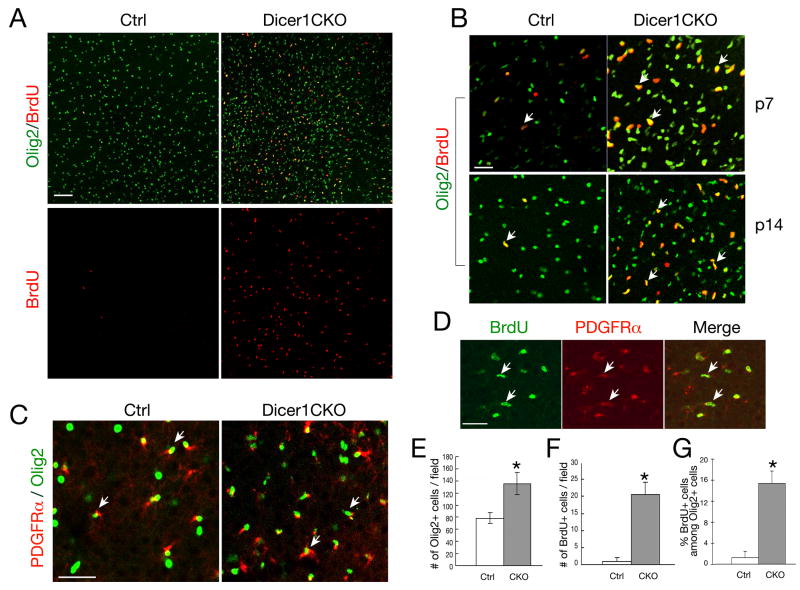
The dysmyelinating phenotype detected in Dicer1CKO mice could be due to increased cell death, inhibited proliferation and/or arrested differentiation. In the CNS of Dicer1CKO mice, we did not detect any significant increase of apoptotic cells by examining the presence of activated Caspase 3 and by TUNEL assay (data not shown). To examine the fate of OPCs, we performed BrdU pulse labeling experiments and analyzed OPC proliferation. In the developing cortex of Dicer1CKO mice, we observed an increase of BrdU+ and Olig2+ cells (Figure 2A-B, E-F). A majority of Olig2+ cells are PDGFRα-positive (Figure 2C) at perinatal stages and essentially all BrdU+ cycling cells are PDGFRα-expressing OPCs (Figure 2D), but not neurons or astrocytes (data not shown). Cycling OPCs (Brdu+/Olig2+) were significantly increased in the cortex (Figure 2G). These observations suggest that OPCs with Dicer1 deletion undergo extensive proliferation without further differentiation into mature oligodendrocytes.
Figure 2. Oligodendrocyte Precursor Proliferation in the Cortex of Dicer1 Mutants.
A-C) Immunocytochemistry using antibodies to Olig2, BrdU and PDGFRα on frozen sections of P7 (B) and P14 (A-C) brains from Olig1Cre;Dicer1lox/+ (Ctrl) and Olig1Cre;Dicer1lox/lox (Dicer1CKO) mice.
D) Immunocytochemistry using antibodies to BrdU and PDGFRα on frozen sections of P14 brains from Olig1CreDicer1lox/+ (Ctrl) mice.
Scale bars: 100 μm. Arrows in B-D indicate co-labeling cells.
E-F) Histograms depict the quantification of Olig2 and BrdU expressing cells. Error bars indicated mean± S.D. * P < 0.001.
G) Histogram depicts the percentage of BrdU expressing cells among Olig2 expressing cells. Error bars indicated mean± S.D. * P < 0.001.
Identification of miR-219 and miR-338 as Oligodendrocyte-specific miRNAs
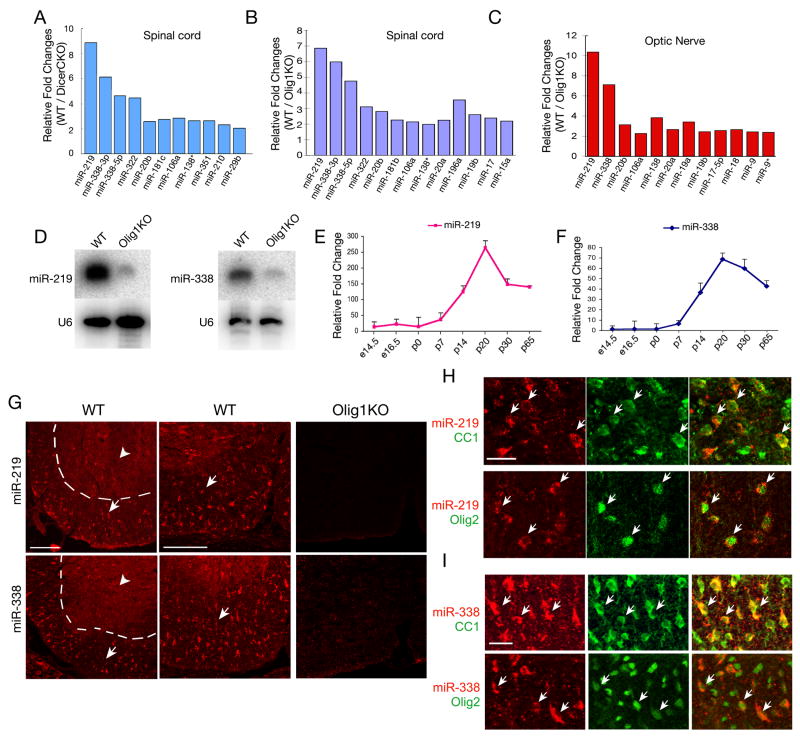
In light of the requirement of miRNA maturation for oligodendrocyte differentiation, we searched for miRNAs that are responsible for oligodendrocyte maturation. We screened for miRNAs downregulated in the spinal cord of dysmyelinating Dicer1CKO mice by using miRNA microarray analysis. The optic nerve and spinal cord tissues of myelin-deficient Olig1 null mice (Xin et al., 2005) were included for analysis because Dicer1 deletion may block the majority of, if not all, miRNA maturation. Among those miRNAs downregulated in both Dicer1 and Olig1 mutants, expression of miR-219 and miR-338 was the most significantly reduced compared to wildtype (Figure 3A-C, Table S1). This is in keeping with previous data showing that these two miRNAs are highly enriched in mature oligodendrocytes (Lau et al., 2008). Northern blot analysis confirmed that both miR-219 and miR-338 were essentially absent in the spinal cord of Olig1 null or Dicer1CKO mice (Figure 3D; data not shown).
Figure 3. Identification of miR-219 and miR-338 as Oligodendrocyte-Specific miRNAs in the Spinal Cord.
A-C) miRNAs isolated from spinal cord and optic nerve tissues from WT, Dicer1CKO or Olig1 null mice were subjected to miRNA microarray analysis. The log2 transformation of relative fold changes in the miRNA expression level from WT versus Dicer1CKO or Olig1KO mice are indicated.
D) Expression of miR-219 and miR-338 in WT and Olig1 null spinal cord was analyzed by Northern blot. U6 RNA is used as a loading control.
E-F) miR-219 (E) and miR-338 (F) expression was analyzed by qRT-PCR from WT mouse spinal cords at stages indicated. U6 RNA was used as internal control.
G) Expression of miR-219 and miR-338 was examined in the spinal cord of WT and Olig1KO mice at P14 by in situ hybridization as indicated. Arrows indicate the miRNA expressing cells in the white matter region (left part of dashed lines); arrowheads indicate the gray matter. Left panels show at lower magnification, while middle and right panels show at higher magnification.
