Abstract
RNA interference is currently envisioned as the basis of gene function and drug target validation studies. This novel technology has the advantage of providing a remarkably faster tool for gene silencing than traditional transgenic animal methodologies. In vivo administration of short interfering RNA (siRNA) typically results in reduced target gene expression for approximately 1 week. Viral vectors offer the possibility to express constitutive levels of short hairpin RNA (shRNA) so that the effects of knocking down the target gene can be studied for a few weeks, rather than a few days. Helper-dependent vectors have a significant advantage over previous generations of adenoviral vectors because of their much higher cloning capacity, potential for long-term transgene expression, and enhanced safety profiles on administration in vivo. Therefore, this advanced type of vector is an excellent tool to carry out in vivo studies directed at constitutive expression of shRNA. Here we show it is possible to obtain more than 90% target gene knockdown in an animal model of type 2 diabetes for several weeks, thereby consolidating this technology as an alternative to generating liver-specific knockout animals.
Introduction
RNA interference (RNAi) has become the cornerstone of gene function studies, circumventing the limitations of knockout animals and shortening the otherwise long process of target identification and validation. In some instances, gene ablation is incompatible with survival through the embryo stages, or gene compensatory mechanisms occur. One example is the transcription factor sterol regulatory element-binding protein-1c (SREBP-1), a central mediator of insulin action on lipid and carbohydrate metabolism in the liver (Horton et al., 2003). SREBP-1 activity is enhanced in animal models of sucrose-induced hepatic insulin resistance, obesity, and type 2 diabetes (Shimomura et al., 1999; Nagai et al., 2002). Approximately 85% of SREBP-1 null mice die in utero. RNAi has the potential to overcome these limitations because it allows the conduction of experiments in adult animals, thereby bypassing developmental stages.
Helper-dependent or high-capacity adenoviral vectors have emerged as a valuable tool for gene delivery to the liver (Morral et al., 1998, 1999; Schiedner et al., 1998). This advanced type of vector retains the inverted terminal repeats and packaging signal, but does not contain any viral coding sequences (Mitani et al., 1995; Clemens et al., 1996; Fisher et al., 1996; Haecker et al., 1996; Kochanek et al., 1996; Kumar-Singh and Chamberlain, 1996; Lieber et al., 1996; Parks et al., 1996; Alemany et al., 1997; Chen et al., 1997; Hardy et al., 1997). A single intravenous injection results in lifelong expression of the transgene in mice and rats (Kim et al., 2001; Oka et al., 2001; Toietta et al., 2005). Furthermore, administration of high doses is well tolerated, without inducing major alterations in hepatic function markers (Morral et al., 1998).
To provide the proof of concept that helper-dependent adenoviral vector-mediated short hairpin RNA (shRNA) expression represents an alternative strategy to traditional knockout strategies and allows study of the effects of inhibiting gene activity in liver, we have generated a vector expressing an shRNA to target SREBP-1. A potential problem to generating vectors containing shRNA expression cassettes is the possibility of recombination events that eliminate the hairpin sequence during vector propagation (Narvaiza et al., 2006). In addition to the successful knockdown of SREBP-1, we observe that genomes of helper-dependent vectors containing shRNA expression cassettes are not more susceptible to recombination events than transgene-encoding vectors.
Materials and Methods
Expression cassette design and shRNA testing in HEK-293 cells
Plasmid pAlb, containing the albumin promoter/enhancer, was a generous gift from R. Palmiter (Howard Hughes Medical Institute, University of Washington, Seattle, WA) (Pinkert et al., 1987). Plasmid pRES (see Helper-Dependent Adenoviral Vector Production, below) was cut with XbaI and AscI, and the resulting fragment was cloned into the NotI site of pAlb. Plasmid pRES was then cut with XhoI and AscI and the resulting 800-bp fragment was cloned into pRES-Alb. The fatty acid-binding protein-5 (FABP5, a.k.a mal1) cDNA with a bovine growth hormone poly(A) sequence has been previously described (Witting et al., 2008), and was introduced into the EcoRV site of pRES-Alb after digestion with HindIII and XhoI and blunting with Klenow fragment. Plasmid pRES-Alb-FABP5 was used to rescue a helper-dependent adenoviral vector.