H-I) Expression of miR-219 and miR-338 was examined in spinal cord of P14 WT mice by in situ hybridization, followed by immunostaining with CC1 and Olig2. Arrows indicate colabeling of miR-219 or miR-338 with oligodendrocyte markers CC1 and Olig2 in the same cells. Scale bars, in G, 100 μm; in H and I, 50 μm.
qRT-PCR analysis of the time-course of miR-219 and miR-338 expression indicated that expression of these miRNAs increased during spinal cord development (Figure 3E, F). A sharp increase of miR-219 and miR-338 at perinatal stages coincides with the onset of oligodendrocyte maturation at these stages (Figure 3E, F). Consistently, expression of Dicer1 increases during differentiation of OPCs into mature oligodendrocytes (Figure S1). To determine the cell type specificity of miR-219 and miR-338, we carried out in situ hybridization in the P14 spinal cord using fluorescein-labeled miRNA probes that targeted to the mature form of miRNAs. miR-219 and miR-338 positive cells were detected mainly in the white matter of spinal cord (Figure 3G). In contrast to specific expression pattern of miR-219 and miR-338 in the spinal white matter, scrambled control probe did not detect any specific signal (data not shown). Consistently, expression of these two miRNAs was undetectable in the P14 spinal cord of Olig1 null mice, where there is a specific loss of mature myelinating oligodendrocytes (Figure 3G). Furthermore, we demonstrated that miR-219 and miR-338 were co-labeled with oligodendrocyte markers CC1 and Olig2 (Figure 3H, I), but not with a neuronal maker NeuN (data not shown). These data suggest that miR-219 and miR-338 are specifically expressed in oligodendrocytes in the spinal cord.
miR-219 and miR-338 Promote OPC Maturation in vitro
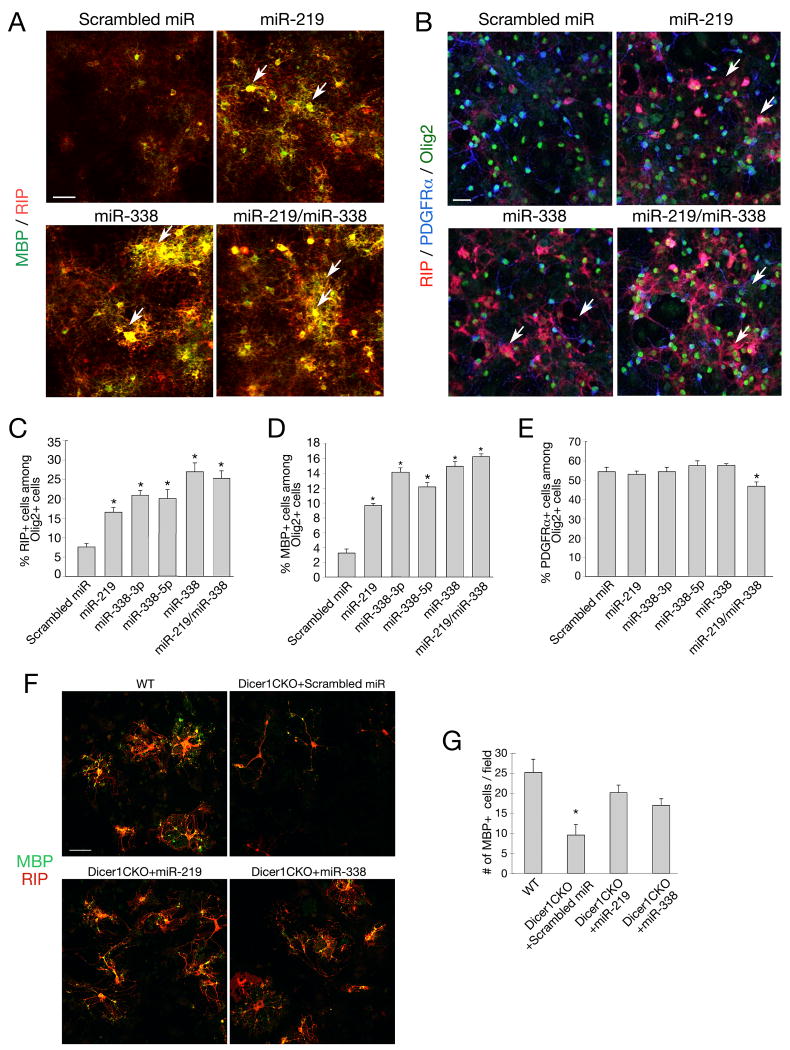
To examine the role of these miRNAs in oligodendrocyte differentiation, we first transfected oligodendrocyte precursor-enriched cultures (Chen et al., 2007) with miR-219 and miR-338 mimics individually or together. Four days after treatment, the cells were subjected to immunostaining for stage-specific oligodendrocyte markers. Enforced expression of miR-219, miR-338 or both resulted in a significant increase in the number of early differentiated RIP+ oligodendrocytes and mature MBP+ oligodendrocytes (Figure 4A-D). A slight reduction of PDGFRα–expressing OPCs was observed after treatment of both miRNA mimics (Figure 4B, E). These results suggest that enforced miR-219 and miR-338 expression promotes oligodendrocyte differentiation from their precursors.
Figure 4. Enforced miR-219 and miR-338 Expression Promotes Oligodendrocyte Maturation in vitro.
A-B) Mouse oligodendrocyte precursor-enriched cultures were transfected with miR-219, miR-338(5p+3p) mimics and scrambled miR control as indicated. The precursor cells were cultured in oligodendrocyte growth medium for another 4 days and subjected to immunostaining with PDGFRα (blue), RIP (red) and MBP (green). Scale bars: 50 μm.
C-E) Histograms depict the percentage of RIP+ (C), MBP+ (D) and PDGFRα+ (E) cells among Olig2+ oligodendrocyte lineage cells, which represents all stages of oligodendrocytes. Data represent mean ± S.D. from three independent experiments. * P < 0.01.
F) Oligodendrocyte precursor-enriched cultures from Dicer1CKO mice were transfected with miR-219, miR-338 (5p+3p) mimics and scrambled miR control as indicated. Cells were cultured in oligodendrocyte differentiation medium without mitogens for 3 days and subjected to immunostaining with RIP (red) and MBP (green). Scale bars: 50 μm.
G) Histogram depicts the number of MBP+ cells per defined area (0.14 mm2). Data represent mean ± S.D. from three independent experiments. * P < 0.01.
To further determine whether overexpression of miR-219 and miR-338 could rescue the defects of oligodendrocyte differentiation due to Dicer1 deletion, we transfected primary oligodendrocyte progenitor-enriched culture isolated from Dicer1CKO embryos with miR-219 or miR-338 mimics. In contrast to the treatment of scrambled control, miR-219 or miR-338 mimics could enhance oligodendrocyte maturation and increase the number of mature MBP+ oligodendrocytes with complex morphologies (Figure 4F, G). This result suggests that overexpression of miR-219 and miR-338 is sufficient, at least partially, to rescue the deficits in oligodendrocyte differentiation caused by Dicer1 deletion.
miR-219 and miR-338 Induce Oligodendrocyte Differentiation in vivo
To examine their in vivo role in oligodendrocyte development, we first carried out an in ovo gain-of-function study in the embryonic chick neural tube. Expression vectors for miR-219 and miR-338 were electroporated into the neural tube of E2.5 chick embryos. The embryos were harvested at E5.5, at which stage the differentiation of endogenous oligodendrocytes had not yet occurred. Expression of miR-219 and miR-338 from expression vectors was confirmed by Northern blot and in situ hybridization analyses (Figure S2 A,B). Overexpression of miR-219 was able to promote ectopic formation of OPCs expressing PDGFRα or Olig2 in the electroporated side of neural tube (Figure 5A), however, we did not observe expression of genes characteristic of MBP+ mature oligodendrocytes (data not shown). In contrast, overexpression of miR-338 not only led to ectopic formation of OPCs expressing Olig2 but also promoted precocious formation of MBP+ mature oligodendrocytes (Figure 5B). These results suggest that overexpression of miR-219 and miR-338 promotes oligodendrocyte specification and differentiation in the chick neural tube.
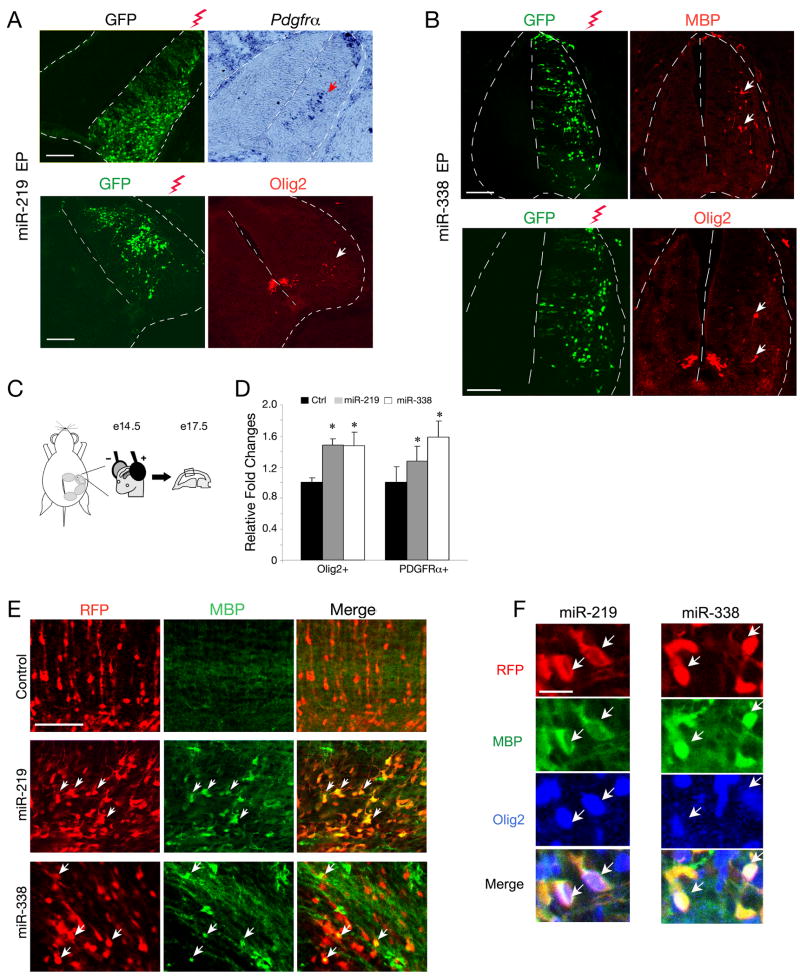
Figure 5. Ectopic Expression of miR-219 and miR-338 Promotes Precocious Oligodendrocyte Differentiation in Developing Chick Neural Tube and Embryonic Mouse Cortex.
A-B) Chick neural tubes were electroporated with pCIG expression vectors for miR-219 (A) and miR-338 (B) into the neural tube at E2.5 and harvested at E5.5. The spinal cord sections were analyzed by in situ hybridization with probes to Pdgfrα and by immunostaining with antibody against Olig2 or MBP. GFP expression indicates the electroporated side. Arrows indicate ectopic expression of Pdgfrα, Olig2 and MBP detected on the electroporated side of chick neural tubes, respectively. Scale bars, 100μm.
C) Schematic diagram depicts in utero electroporation in the mouse developing cortex at e14.5. Electroporated embryonic cortices (n =5) were harvested and analyzed at e17.5, at which stage there is absent of endogenous MBP expression.
D) Quantification of the number of Olig2+ or PDGFRα+ cells on the electroporated cortices at a defined cortical area (0.1 mm2). Y axis indicated the ratio of the number of Olig2+ or PDGFRα+ cells in miR-219 or miR-338 overexpressing cortices to that in the control. Error bars indicated mean± S.D. * P < 0.05.
E) Mouse embryos at e14.5 were electroporated with expression vectors for miR-219, miR-338 and RFP, respectively, and harvested at e17.5. The sections of electroporated cortices were analyzed by immunostaining with antibodies to MBP. Arrows indicate the electroporated cells expressing MBP.
F) Triple immunostaining revealed that miR-219 or miR-338 transfected cells express MBP and Olig2 in the electroporated cortices. Arrows indicate co-labeling cells. Scale bars, in E, 100μm; in F, 25 μm.
The lack of mature oligodendrocyte formation promoted by miR-219 in the chick neural tube could be due to the difference in species and/or the cellular context. To determine effects of miR-219 overexpression on oligodendrocyte development in mice, we electroporated a miR-219 expressing vector into the neocortical ventricular zone of developing embryos at E14.5 (Figure 5C). The cortices from electroporated embryos were collected 3 days later (E17.5), at which stage no MBP+ oligodendrocytes can be observed in the cortex. Overexpression of miR-219 resulted in an increase of Olig2+ and PDGFRα+ OPCs in the cortex (Figure 5D; Figure S2C). Strikingly, we detected precocious formation of MBP-expressing cells in the electroporated cortex (Figure 5E). Similar results were also observed in the cortex with ectopic miR-338 overexpression (Figure 5E). Triple immunolabeling indicated that the ectopic MBP-expressing cells also express an oligodendrocyte marker Olig2 (Figure 5F). These observations suggest that overexpression of either miR-219 or miR-338 can promote oligodendrocyte formation in the developing mouse cortex.
Knockdown of Endogenous miR-219 and miR-338 Inhibits Oligodendrocyte Maturation
To investigate whether endogenous miR-219 or miR-338 is necessary for oligodendrocyte formation, we treated oligodendrocyte precursor cultures with miRNA inhibitors to down-regulate miR-219 and miR-338 activities. In contrast to scrambled control, treatment of miRNA antisense inhibitors for miR-219 or miR-338 under differentiation condition resulted in significant reduction of MBP+ oligodendrocyte formation (Figure 6A, B). Knockdown of these miRNAs appeared to inhibit oligodendrocyte process formation and maturation (Figure 6A). These observations suggest that inhibition of miR-219 and/or miR-338 blocks oligodendrocyte differentiation and maturation.
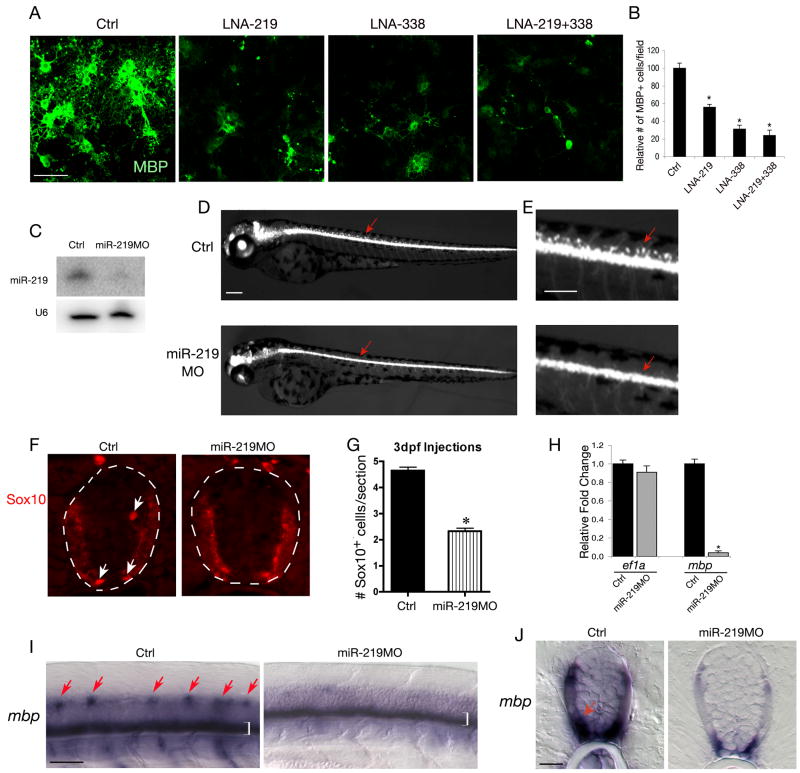
Figure 6. Inhibition of miR-219 and miR-338 Blocks Oligodendrocyte Maturation.
A) Purified rat oligodendrocyte precursor cells were transfected with LNA knockdown antisense miRNAs and scrambled control oligonucleotides as indicated and cultured in oligodendrocyte differentiation medium. Four days after transfection, cultures were subjected to MBP immunostaining.
B) Histogram depicts the relative number of MBP+ cells in per defined area (0.14 mm2) treated with LNA-anti-219 and LNA-anti-338(5p+3p) compared with that treated with scrambled control oligonucleotide. Data represent mean ± S.D. from three independent experiments, *P<0.01.
C) Expression of miR-219 in WT and miR-219MO injected larvae was analyzed by Northern blot. U6 RNA is used as loading control.
D-E) Stereomicroscope images showing a lateral view of living Tg(olig2:egfp) zebrafish larvae at 3 dpf with or without morpholino miR-219 injection. E is shown at larger magnification to show the OPCs in the dorsal spinal cord in control larvae but their absence in the miR-219 MO-injected larvae as indicated by arrows.
F) Transverse sections through the spinal cord of larvae at 3 dpf were immunostained with antibody to Sox10. Sox10+ OPCs are indicated by arrows.
G) Quantification of the average number of Sox10+ OPCs per transverse section in miR-219MO injected and control zebrafish (*P<0.01). Error bars indicated mean± SD.
H) qRT-PCR analysis of expression of mbp and ef1a encoding eukaryotic elongation factor 1 from RNAs isolated from miR-219 MO injected and control zebrafishes. rpl13 encoding a ribosomal protein was used as an internal control. *P<0.01.