To generate constructs containing shRNA hairpins, 19- or 21-bp sequences were selected as described (Witting et al., 2008) and cloned into pENTR of the BLOCK-iT U6 RNAi entry vector kit (Invitrogen, Carlsbad, CA) or pRNA-H1.1/Neo (GenScript, Piscataway, NJ ), in accordance with the manufacturer's instructions. The first system (Invitrogen) uses the human U6 promoter to drive shRNA expression. The following sequences were targeted: fatty acid-binding protein-5 (FABP5), 5′-GGA-GAGAAGTTTGATGAAACG-3′ (previously described as sh242; Witting et al., 2008); scrambled control (SCR), 5′-GAG-AGTATAAGGAGGTCAAGT-3′ (Witting et al., 2008); LacZ (5′-CTACACAAATCAGCGATTT; sequence provided in the BLOCK-iT U6 kit); and sterol regulatory element-binding protein-1 (SREBP-1), 5′-GGTTGTGGACACAGACAAACT-3′, 5′-GCTTCTAACCTGGCACTAAGT-3′, 5′-GGAGGACATCTT-GCTGCTTCT-3′, and 5′-GCTGGCCAATGGACTACTAGT-3′. The second system (GenScript) uses the H1 promoter. Three constructs were generated to target SREBP-1: 5′-TTGGTTG-TGGACACAGACAAA-3′, 5′-GATATCTGCAGTTGCTAAA-TA-3′, and 5′-ACCAACTGGCAGTTCCATTGA-3′, and a scrambled sequence was used as control: 5′-ATAGTTGTCCG-TTGCACTCGA-3′.
To test the efficacy of the SREBP-1 shRNA constructs, a plasmid expressing the target gene was generated. Mouse SREBP-1 cDNA was obtained from the American Type Culture Collection (Manassas, VA) and cloned into plasmid pEF1α-SREBP1, containing the elongation factor-1α promoter and bovine growth hormone polyadenylation signal (Morral et al., 2002), by blunt-end ligation. HEK-293 cells were obtained from Microbix Biosystems (Toronto, ON, Canada) and cultured in MEMα supplemented with 10% fetal bovine serum. To test shRNA efficacy, HEK-293 cells were seeded in 6-well plates at 1.0 × 106 cells per well and the next day were transfected with 0.75 μg of pEF1α-SREBP1 and 1.5 μg of shRNA-expressing plasmid. METAFECTENE reagent (Biontex Laboratories, Martinsried/Planegg, Germany) was used at a DNA-to-lipid ratio of 1:3. Cells were harvested after 24 hr and lysed in modified radioimmunoassay (RIPA) buffer containing protease inhibitors (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 0.25% [w/v] deoxycholate, 1% Triton X-100, 1 μM phenylmethylsulfonyl fluoride [PMSF], aprotonin/leupeptin/pepstatin [1 μg/ml each]; pH 7.5). Western blot analysis was carried out to determine the level of SREBP-1 silencing (data not shown). Sequences 5′-GGTTGTGGACA-CAGACAAACT-3′ (U6 promoter) and 5′-ACCAACTGGC-AGTTCCATTGA-3′ (H1 promoter) gave the highest level of knockdown. The expression cassette was amplified by polymerase chain reaction (PCR), using primers binding the flanking regions, and subsequently cloned into the AscI restriction site of plasmid pRES. Clones were sequenced before being used for rescue of helper-dependent adenoviral vectors (see the next section).
Helper-dependent adenoviral vector production
Helper-dependent adenoviral vectors were generated with a Cre–loxP system (Microbix Biosystems) (Chen et al., 1996; Parks et al., 1996; Sandig et al., 2000). Cloning of expression cassettes into plasmid pC4HSU was carried out by homologous recombination in Escherichia coli BJ5183 (Youil et al., 2001). Briefly, plasmid pRES was created by cloning a 3.2-kb HindIII fragment from pC4HSU into pBluescript II-SK (Stratagene, La Jolla, CA). All expression cassettes were cloned into pRES, as described previously, and the resulting plasmids were digested with HindIII. Simultaneously, plasmid pC4HSU (Sandig et al., 2000) was digested with AscI. Escherichia coli BJ5183 cells were cotransformed with the linearized plasmid (100 ng) and each of the HindIII fragments (300 ng), as described previously (Youil et al., 2001). Helper-dependent adenovirus plasmids containing expression cassettes were generated through recombination between homologous regions.