I-J) In situ hybridization with an mbp probe in zebrafish embryos with or without miR-219MO injection. Arrows in I (lateral view) indicate the formation of mature oligodendrocytes in the dorsal spinal cord. Brackets indicate mbp expression in ventral spinal cord. Arrows in J (cross-section) indicate mbp expression.
Scale bars in A, 50μm; C-D40 μm; E, 20 μm, G, 40 μm.
To examine the function of these miRNAs in oligodendrocyte development in vivo, we chose to examine the knockdown effect in zebrafish, a model of vertebrates, because the mature sequences for these miRNAs are identical between zebrafish and mouse. We injected antisense morpholino oligonucleotides (MO) designed to hybridize to miR-219 or miR-338 into Tg(olig2:egfp) zebrafish embryos, which express EGFP in oligodendrocyte lineage cells under control of olig2 regulatory element (Shin et al., 2003). Embryos injected with either MO developed normally through 3 days post fertilization (dpf), with few consistent morphological defects. The downregulation of endogenous miR-219 expression by miR-219MO was verified by Northern blot analysis (Figure 6C). At 3 dpf, dorsally migrated OPCs were evident in the spinal cord of control larvae, whereas all miR-219 MO-injected larvae (n=46) had deficits of dorsal OPCs (Figure 6D-E). We confirmed the deficit of OPCs using Sox10 immunocytochemistry, which revealed an approximately 2-fold reduction in their number (Figure 6F,G). We did not detect a significant alteration in OPC number or distribution in miR-338 MO injected larvae (data not shown), which might reflect a relatively low level of miR-338 in the CNS compared to miR-219 (Wienholds et al., 2005). Furthermore, formation of MBP+ mature oligodendrocytes in the dorsal spinal cord was severely compromised in miR-219MO injected zebrafish embryos (Figure 6I-J). Significant downregulation of Mbp, but not the internal control Ef1a, in miR-219MO injected embryos was further confirmed by qRT-PCR assay (Figure 6H). These observations suggest that miR-219 is essential for oligodendrocyte maturation in the zebrafish.
Oligodendrocyte Differentiation Inhibitors as the Targets of miR-219 and miR-338
To identify potential physiological targets of miR-219 and miR-338, we performed the analysis of computationally predicted targets using TargetScan, PicTar, miRanda and mirBase prediction algorithms (Bartel, 2009; Grimson et al., 2007; Krek et al., 2005) to search for evolutionarily conserved targets for these miRNAs. Despite the lack of sequence homology between miR-219 and miR-338, both miRNAs were predicted to target a disproportionately large number of the same genes involved in negative regulation of oligodendrocyte differentiation (Figure S3 and Table S2).
Among these targets, we identified Sox6 and Hes5 as known inhibitors of OPC differentiation. Sox6 is a SRY-box transcription factor that inhibits oligodendrocyte maturation (Stolt et al., 2006). Hes5 is a member of the bHLH family of transcription factors that acts as critical downstream effector of the Notch signaling pathway. It has been shown to inhibit myelin gene expression and oligodendrocyte differentiation (Kondo and Raff, 2000; Liu et al., 2006; Wang et al., 1998). Consistent with these findings, expression of Sox6 was mainly observed in OPCs but not in differentiated oligodendrocytes (Figure S3C,D), which are enriched with miR-219 and miR-338 (Lau et al., 2008).
We tested Sox6 and Hes5 for miR-219 and miR-338 mediated repression by placing their 3′ UTR segments downstream of a cytomegalovirus (CMV)-driven luciferase reporter and performed reporter assays in COS7 cells transfected with expression plasmids for miR-219, miR-338 or a control plasmid encoding the reverse sequence of corresponding miRNAs. Overexpression of miR-219 and miR-338 was confirmed by Northern blot analysis with miRNA specific probes (Figure S2A). The repression was specifically correlated with the presence of predicted target sites for these miRNAs (Figure 7A). miR-219 and miR-338 overexpression significantly decreased the luciferase activity of reporters carrying Sox6 or Hes5 3′UTR segments with their predicted binding sites, respectively (Figure 7B, C). Mutation of the “seed” sequence of miR-219 and miR-338 abolished their repressive activities on above corresponding luciferase reporters (Figure 7B, C). In addition, mutation of either predicted miR-219 or miR-338 binding sites in the Sox6 UTR-I or UTR-II segment resulted in a significant reduction of responses to corresponding miRNA overexpression (Figure 7D,E; data not shown). Thus, the mutagenesis studies suggest that these predicted binding sites are functional binding sites at least based on the in vitro assay and that there is a direct binding of these miRNAs with their targets.
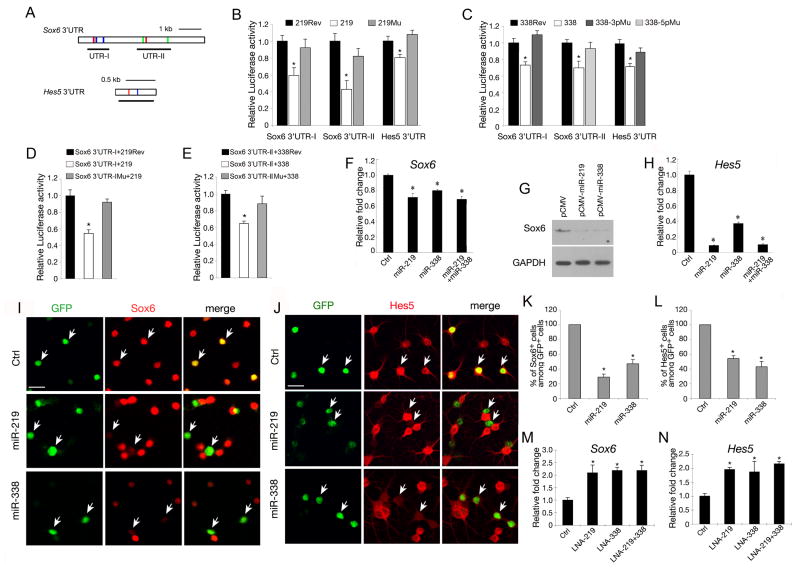
Figure 7. miR-219 and miR-338 Target Oligodendrocyte Differentiation Inhibitors.
A) Sequence analysis of 3′UTR of mouse Sox6 and Hes5 transcripts. miR-219, miR-338-5p and miR-338-3p recognition sites are indicated by red, green and blue bars, respectively. Black lines underneath depict the regions (UTR-I or UTR-II) of 3′ UTRs that cloned into pMIR-REPORTER: Sox6 3′UTR-I, 863bp; Sox6 3′UTR-II, 1380bp; Hes5 3′UTR, 630bp.
B-C) Luciferase reporter assays for the effects of expression of miR-219, miR-338-3p or -5p and their mutant forms with corresponding “seed” sequence mutations on activities of reporters carrying the 3′UTR segments of Sox6 or Hes5. The histogram shows the ratio of the luciferase activity normalized to control expression vector carrying the reverse sequence of miR-219 and miR-338-3p and miR-338-5p, respectively. Data represent mean ± SDs from three independent experiments. * P < 0.01.
D-E) Luciferase reporter assays for the effects of expression of miR-219 and miR-338 on activities of reporters carrying WT and mutant 3′UTR of Sox6 and Hes5. Sox6-3′UTR-IMu and Sox6-3′UTR-IIMu represent Sox6-3′UTR reporters with mutations of a miR-219 binding site in UTR-I and two miR-338-5p binding sites in UTR-II, respectively, as shown in A. The histogram shows the ratio of the luciferase activity normalized to control expression vectors carrying miR-219rev and miR-338rev, respectively. Data represent mean ± SDs from three independent experiments. * P < 0.01.
F) qRT-PCR analysis of Sox6 expression from RNAs isolated from mouse primary OPC cultures 4 days after transfection with miR-219, miR-338 (5p+3p) mimics or both. Scrambled miR transfection was included as control. * P < 0.01.
G) Western blot analysis shows that Sox6 protein was downregulated by miR-219 or miR-338 overexpression compared with vector-transfected cells. GAPDH was included as an internal control.
H) qRT-PCR analysis of Hes5 expression from RNAs isolated from mouse primary OPC cultures transfected with miR-219, miR-338 (5p+3p) mimics or both. Scrambled miR transfection was included as control. * P < 0.01.
I-J) Expression of Sox6 and Hes5 in rat hippocampal neural precursor cells was examined by immunostaining after 3 days transfection with miR-219 or miR-338-expressing and control vectors. Arrows indicate the transfected cells. Scale bars: 50μm.
K-L) Quantification of Sox6 (K) and Hes5 (L) positive cells among transfected cells (GFP+) from three independent above experiments. * P < 0.01.
M-N) qRT-PCR analyses of Sox6 (M) and Hes5 (N) expression using RNAs isolated from mouse primary OPC cultures 4 days after transfection with LNA-miR-219, LNA-miR-338 (5p+3p) or both knockdown probes as well as the scrambled oligonucleotide control (ctrl) as indicated. Histograms indicated that knockdown of these miRNAs led to significant upregulation of Sox6 and Hes5. * P < 0.01. GAPDH was included as an internal control.
To determine whether expression of Sox6 and Hes5 can be negatively regulated by miR-219 and/or miR-338, we transfected primary oligodendrocyte precursor-enriched cultures with miR-219 and/or miR-338 mimics. Four days after transfection, total RNAs isolated from control and miRNA-mimics treated cells were subject to qRT-PCR measurement. Although the mRNA level of Sox6 was downregulated approximately 25% and 20% by miR-219 and miR-338 mimics, respectively (Figure 7F), Sox6 protein expression was reduced significantly ∼75% after transfection of miR-219 or miR-338 mimics as shown by Western blot analysis (Figure 7G), suggesting a post-transcriptional inhibition by these miRNAs. In contrast, mRNA expression of Hes5 was drastically decreased after treatment of miR-219 or miR-338 mimics (Figure 7H). In addition, we transfected expression vectors for miR-219 and miR-338 into adult hippocampal derived neural progenitors and induced them to differentiate into oligodendrocytes with IGF-1 (Hsieh et al., 2004). Cells transfected with miR-219 or miR-338 exhibited a substantial reduction of Sox6 and Hes5 expression compared to control (Figure 7I-L). Conversely, knockdown miR-219 and/or miR-338 by miRNA inhibitors resulted in an upregulation of Sox6 and Hes5 expression in primary oligodendrocyte precursor culture (Figure 7M, N). Thus, these observations suggest that expression of Sox6 and Hes5 can be negatively regulated by miR-219 and miR-338.
Progression from neural progenitor cells into the oligodendroglial lineage requires downregulation of factors directing other neural cell lineages such as neurons. Consistent with this, miR-219 and miR338 also target to a number of genes known in regulating neuronal differentiation such as pro-neurogenic factors NeuroD1, Isl1 and Otx2 (Lee et al., 1995; Pfaff et al., 1996; Vernay et al., 2005) (Table S2). Both miR-219 and miR-338 can specifically target to the 3′UTR of Zfp238/RP58, which is required for cortical neurogenesis (Okado et al., 2009) (Figure S4). Zfp238 appears to be an inhibitor of oligodendrocyte differentiation since overexpression of Zfp238 in neural progenitor cells repressed expression of Olig2, an essential factor for oligodendrocyte development, and inhibited formation of RIP+ mature oligodendrocytes (Figure S4). Collectively, our data suggest that miR-219 and miR-338 control oligodendrocyte maturation by negatively regulating oligodendrocyte differentiation inhibitors and perhaps pro-neuronal differentiation factors.
Discussion
miRNAs are Important Regulators of Oligodendrocyte Differentiation and Myelination
Over the past few years, miRNAs have been implicated in many biological processes. Our work presented here provides the first evidence, to the best of our knowledge, that miRNAs, particularly miR-219 and miR-338, are critical regulators of oligodendrocyte differentiation and myelination in the vertebrate CNS. Our studies demonstrate that miRNAs are not only essential for the formation of myelinating oligodendrocytes but also sufficient to promote the differentiation from their precursors. These results challenge the view that miRNAs mainly play fine-tuning roles in developmental processes (Bartel, 2004; Schratt, 2009; Tsai and Yu, 2009).
Inhibition of miRNA maturation by selectively deleting the miRNA processing enzyme Dicer1 in oligodendrocyte lineage cells results in severe myelination deficits in the CNS. In the absence of Dicer1, oligodendrocyte progenitors exhibit an increase of proliferation but they fail to further differentiate into mature myelinating oligodendrocytes. These observations suggest that miRNAs normally inhibit OPC proliferation, while promoting their differentiation. Thus, miRNAs generated after Dicer1-mediated processing are the key regulators for the transition of OPCs into mature myelinating oligodendrocytes.
The significant reduction of Dicer1 expression in the optic nerve of Dicer1CKO mutants as compared to control mice suggests that the excision of the floxed exon 23 occurred in oligodendrocyte lineage cells mediated specifically by Olig1-Cre. In addition, it also suggests that a majority of Dicer1 is expressed in mature oligodendrocytes rather than in their precursors and other cell types in the optic nerve. This is consistent with data from cultured oligodendrocytes and OPCs (Figure S1).
The present Dicer1CKO mutant phenotype is perinatal-lethal, unlike the parallel study from Dugas et al. in which Dicer1flox/flox;Olig2-Cre mice show a developmental delay but ultimate recovery of myelination (see the companion paper). This apparent discrepancy is most likely the result of differences in the efficiency of Cre-mediated Dicer1 excision and the differences of temporal and spatial expression of Cre recombinase driven by Olig1 and Olig2 promoters as used in our two studies respectively. In the Dugas et al. study, the authors observed a recovery of myelination in the older Dicer1flox/flox;Olig2-Cre animals, however, they also demonstrated that functional Dicer1 expression in mature oligodendrocytes recovered in these older animals. The most likely explanation is that the floxed Dicer1 allele is inefficiently removed in cells where Cre expression is driven by the Olig2 promoter, and that the few immature cells in which Dicer1 expression is not disrupted eventually expand and restore near normal levels of myelin in the older mutant animals. In the Dicer1CKO mice, we observed a more complete deletion of Dicer1 expression mediated by Olig1-Cre, which corresponds with the eventual postnatal lethality of Dicer1CKO mutant mice.
Oligodendrocyte Differentiation Induced by miR-219 and miR-338
We show at a single-cell resolution that miR-219 and miR-338 are mainly expressed in CC1+ differentiated oligodendrocytes in the developing spinal cord. The fact that their expression is essentially absent in the myelin-specific deficient Dicer1 and Olig1 mutant mice supports the notion that these miRNAs are primarily, if not exclusively, expressed in oligodendrocytes in the spinal cord. Our in vitro and in vivo functional studies demonstrate that miR-219 and miR-338 are both necessary and sufficient for oligodendrocyte differentiation, although we do not exclude the possibility that additional miRNAs may also contribute to oligodendrocyte specification and differentiation.
By manipulating the expression level of miR-219 and miR-338 in oligodendrocyte precursor culture, we found that miR-219 and miR-338 can promote oligodendrocyte differentiation. The combined treatment of miR-219 and miR-338 mimics promote oligodendrocyte differentiation while resulting in a slight reduction of the number of PDGFRα+ OPCs, although we did not detect significant change of the number of PDGFRα+ OPCs treated with individual miRNA mimics (Figure 4E). The exact reason for this difference is not clear. Based on in vivo data that both miR-219 and miR-338 can promote PDGFRα+ OPC formation from neural progenitors, it is possible that miR-219 and miR-338 mimics may have an effect on promoting OPC formation from early progenitors, which may compensate the reduction of OPCs due to their differentiation into mature oligodendrocytes. The treatment of both miR-219 and miR-338 mimics may have more potent effects than an individual miRNA mimic on promoting PDGFRα+ OPC differentiation into mature oligodendrocytes. In addition, by introducing miR-219 and miR-338 in OPC cultures derived from Dicer1CKO mice, we observed that both of miRNAs could compensate, at least partially, for the defects in oligodendrocyte differentiation due to the loss of Dicer1, suggesting a crucial role of these miRNAs in promoting oligodendrocyte maturation.
In developing chick neural tube, ectopic expression of these miRNAs promotes oligodendrocyte specification from neural progenitor cells. miR-338 appears to be more potent than miR-219 in promoting mature oligodendrocyte formation since no MBP+ cells were detected in neural tube with ectopic miR-219 expression. However, overexpression of either miR-219 or miR338 is able to promote precocious oligodendrocyte differentiation in the embryonic mouse cortex, suggesting a species- or context-dependent function of these miRNAs.