To rescue the helper-dependent adenoviral vectors, pC4HSU plasmids containing expression cassettes were digested overnight with PmeI to remove the bacterial sequence, and DNA was purified with QIAEX II (Qiagen, Valencia, CA). Five micrograms of PmeI-digested plasmid DNA was used to transfect 293Cre4 cells, so that approximately 40–50% cells were positive. This minimized the number of passages needed to reach plateau level (Parks et al., 1996). To monitor the amplification process, a pC4HSU construct expressing green fluorescent protein (pC4HSU-GFP) was used in parallel. Twenty-four hours later, helper H14 was added at a multiplicity of infection (MOI) of 3. This MOI was determined in a separate experiment to result in >95% transduction and a low level of helper contamination (<2.0 × 106 plaque-forming units [PFU]/ml) in cell lysates harvested at the onset of >95% cytopathic effect (CPE). To monitor the percentage of cells producing vector at each passage, we used the vector expressing GFP. We rationalized that maximal vector production occurs when all cells are infected with at least one vector and one helper particle. After three consecutive passages in 6-cm dishes (5 × 106 cells per dish), >95% cells expressed GFP, indicating that maximal vector production was achieved. For large-scale vector amplification 15-cm dishes (2.0 × 107 cells per dish) were used followed by 500-cm2 flasks (∼1 × 109 cells total). A critical point was to use the appropriate dilution factor when escalating to larger dishes, as diluting the lysate excessively resulted in low yields or no vector production (data not shown). To confirm that >95% of 293Cre4 cells produced the vector, separate dishes were used to monitor GFP expression. The virus was purified by one-step gradient centrifugation (CsCl at 1.25 and 1.40 g/ml; two SW28 tubes) for 2 hr at 22,500 rpm, followed by one isopycnic separation (CsCl at 1.33 g/ml; four SW40 tubes) at 35,000 rpm for 36–42 hr. The helper-dependent adenovirus band (upper) was collected, placed in dialysis cassettes (Pierce Biotechnology, Rockford, IL), and dialyzed in TMNG buffer (10 mM Tris-HCl [pH 7.5], 1 mM MgCl2, 150 mM NaCl, 10% glycerol) for 4–5 hr at 4°C (dialysis buffer was replaced with fresh cold buffer every hour). Total particle counts were determined spectrophotometrically after particle disruption with 0.1% sodium dodecyl sulfate (SDS) (absorbance at 260 nm [A260] = 1 corresponds to 1.1 × 1012 particles/ml) (Maizel et al., 1968). The level of contamination with helper was determined by plaque assay.
Animals
Male 8-week-old C57BL/6J and db/db mice were obtained from The Jackson Laboratory (Bar Harbor, ME). Animal care guidelines set forth by the Indiana University School of Medicine (Indianapolis, IN) were followed. Mice were kept in a BSL2 facility and had free access to a standard chow diet and water at all times. At the time points indicated in text, the animals were killed and tissues were collected and snap-frozen in liquid nitrogen or fixed in 10% buffered formalin for histological analysis.
Western blotting
Lysed HEK-293 cells were centrifuged at 12,000 × g and the supernatant was used for Western blot analysis, as previously described (Witting et al., 2008). To generate liver protein extracts, 150–200 mg of frozen liver was homogenized in modified RIPA buffer containing protease inhibitors. Liver extracts were centrifuged at 12,000 × g, the fat layer was carefully aspirated, and the supernatant was collected for use in Western blotting. Approximately 40 μg were separated in 4–15% SDS–PAGE Criterion gels (Bio-Rad) and transferred to 0.2-mm polyvinylidene difluoride (PVDF ) membranes (Bio-Rad, Hercules, CA). Antibodies to SREBP-1 were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA). Blots were developed with Immun-Star (Bio-Rad) and exposed to enhanced chemiluminescence (ECL) film (GE Healthcare, Piscataway, NJ ). Densitometric analysis was carried out with LabWorks 4.6 software (UVP, Upland, CA).
Histology
Four-micron-thick sections were cut from routinely processed paraffin-embedded tissue and stained with hematoxylin and eosin for histological analysis. Processing and staining were performed at the Immunohistochemistry Core (IHC) at Indiana University Medical Center (Indianapolis, IN).
Liver enzymes
Levels of aspartate aminotransferase (AST) in serum were measured with a kit from Pointe Scientific (Canton, MI), in accordance with the manufacturer's instructions.
Real-time PCR
Amplification of helper-dependent vector genomes was carried out by real-time PCR using primer pair pSHL-F (5′-GCGTGCCAGACAAAAGGAAAG-3′) and pSHL-B (5′-CACTCCAGCAGCCCAGAATC-3′). The glucokinase gene was used as loading control, using primer pair GK-F (5′-GTTTCGGAGGGACTGCATGG-3′) and GK-B (5′-ACGAG-CCTGAGCAGCACAAG-3′). Real-time PCR was performed with an ABI PRISM 7500 instrument (Applied Biosystems, Foster City, CA), and the ABI Power SYBR Green kit, in accordance with the manufacturer's protocol and using a 0.5 mM concentration of each primer. Primer pairs were designed to amplify a fragment of approximately 350–375 bp and were first tested to yield a single PCR product based on the melting curve and confirmation by agarose gel electrophoresis. A standard curve was generated by spiking DNA, isolated from a mouse treated with vehicle, with known amounts of pSHL-GFP plasmid to generate a standard curve with 0.01, 0.1, 1, 10, and 100 copies per cell. DNA levels in standards and test samples were measured by analyzing 30 ng of DNA, in duplicate.
Results and Discussion
Production of shRNA-expressing helper-dependent adenoviral vectors
To generate reproducible yields with each of the vectors rescued, we used the protocol described in Materials and Methods. Using this strategy, titers of the first large-scale preparation were 3–7 × 1011 viral particles (VP)/ml, and overall yields were 7–28 × 1011 VP, similar to those obtained for transgene-expressing vector gAd.Alb-mal1 (Table 1). None of these preparations showed the presence of rearrangements, based on DNA analysis using restriction enzymes (Fig. 1).
Table 1.
Helper-Dependent Vector Production
| Vector | Titer (particles/ml) | Total yield (particles) | Helper Contamination (PFU/ml) | Lot used for infection |
|---|---|---|---|---|
| gAd.shFABP5-1 | 2.8 × 1011 | 7.7 × 1011 | N/D | |
| gAd.shFABP5-2 | 5.2 × 1011 | 1.9 × 1012 | N/D | Lot 1 |
| gAd.shFABP5-3 | 4.1 × 1011 | 1.4 × 1012 | 7.8 × 106 | Lot 1 |
| gAd.shFABP5-4 | 6.1 × 1011 | 1.8 × 1012 | 7.8 × 106 | Lot 1 |
| gAd.shFABP5-5 | 3.4 × 1011 | 9.4 × 1011 | N/D | Lot 3 |
| gAd.shFABP5-8 | 7.4 × 1011 | 1.3 × 1012 | N/D | Lot 3 |
| gAd.shFABP5-9 | 1.3 × 1012 | 4.1 × 1012 | N/D | Lot 8 |
| gAd.shFABP5-10 | 7.2 × 1011 | 2.2 × 1012 | N/D | Lot 8 |
| gAd.shSCR-1 | 6.7 × 1011 | 2.5 × 1012 | 8 × 108 | |
| gAd.shSCR-2 | 8.6 × 1011 | 2.4 × 1012 | 8 × 108 | Lot 1 |
| gAd.shSCR-3 | 5.0 × 1011 | 1.7 × 1012 | N/D | Lot 1 |
| gAd.shSCR-4 | 8.8 × 1011 | 3.5 × 1012 | N/D | Lot 1 |
| gAd.shSCR-5 | 9.7 × 1011 | 2.4 × 1012 | N/D | Lot 4 |
| gAd.shSCR-6a | 5.0 × 1011 | 1.3 × 1012 | N/D | Lot 4 |
| gAd.shSCR-12 | 7.4 × 1011 | 2.3 × 1012 | N/D | Lot 11 |
| gAd.shSCR-13 | 6.9 × 1011 | 2.1 × 1012 | N/D | Lot 11 |
| gAd.shSCR-14 | 5.8 × 1011 | 1.3 × 1012 | N/D | Lot 11 |
| gAd.shLacZ-1 | 3.3 × 1011 | 1.3 × 1012 | 3.3 × 108 | |
| gAd.shLacZ-2 | 5.6 × 1011 | 1.4 × 1012 | 2.6 × 107 | Lot 1 |
| gAd.shLacZ-3a | 4.7 × 1011 | 8.0 × 1011 | N/D | Lot 2 |
| gAd.shLacZ-5 | 7.4 × 1011 | 1.6 × 1012 | 4.5 × 107 | Lot 2 |
| gAd.shLacZ-6 | 4.6 × 1011 | 7.3 × 1011 | 4.5 × 107 | Lot 2 |
| gAd.shSREBP-1 | 6.655 × 1011 | 2.86 × 1012 | N/D | |
| gAd.shSREBP-2 | 1.265 × 1012 | 4.048 × 1012 | N/D | Lot 1 |
| gAd.shSREBP-3 | 7.205 × 1011 | 2.52 × 1012 | N/D | Lot 2 |
| gAd.shSREBP-4 | 4.29 × 1011 | 1.54 × 1012 | N/D | Lot 3 |
| gAd.H1shSREBP-1 | 3.5 × 1011 | 5.3 × 1012 | 3.2 × 107 | |
| gAd.H1shSREBP-2 | 1.0 × 1012 | 3.0 × 1012 | 1.2 × 108 | Lot 1 |
| gAd.H1shSCR-1 | 3.1 × 1011 | 1.7 × 1012 | 7.8 × 107 | |
| gAd.H1shSCR-2 | 1.6 × 1012 | 3.5 × 1012 | 1.1 × 109 | Lot 1 |
| gAd.Albmal1-1 | 2.5 × 1011 | 4.8 × 1011 | N/D | |
| gAd.Albmal1-2 | 7.5 × 1011 | 2.6 × 1012 | N/D | Lot 1 |
| gAd.Albmal1-3 | 1.7 × 1012 | 6.0 × 1012 | N/D | Lot 2 |
Abbreviation: N/D, not determined.
shRNA expression cassette from this preparation was sequenced.
FIG. 1.