A Critical Role of miR-219 and miR-338 in Oligodendrocyte Maturation
Blocking endogenous activities of miR-219 and miR-338 individually or in combination inhibits OPC differentiation and maturation in culture. In addition, loss of function of miR-219 results in an impairment of Olig2+ OPC migration and differentiation in zebrafish, suggesting an essential role of miR-219 in oligodendrocyte development. Intriguingly, despite the robust activity of miR-338 in promoting oligodendrocyte differentiation in developing chick neural tube and embryonic mouse cortex, knockdown of miR-338 in zebrafish did not have detectable effects on OPC differentiation. Despite the evolutionary conservation of miR-338, tissue specificity may have changed in different species. In zebrafish, miR-338 expression is barely detectable in the CNS (Wienholds et al., 2005), whereas in mice, miR-338 is mainly expressed in oligodendrocytes in the spinal cord. Thus, specific cell type expression or perhaps, target selection, might also provide an explanation for the major differences of phenotype observed between zebrafish and mouse.
miR-219 was first described as a clock- and light-regulated gene that modulates the circadian clock located in the suprachiasmatic nucleus in the adult mouse brain (Cheng et al., 2007). Recently, miR-219 was found to participate in NMDA receptor signaling by targeting CaMKII gamma to modulate behavioral responses (Kocerha et al., 2009) and ELOVL7 encoding a lipid synthesis enzyme (Shin et al., 2009). miR-338 is an intronic miRNA, transcribed together with a host gene encoding an apoptosis-associated tyrosine kinase (Barik, 2008). It may participate in neuronal differentiation and function (Kim et al., 2004; Raghunath et al., 2000). Although miR-219 and miR-338 could be expressed in a population of neurons, we found that these miRNAs are mainly expressed in oligodendrocytes in the spinal white matter at postnatal stages. Thus, miR-219 and miR-338 may have pleiotropic effects on other biological processes, while promoting oligodendrocyte differentiation in a context- and temporally specific manner.
Downregulation of Differentiation Inhibitors by miRNAs is a Critical Event for Oligodendrocyte Terminal Differentiation
The timing of oligodendrocyte differentiation is controlled by the balance of activities between positive and negative regulators. Sox6 and Hes5 could be physiological targets of miR-219 and miR-338 because both miRNAs have multiple target recognition sites in their 3′UTR. We validated Sox6 and Hes5 to be bona fide targets by demonstrating that their expression levels can be directly controlled by miR-219 and miR-338 through their 3′ UTR. Identification of Sox6 and Hes5 as functional targets of miR-219 and miR-338 is consistent with their inhibitory functions in oligodendrocyte differentiation (John et al., 2002; Kondo and Raff, 2000; Liu et al., 2006; Stolt et al., 2006). Loss of Sox6 or Hes5 in mice results in upregulation of myelin gene expression (Kondo and Raff, 2000; Liu et al., 2006; Stolt et al., 2006). Sox6, a member of the SoxD family, competes and counters the function of SoxE proteins such as Sox10, which promotes oligodendrocyte terminal differentiation (Stolt et al., 2006). Notably, Sox6 transcript is mainly present in PDGFRα+ oligodendrocyte precursors, but not in the MBP+ mature oligodendrocytes, consistent with its role as a negative regulator of oligodendrocyte maturation (Figure S3). Hes5, another potential target of these miRNAs, negatively regulates oligodendrocyte differentiation and myelin gene expression by repressing the differentiation activators Sox10 and Mash1 (Kondo and Raff, 2000; Liu et al., 2006; Wang et al., 1998).
Regulation of differentiation inhibitors by miRNAs suggests a common theme by which oligodendrocyte differentiation can be executed through the combinatorial actions of miRNAs and transcriptional regulators. Unlike pro-neuronal genes, which are downregulated after neuronal differentiation, oligodendrocyte differentiation-promoting factors, including Olig1/2, Sox10 and MRF (Emery et al., 2009; Wegner, 2008), are normally maintained in differentiated oligodendrocytes. Our observations suggest that an active repression of a cohort of differentiation inhibitors such as Sox6 and Hes5 and perhaps pro-neuronal differentiation factors by miRNAs can be one of critical steps necessary for allowing OPCs to become terminal differentiated oligodendrocytes (Figure 8).
Figure 8. Schematic Diagram for the Potential Function of miR-219 and miR-338 in Regulating Oligodendrocyte Differentiation.
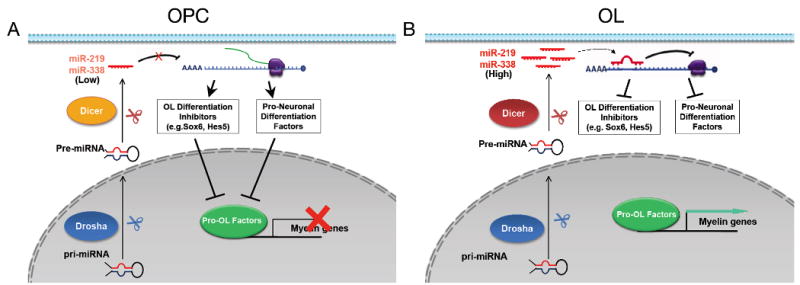
A) In oligodendrocyte precursor cells (OPCs), mature miRNAs (e.g. miR-219 and miR-338) and their processing enzyme Dicer1 are present at a relatively low level. The amount of miRNAs in OPCs is not sufficient to repress the expression of oligodendrocyte differentiation inhibitors (e.g. Sox6 and Hes5) and/or pro-neuronal differentiation factors (e.g. NeuroD1 and Zfp238), which thereby prevent OPC differentiation.
B) During oligodendrocyte differentiation, expression of mature miRNAs, particularly miR-219 and miR-338, and Dicer1 increases substantially. miR-219 and miR-338 permit or activate pro-OL factors for oligodendrocyte differentiation and maturation by downregulating a cohort of oligodendrocyte differentiation inhibitors, and repressing genes involved in neuronal differentiation. An important future study would be to identify the molecules that regulate temporal expression of these miRNAs during oligodendrocyte lineage progression. OPCs, oligodendrocyte precursor cells. OL, oligodendrocytes.
miR-219 and miR-338 may be used in future investigations as a tool to dissect the details of extracellular and intracellular pathways that are involved in oligodendrocyte differentiation. Because both transcriptional and posttranscriptional mechanisms exist for modulating miRNA expression (Bartel, 2004; Thomson et al., 2006), there is a possibility that a feedback regulatory loop of Hes5 and Sox6 may exist to regulate the expression of miR-219 and miR-338, as there are multiple consensus sites recognized by these transcription factors that are present in their putative promoter region (data not shown).
The discovery of miRNAs as important modulators of oligodendrocyte differentiation processes can also provide unanticipated insights into demyelinating disease mechanisms through the identification of the cellular effectors of miRNA action. It is conceivable that the manipulation of specific miRNAs such as miR-219 and miR-338 might have therapeutic value in promoting oligodendrocyte remyelination after injury.
Experimental Procedures
Animals, Tissue Processing and Immunohistochemistry
Olig1-Cre heterozygous mice were crossed with Dicer1lox/lox mice to generate Olig1Cre+/-; Dicerlox/+ mice, which were then bred with Dicer1lox/lox mice to produce Dicer1CKO offsprings (Olig1Cre+/-; Dicerlox/lox). Olig1Cre+/-; Dicerlox/+ mice developed and behaved the same as WT. Dicer1lox/lox mice were purchased from the Jackson laboratory (strain name, B6.Cg-Dicer1tm1Bdh/J) (Harfe et al., 2005). All animal use and studies were approved by the Institutional Animal Care and Use Committee of the University of Texas Southwestern Medical Center at Dallas, USA and Sichuan University at Chengdu, P.R. China.
For immunohistochemistry, mouse brains or spinal cord were short-fixed 1 hr in 4% paraformaldehyde and processed for vibratome-section. For BrdU pulse labeling, animals were injected intraperitoneally with 100 mg BrdU/kg body weight 3 hrs prior to sacrifice. We used antibodies to Olig2 (gift of C. Stiles, Harvard Medical School), Sox6 (gift of M. Wegner), PDGFRα (BD Bioscience, 558774), CC1 (Oncogene Research, OP80), MBP (Covance, SMI-94R), BrdU (BD Bioscience, 555627), GFAP (Sigma, G3893) and Hes5 (Santa Cruz, sc-13859). Monoclonal antibody to RIP were obtained from the Developmental Studies Hybridoma Bank at the University of Iowa under the auspices of the National Institute of Child Health and Human Development. The protocols of miRNA in situ hybridization and immunostaining following miRNA in situ hybridization are provided in detail in supplemental methods.