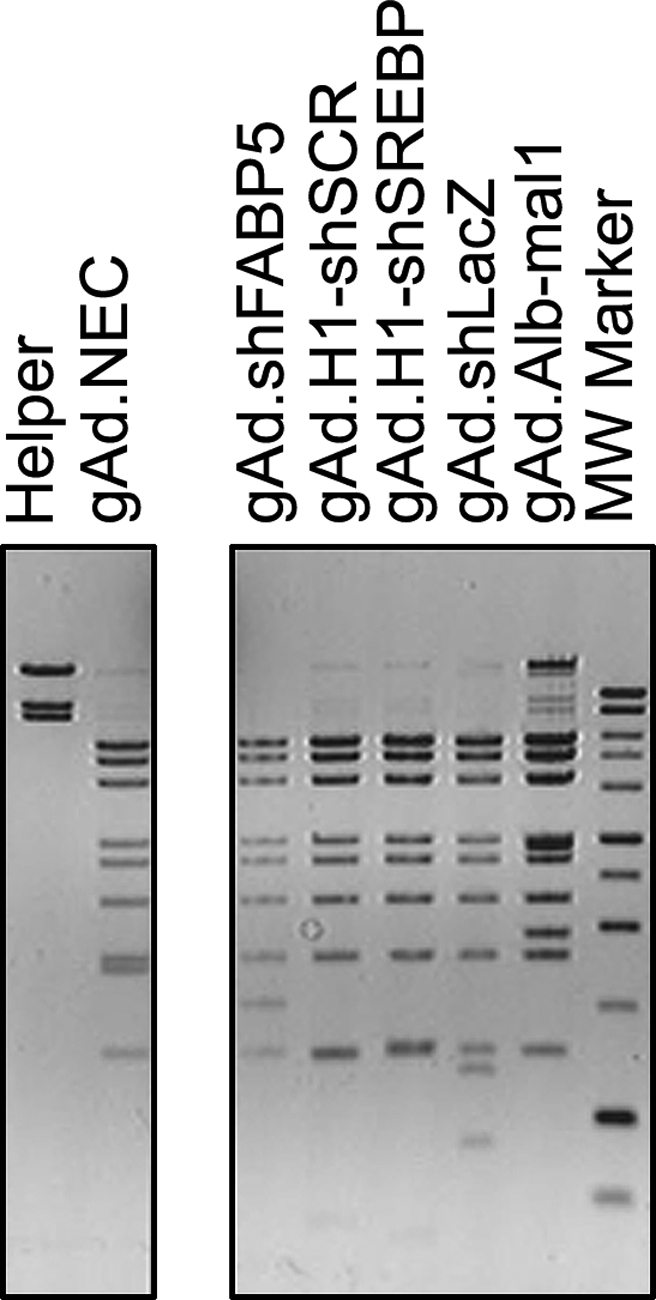
DNA structure of helper-dependent vectors. To confirm the molecular structure of the viral particles, DNA was isolated and digested with the restriction enzyme BamHI. Bands corresponding to the pattern of helper H14 DNA were barely seen in the vectors, suggesting the level of helper contamination was low. Vector gAd.Alb-mal1 had slightly higher levels of H14 because its expression cassette is approximately 3 kb larger than the shRNA cassette, resulting in a construct with a DNA molecular mass closer to that of the helper (∼31 and 36 kb, respectively). After isopycnic centrifugation the vector and helper bands were closer, making collection of the helper-dependent vector band more difficult.
To confirm that the shRNA sequence is stable during vector propagation, the first preparation (lot 1) was used as a stock to generate subsequent lots. The yield obtained from lot 1 was similar to or slightly lower than that obtained in preparations generated by infection of cells with purified virus, suggesting that a high yield can be obtained from the first large-scale preparation (Table 1). The stability of the vector genome was maintained in preparations generated with higher lot numbers, such as lots 4 and 8 (gAd.shSCR and gAd.shFABP5, respectively) (Table 1). No rearrangements have been observed in any of these preparations (data not shown). Helper contamination in all preparations tested was 0.1% or lower (PFU:VP). Sequencing of selected vector preparations confirmed that no alterations of the shRNA expression cassette were introduced during vector production (Table 1).
In vivo administration of shRNA-expressing vectors
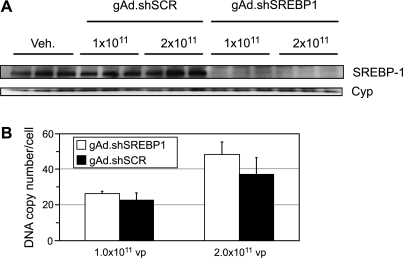
To determine the efficacy at knocking down the target gene in vivo, mice were intravenously injected with 2.0 × 1011 or 1.0 × 1011 VP of gAd.shSREBP1 (to target SREBP-1) or gAd.shSCR (expressing a scrambled sequence). One week after vector administration >90% gene silencing was observed (Fig. 2A), regardless of the dose of gAd.shSREBP1 received. The amount of vector DNA present in the liver was estimated by real-time PCR. Approximately 40 and 20 copies per cell were present in the liver of mice that received 2.0 × 1011 and 1.0 × 1011 VP, respectively (Fig. 2B). We have previously observed that there is a limit to the level of gene silencing that can be achieved in liver, and that once it is reached, increasing the dose of vector does not translate into a higher level of knockdown (Witting et al., 2008). The current data confirm this observation with an independent gene target. The presence of unnecessarily high shRNA levels may activate the interferon response and induce toxic effects (Witting et al., 2008). Thus, when conducting silencing studies in vivo, it is advisable to carry out pilot dose–response experiments to determine the lowest vector dose that results in the highest gene silencing.