For electron microscopy, spinal cord and optic nerves were dissected and fixed in 2% glutaraldehyde and 4% paraformaldehyde in 0.1M cacodylate buffer (pH7.2) for 24 hrs and processed as previously described (Xin et al., 2005).
RNA Extraction and qRT-PCR
Total RNAs were purified from tissues or cell cultures using TRizol reagent according to the manufacturer's instructions (Invitrogen). RNA was transcribed to cDNA with First-Strand cDNA Synthesis Kit (GE Healthcare). Quantitative real-time PCR was performed using the ABI Prism 7700 Sequence Detector System (Perkin-Elmer Applied Biosystems) and the relative gene expression was normalized to internal control such as Gapdh or Rpl13a. Primer sequences for SybrGreen probes of target genes are as follows: mouse Sox6: “tcagagcaatcaccacaccagaca” and “aaggttgaatgtcagggcaaaggc”; mouse Hes5: “tacctgaagcacagcaaagccttc” and “taaagcagcttcatctgcgtgtcg”; mouse Dicer1: “accagcgcttagaattcctgggag” and “gctcagagtccattccttgc”; mouse GAPDH: “gtgtgaaccacgagaaata” and “gttgtcatggatgacctt”; zebrafish mbp: “atcttcaacctgggagaaagccga” and “tgcttcccgtccatttcactctct”; zebrafish ef1a: “tcaagcctggtatggttgtgacct” and “acggatgtccttga cagacacgtt”; zebrafish rpl13a: “tcctccgcaagagaatgaacacca” and “tcaaacaccttcagc ctgtccaga”. miRNA-specific primers were purchased from Applied Biosystems, Inc. and miRNA qRT-PCR was performed according to manufacturer's instruction.
miRNA Microarray, Northern blot and Western blot analyses
miRNAs microarray assays were performed by using a service provider (LC Sciences). The assay started from 10 μg of total RNA samples on Dual sample chips with Rodentia miRNA Array based on Sanger miRBase Release 13.0.
For miRNA Northern blot analysis, RNA extracted from spinal cord was electrophoresed on a 20% polyacrylamide (7.6 M urea) gel in 1× TBE. Ten micrograms of RNA was denatured for 5 min at 70°C in a buffer containing 50% formamide and 10 mM EDTA (pH 8.0) before loading. After electrophoresis, RNA was then transferred onto a Zeta probe membrane (Biorad) in 0.5× TBE buffer at 80 V for 1 h. Hybridization was performed at 37°C according to a standard protocol. 32P-labeled Star-Fire oligonucleotide probes (IDT) against mature miR-219, miR-338-5p, and U6 were purified on Sephadex G-25 microspin columns (BioRad) and used in the hybridization. For Western blot analysis, protein lysates were resolved by SDS-PAGE and blotted using standard procedures. Antibodies used were as follows: Sox6 (Santa Cruz) and GAPDH (Abcam). Signals were revealed by using chemiluminescence with the ECL kit (Pierce) according to the manufacturer's instruction.
Culture of Rodent Oligodendroglial Precursor Cells and Rat Hippocampus-Derived Adult Neural Progenitor Cells
For mouse primary OPC-enriched cell cultures, cortical precursors were isolated from E15.5 as described previously (Chen et al., 2007). Oligodendrocyte precursors were enriched in serum-free oligodendrocyte growth medium supplemented with bFGF and PDGF-AA. For miRNA tranfection study, primary OPCs were seeded in 24-well plates in oligodendrocyte growth medium and transfected with 50 nM of miRNA mimics (Pre-miRNAs, Ambion) using Lipofectamine 2000 (Invitrogen). A Cy3 labeled scrambled miRNA (Pre-miR™ miRNA Precursor Negative Control, Ambion Inc.) transfection was included as negative control and the transfection efficiency is >50% in OPC culture. Four days post-transfection, cultures were harvested for immunocytochemistry and qRT-PCR assay. In knockdown study, OPC-enriched cultures were transfected with 25 nM miRCURY LNA™ miRNA knockdown probes (Exiqon, Vedbaek, Denmark. mmu-miR-219, 139113-04; mmu-miR-338-5p, 139531-04; mmu-miR-338-3p, 139532-04) using Lipofectamine 2000, and control LNA knockdown probe was included as negative control (Exiqon, Product # 199002-04). The control knockdown probe has similar probe length as LNA knockdown probes, and it has no homology to any known miRNA or mRNA sequences in mouse, rat or human. Cells were then cultured in oligodendrocyte differentiation medium without mitogens and four days after transfection, cultures were harvested for analysis.
Isolation and culture of rat OPCs followed the protocol as previously described (Chen et al., 2007). The rat hippocampus-derived adult neural progenitor cells (HCN) were originally isolated from adult female Fisher 344 rats. They give rise to homogeneous populations of oligodendrocytes on IGF-1 stimulation (Hsieh et al., 2004). The progenitor cells expressed Olig2 when cultured in N2-FGF media (DMEM:F12 with N2 supplement and bFGF). HCN progenitor cells were grown in N2-FGF media, transfected with miRNAs or plasmids using Amaxa electroporator according to manufacturer's protocol and assayed for immunocytochemistry and qRT-PCR analysis.
miRNA Expression Vectors and Luciferase Reporter Assays
miR-219 locus on chromosome 2 and mir-338 locus on chromosome 11 with their ∼200 bp flanking sequences were PCR amplified from mouse genomic DNA and inserted into pCMV6 vector or pCIG vector (Megason and McMahon, 2002). For generating miRNA mutant constructs, the seeding sequence for miR-219 was changed from “GATTG” to “CTAAC”, seeding sequences for miR-338-5p and miR-338-3p were changed from “ACAAT” to “TGTTA” and from “CAGCA” to “GTCGT”, respectively.
Segments carrying putative miR-219 and miR-338 binding sites in 3′UTR of Sox6, Hes5 and Zfp238 were cloned into pMIR-REPORT vector (Ambion, Inc). For mutagenesis of 3′UTR for Sox6, the predicted binding site in pMir-reporter-Sox6UTR-I for miR-219 was changed from “ACAAT” to “GTCGA”, the two predicted binding sites in pMir-reporter-Sox6UTR-II for miR-338-3p was changed from “ATGCTG” to “GTCGAC” and from “ATGCTG” to “GTCGAC”, respectively by using QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene, San Diego, CA).
Luciferase reporter constructs were cotransfected with vectors expressing miRNAs, their reverse sequences (219rev, 338rev) or mutant miRNAs (219Mu, 338Mu) into COS7 cells by Lipofectamine 2000. The pRSV-renilla luciferase plasmid was included as a control for transfection efficiencies. Luciferase activity was assayed 48 hours after transfection using the dual-luciferase reporter assay system (Promega). At least three transfection assays were performed to obtain statistically significant data. Statistic analyses were performed as previously described (Ye et al., 2009).
In ovo and in utero Electroporation
Chicken eggs were incubated at 38 °C. Approximately 1 ul (3 μg/μl) of pCIG expression vectors carrying mir-219 or miR-338 was injected into a chicken embryo neural tube at stage HH 13–15 (E2.5) with the aid of Picospritzer III (Parker Hannifin). In situ hybridization or immunohistochemistry were performed as perviously described (Ye et al., 2009).
For in utero electroporation, DNA solution (∼2 μl) in PBS containing 0.01% fast green was injected into the lateral ventricle of the embryos at e14.5. After injection, electroporation (five 50 ms square pulses of 40 V with 990 ms intervals) was carried out. Plasmid DNAs (3 μg/μl) used for electroporation were pCMV-miR219 and pCMV-miR338 and control pRFP. Embryos were harvested 72 hrs after electroporation and processed for immunohistology. At least five embryos with expression of each transgene were analyzed and characterized.
Zebrafish Morpholino Injections
Approximately 1nl of morpholinos at 0.12 mM concentration for miR-219 and miR-338 (Gene Tools, LLC) was injected individually into Tg(olig2:egfp) zebrafish embryos at the one-cell stage. The morpholino sequences used were miR219: 5′ acagatgtccaggcacaattctt gg-3′ and miR338: 5′- caacaaaatcactgatgctggagtg-3′. Injected embryos at 3 dpf were paraformaldehyde-fixed, cryosectioned and immunostained using an anti-Sox10 antibody or processed for in situ RNA hybridization using probe for Mbp.
Supplementary Material
Acknowledgments
Authors would like to thank Qinjie Weng, Zhangyan Ma, Linan Chen, Wei Liu, Christina Kearns and Sarah Casper for technical support. We thank Drs. Jonah Chan and Jianrong Li for a critical reading of the manuscript, Jason Dugas and Ben Barres for communicating unpublished results, Michael Wegner for Sox6 antibody, Fiona Doetsch for miRNA in situ protocol, James Amatruda for PCR primers and Ellen Lu for graph drawing. This study was funded by grants from the US National Multiple Sclerosis Society (RG3978 and PP0144) and the US National Institutes of Health (NS050389 to QRL). QRL is a Harry Weaver Neuroscience Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barik S. An intronic microRNA silences genes that are functionally antagonistic to its host gene. Nucleic Acids Res. 2008;36:5232–5241. doi: 10.1093/nar/gkn513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J, Moser HW, Forss-Petter S. Leukodystrophies: recent developments in genetics, molecular biology, pathogenesis and treatment. Curr Opin Neurol. 2001;14:305–312. doi: 10.1097/00019052-200106000-00007. [DOI] [PubMed] [Google Scholar]
- Chen Y, Balasubramaniyan V, Peng J, Hurlock EC, Tallquist M, Li J, Lu QR. Isolation and culture of rat and mouse oligodendrocyte precursor cells. Nat Protoc. 2007;2:1044–1051. doi: 10.1038/nprot.2007.149. [DOI] [PubMed] [Google Scholar]
- Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, et al. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copray S, Huynh JL, Sher F, Casaccia-Bonnefil P, Boddeke E. Epigenetic mechanisms facilitating oligodendrocyte development, maturation, and aging. Glia. 2009 doi: 10.1002/glia.20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B, Agalliu D, Cahoy JD, Watkins TA, Dugas JC, Mulinyawe SB, Ibrahim A, Ligon KL, Rowitch DH, Barres BA. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell. 2009;138:172–185. doi: 10.1016/j.cell.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. Common logic of transcription factor and microRNA action. Trends Biochem Sci. 2004;29:462–468. doi: 10.1016/j.tibs.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Aimone JB, Kaspar BK, Kuwabara T, Nakashima K, Gage FH. IGF-I instructs multipotent adult neural progenitor cells to become oligodendrocytes. J Cell Biol. 2004;164:111–122. doi: 10.1083/jcb.200308101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John GR, Shankar SL, Shafit-Zagardo B, Massimi A, Lee SC, Raine CS, Brosnan CF. Multiple sclerosis: Re-expression of a developmental pathway that restricts oligodendrocyte maturation. Nat Med. 2002;8:1115–1121. doi: 10.1038/nm781. [DOI] [PubMed] [Google Scholar]
- Kawase-Koga Y, Otaegi G, Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev Dyn. 2009;238:2800–2812. doi: 10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci U S A. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocerha J, Faghihi MA, Lopez-Toledano MA, Huang J, Ramsey AJ, Caron MG, Sales N, Willoughby D, Elmen J, Hansen HF, et al. MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc Natl Acad Sci U S A. 2009;106:3507–3512. doi: 10.1073/pnas.0805854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Raff M. Basic helix-loop-helix proteins and the timing of oligodendrocyte differentiation. Development. 2000;127:2989–2998. doi: 10.1242/dev.127.14.2989. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci. 2008;28:11720–11730. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- Leucht C, Stigloher C, Wizenmann A, Klafke R, Folchert A, Bally-Cuif L. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat Neurosci. 2008;11:641–648. doi: 10.1038/nn.2115. [DOI] [PubMed] [Google Scholar]
- Liu A, Li J, Marin-Husstege M, Kageyama R, Fan Y, Gelinas C, Casaccia-Bonnefil P. A molecular insight of Hes5-dependent inhibition of myelin gene expression: old partners and new players. Embo J. 2006;25:4833–4842. doi: 10.1038/sj.emboj.7601352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Megason SG, McMahon AP. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development. 2002;129:2087–2098. doi: 10.1242/dev.129.9.2087. [DOI] [PubMed] [Google Scholar]
- Miller RH. Regulation of oligodendrocyte development in the vertebrate CNS. Prog Neurobiol. 2002;67:451–467. doi: 10.1016/s0301-0082(02)00058-8. [DOI] [PubMed] [Google Scholar]
- Okado H, Ohtaka-Maruyama C, Sugitani Y, Fukuda Y, Ishida R, Hirai S, Miwa A, Takahashi A, Aoki K, Mochida K, et al. The transcriptional repressor RP58 is crucial for cell-division patterning and neuronal survival in the developing cortex. Dev Biol. 2009;331:140–151. doi: 10.1016/j.ydbio.2009.04.030. [DOI] [PubMed] [Google Scholar]
- Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;84:309–320. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- Raghunath M, Patti R, Bannerman P, Lee CM, Baker S, Sutton LN, Phillips PC, Damodar Reddy C. A novel kinase, AATYK induces and promotes neuronal differentiation in a human neuroblastoma (SH-SY5Y) cell line. Brain Res Mol Brain Res. 2000;77:151–162. doi: 10.1016/s0169-328x(00)00048-6. [DOI] [PubMed] [Google Scholar]
- Rowitch DH. Glial specification in the vertebrate neural tube. Nat Rev Neurosci. 2004;5:409–419. doi: 10.1038/nrn1389. [DOI] [PubMed] [Google Scholar]
- Schratt G. Fine-tuning neural gene expression with microRNAs. Curr Opin Neurobiol. 2009;19:213–219. doi: 10.1016/j.conb.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Shen S, Casaccia-Bonnefil P. Post-translational modifications of nucleosomal histones in oligodendrocyte lineage cells in development and disease. J Mol Neurosci. 2008;35:13–22. doi: 10.1007/s12031-007-9014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Shin JY, McManus MT, Ptacek LJ, Fu YH. Dicer ablation in oligodendrocytes provokes neuronal impairment in mice. Ann Neurol. 2009;66:843–857. doi: 10.1002/ana.21927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Park HC, Topczewska JM, Mawdsley DJ, Appel B. Neural cell fate analysis in zebrafish using olig2 BAC transgenics. Methods Cell Sci. 2003;25:7–14. doi: 10.1023/B:MICS.0000006847.09037.3a. [DOI] [PubMed] [Google Scholar]
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Schlierf A, Lommes P, Hillgartner S, Werner T, Kosian T, Sock E, Kessaris N, Richardson WD, Lefebvre V, et al. SoxD proteins influence multiple stages of oligodendrocyte development and modulate SoxE protein function. Dev Cell. 2006;11:697–709. doi: 10.1016/j.devcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp BD, Peterson J, Ransohoff RM, Rudick R, Mork S, Bo L. Axonal transection in the lesions of multiple sclerosis. N Engl J Med. 1998;338:278–285. doi: 10.1056/NEJM199801293380502. [DOI] [PubMed] [Google Scholar]
- Tsai LM, Yu D. MicroRNAs in common diseases and potential therapeutic applications. Clin Exp Pharmacol Physiol. 2009 doi: 10.1111/j.1440-1681.2009.05269.x. [DOI] [PubMed] [Google Scholar]
- Vernay B, Koch M, Vaccarino F, Briscoe J, Simeone A, Kageyama R, Ang SL. Otx2 regulates subtype specification and neurogenesis in the midbrain. J Neurosci. 2005;25:4856–4867. doi: 10.1523/JNEUROSCI.5158-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sdrulla AD, diSibio G, Bush G, Nofziger D, Hicks C, Weinmaster G, Barres BA. Notch receptor activation inhibits oligodendrocyte differentiation. Neuron. 1998;21:63–75. doi: 10.1016/s0896-6273(00)80515-2. [DOI] [PubMed] [Google Scholar]
- Wegner M. A matter of identity: transcriptional control in oligodendrocytes. J Mol Neurosci. 2008;35:3–12. doi: 10.1007/s12031-007-9008-8. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Xin M, Yue T, Ma Z, Wu FF, Gow A, Lu QR. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci. 2005;25:1354–1365. doi: 10.1523/JNEUROSCI.3034-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, Bu H, Hu T, Taketo MM, van Es JH, Clevers H, et al. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009 doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.