FIG. 2.
SREBP1 silencing in liver of C57BL/6 mice. Animals received vehicle, or gAd.shSREBP1 or gAd.shSCR (2.0 × 1011 or 1.0 × 1011 VP), and were killed 1 week later. (A) Western blot analysis was carried out to determine the level of SREBP1 knockdown. (B) DNA copy number was quantified by real-time PCR.
shRNA-expressing helper-dependent vectors for drug target validation
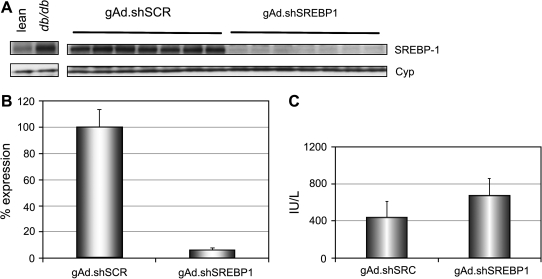
To determine whether robust gene silencing can be achieved for several weeks with helper-dependent adenoviral vectors, we tested the gAd.shSREBP1 vector in an animal model of obesity and type 2 diabetes, the db/db mouse (leptin receptor deficient). SREBP-1 is transcriptionally regulated by insulin, and compared with control lean animals its levels are increased in db/db mice as a result of the hyperinsulinemia developed in these animals (Shimomura et al., 1999, 2000) (Fig. 3A). Reduction of gene expression in adult mice by RNAi should allow study of the metabolic implications of such treatment and may provide the proof of concept that SREBP-1 is a useful drug target. Groups of eight db/db mice received 1.0 × 1011 VP of gAd.shSREBP1 or gAd.shSCR and were killed 3 weeks later. As shown in Fig. 3B, this treatment resulted in > 90% gene silencing, similar to what was observed in C57BL/6 mice after 1 week. Interestingly, despite the difference in body weight between the C57BL/6 mice (21 ± 1 g) and db/db mice (36 ± 2.2 g), and the fact that these mice have higher SREBP-1 levels than lean mice, 1.0 × 1011 VP reduced target gene expression to a similar extent. These data suggest that the increase in body weight in db/db mice, which is largely the result of an increase in adipose tissue mass, does not have an impact on the amount of adenoviral vector that transduces the liver. Moreover, the level of shRNA expression is sufficient to knock down SREBP-1 to a degree comparable to that in lean mice, despite the higher level of transcript in the liver. Concomitant with a decrease in SREBP1 levels, liver weight was reduced (1.8 ± 0.2 vs. 2.6 ± 0.2 g; p < 0.01) and the gain in body weight over the 3-week period was significantly lower for the gAd.shSREBP1 group compared with control-treated animals (4.3 ± 2.6 vs. 8.9 ± 1.8 g; p < 0.01). Metabolic analysis of the impact of reducing hepatic SREBP-1 is reported elsewhere (R. Ruiz, unpublished data).
FIG. 3.
SREBP1 silencing in liver of db/db mice. Mice received 1.0 × 1011 VP of gAd.shSREBP1 or gAd.shSCR, and were killed 3 weeks later. (A) Western blot analysis was used to determine the level of knockdown. Left: db/db mice have higher levels of SREBP-1 compared with lean (C57BKS) mice. Right: db/db mice administered gAd.shSREBP1 vector display a 90% reduction in SREBP-1 levels compared with db/db animals that received the gAd.shSCR vector. (B) Densitometric analysis of Western blot. (C) Aspartate aminotransferase (AST) levels in the serum of mice 3 weeks after gene transfer. No significant difference was observed between the two groups. SREBP-1, sterol regulatory element-binding protein-1; Cyp, cyclophilin.
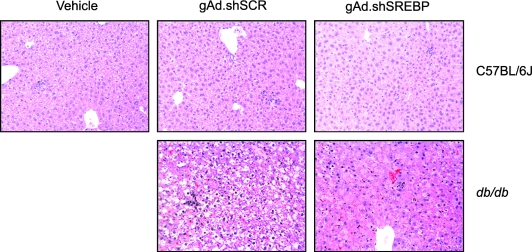
Maintenance of hepatic gene silencing on day 21 is a remarkable observation given that expression of shRNA using first-generation adenoviral vectors has resulted in complete loss of silencing by day 24 (Narvaiza et al., 2006). This would be anticipated on the basis of the fact that this type of vector is a strong activator of cytotoxic immune responses, leading to loss of liver cells within a few weeks (Yang et al., 1994). In addition, adeno-associated virus (AAV)-mediated expression of shRNA has resulted in the death of animals at high doses of vector (Grimm et al., 2006), which did not occur with our system. None of our animals displayed signs of distress or sickness for the duration of the study. Furthermore, no signs of toxicity were observed on the basis of histochemical analysis of liver sections and AST levels (Figs. 3C and 4). Thus, when using helper-dependent vectors, it is possible to achieve robust gene silencing for at least 3 weeks. Similar results were observed in a study using this type of vector to knock down huntingtin expression in the brain (Huang et al., 2007).
FIG. 4.
Liver histology. Mice were treated as described in the captions to Figs. 2 and 3. Four-micron-thick sections were cut from routinely processed paraffin-embedded tissue and stained with hematoxylin and eosin for histological analysis. Livers of mice treated with the gAd.shSREBP1 vector displayed lower levels of glycogen and lipid droplets (R. Ruiz, unpublished data). Processing and staining were performed as described in Materials and Methods. None of the groups had signs of toxicity related to expression of shRNA.
In conclusion, helper-dependent adenoviral vectors containing shRNA expression cassettes can be stably propagated over multiple passages, without genome rearrangements. A critical step in the process to ensure successful rescue and production is to monitor the percentage of cells producing vector by simultaneously amplifying a GFP-expressing vector and infecting >95% cells with vector and helper to ensure efficient production. Developments in helper-dependent vector production will allow for further reductions of helper contamination, minimization of replication-competent adenovirus (RCA) generation, and increases in overall yields (Schiedner et al., 2000; Barjot et al., 2002; Chamberlain et al., 2003; Palmer and Ng, 2003; Sakhuja et al., 2003). Generating transgenic animals for drug development is time-consuming and the breeding process is expensive. In addition, in type 2 diabetes studies silencing gene function in an adult animal (instead of at the stem cell level) provides a model that is more relevant to the human disease. We have provided evidence that by using helper-dependent adenoviral vectors, a high level of silencing is possible for several weeks in an animal model of obesity and type 2 diabetes. This system offers the possibility to conduct experiments in adult animals, representing an alternative to traditional transgenic animal technologies. Thus, a highly valuable use of helper-dependent vectors is in RNAi-mediated functional genomic and target validation studies.
Acknowledgments
The authors thank Drs. Sheila Connelly and P. Seshidhar Reddy for technical advice on plasmid recombination in E. coli BJ5183 cells; the Immunohistochemistry Core (IHC) at Indiana University Medical Center for hematoxylin–eosin staining of liver sections; and Dr. Palmiter for kindly providing the albumin promoter. This work was supported by grants from the NIDDK (DK069432 and DK078595), American Diabetes Foundation, and INGEN (Indiana Genomics Initiative of Indiana University supported in part by Lilly Endowment), and by the American Heart Association (postdoctoral fellowship to S.R.W.).
References
- Alemany R. Dai Y. Lou Y.C. Sethi E. Prokopenko E. Josephs S.F. Zhang W.-W. Complementation of helper-dependent adenoviral vectors: Size effects and titer fluctuations. J. Virol. Methods. 1997;68:147–159. [PubMed] [Google Scholar]
- Barjot C. Hartigan-O'Connor D. Salvatori G. Scott J.M. Chamberlain J.S. Gutted adenoviral vector growth using E1/E2b/E3-deleted helper viruses. J. Gene Med. 2002;4:480–489. doi: 10.1002/jgm.305. [DOI] [PubMed] [Google Scholar]
- Chamberlain J.S. Barjot C. Scott J. Packaging cell lines for generating replication-defective and gutted adenoviral vectors. Methods Mol. Med. 2003;76:153–166. doi: 10.1385/1-59259-304-6:153. [DOI] [PubMed] [Google Scholar]
- Chen H.H. Mack L.M. Kelly R. Ontell M. Kochanek S. Clemens P.R. Persistence in muscle of an adenoviral vector that lacks all viral genes. Proc. Natl. Acad. Sci. U.S.A. 1997;94:1645–1650. doi: 10.1073/pnas.94.5.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. Anton M. Graham F. Production and characterization of human 293 cell lines expressing the site-specific recombinase. Cre. Somat. Cell Mol. Genet. 1996;22:477–488. doi: 10.1007/BF02369439. [DOI] [PubMed] [Google Scholar]
- Clemens P.R. Kochanek S. Sunada Y. Chan S. Chen H.H. Campbell K.P. Caskey C.T. In vivo muscle gene transfer of full-length dystrophin with an adenoviral vector that lacks all viral genes. Gene Ther. 1996;3:965–972. [PubMed] [Google Scholar]
- Fisher K.J. Choi H. Burda J. Chen S.-J. Wilson J.M. Recombinant adenovirus deleted of all viral genes for gene therapy of cystic fibrosis. Virology. 1996;217:11–22. doi: 10.1006/viro.1996.0088. [DOI] [PubMed] [Google Scholar]
- Grimm D. Streetz K.L. Jopling C.L. Storm T.A. Pandey K. Davis C.R. Marion P. Salazar F. Kay M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Haecker S.E. Stedman H.H. Balice-Gordon R.J. Smith D.B.J. Greelish J.P. Mitchell M.A. Wells A. Sweeney H.L. Wilson J.M. In vivo expression of full-length human dystrophin from adenoviral vectors deleted of all viral genes. Hum. Gene Ther. 1996;7:1907–1914. doi: 10.1089/hum.1996.7.15-1907. [DOI] [PubMed] [Google Scholar]
- Hardy S. Kitamura M. Harris-Stansil T. Dai Y. Phipps M.L. Construction of adenovirus vectors through Cre–lox recombination. J. Virol. 1997;71:1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J.D. Shah N.A. Warrington J.A. Anderson N.N. Park S.W. Brown M.S. Goldstein J.L. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B. Schiefer J. Sass C. Landwehrmeyer G.B. Kosinski C.M. Kochanek S. High-capacity adenoviral vector-mediated reduction of huntingtin aggregate load in vitro and in vivo. Hum. Gene Ther. 2007;18:303–311. doi: 10.1089/hum.2006.160. [DOI] [PubMed] [Google Scholar]
- Kim I.H. Jozkowicz A. Piedra P.A. Oka K. Chan L. Lifetime correction of genetic deficiency in mice with a single injection of helper-dependent adenoviral vector. Proc. Natl. Acad. Sci. U.S.A. 2001;98:13282–13287. doi: 10.1073/pnas.241506298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochanek S. Clemens P.R. Mitani K. Chen H.-H. Chan S. Caskey C.T. A new adenoviral vector: Replacement of all viral coding sequences with 28 kb of DNA independently expressing both full-length dystrophin and β-galactosidase. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5731–5736. doi: 10.1073/pnas.93.12.5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar-Singh R. Chamberlain J.S. Encapsidated adenovirus minichromosomes allow delivery and expression of a 14 kb dystrophin cDNA to muscle cells. Hum. Mol. Genet. 1996;5:913–921. doi: 10.1093/hmg/5.7.913. [DOI] [PubMed] [Google Scholar]
- Lieber A. He C. Kirillova I. Kay M.A. Recombinant adenoviruses with large deletions generated by Cre-mediated excision exhibit biological properties compared with first-generation vectors in vitro and in vivo. J. Virol. 1996;70:8944–8960. doi: 10.1128/jvi.70.12.8944-8960.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizel J.V., Jr. White D.O. Scharff M.D. The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology. 1968;36:115–125. doi: 10.1016/0042-6822(68)90121-9. [DOI] [PubMed] [Google Scholar]
- Mitani K. Graham F.L. Caskey C.T. Kochanek S. Rescue, propagation and partial purification of a helper virus-dependent adenovirus vector. Proc. Natl. Acad. Sci. U.S.A. 1995;92:3854–3858. doi: 10.1073/pnas.92.9.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morral N. Parks R. Zhou H. Langston C. Schiedner G. Quinones J. Graham F.L. Kochanek S. Beaudet A.L. High doses of a helper-dependent adenoviral vector yield supraphysiological levels of α1-antitrypsin with negligible toxicity. Hum. Gene Ther. 1998;9:2709–2716. doi: 10.1089/hum.1998.9.18-2709. [DOI] [PubMed] [Google Scholar]
- Morral N. O'Neal W. Rice K. Leland M. Kaplan J. Piedra P.A. Zhou H. Parks R. Velji R. Aguilar-Cordova E. Wadsworth S. Graham F.L. Kochanek S. Carey K.D. Beaudet A.L. Administration of helper-dependent adenoviral vectors and sequential delivery of different vector serotype for long-term liver-directed gene transfer in baboons. Proc. Natl. Acad. Sci. U.S.A. 1999;96:12816–12821. doi: 10.1073/pnas.96.22.12816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morral N. McEvoy R. Dong H. Meseck M. Altomonte J. Thung S. Woo S.L. Adenovirus-mediated expression of glucokinase in the liver as an adjuvant treatment for type 1 diabetes. Hum. Gene Ther. 2002;13:1561–1570. doi: 10.1089/10430340260201653. [DOI] [PubMed] [Google Scholar]
- Nagai Y. Nishio Y. Nakamura T. Maegawa H. Kikkawa R. Kashiwagi A. Amelioration of high fructose-induced metabolic derangements by activation of PPARα. Am. J. Physiol. Endocrinol. Metab. 2002;282:E1180–E1190. doi: 10.1152/ajpendo.00471.2001. [DOI] [PubMed] [Google Scholar]
- Narvaiza I. Aparicio O. Vera M. Razquin N. Bortolanza S. Prieto J. Fortes P. Effect of adenovirus-mediated RNA interference on endogenous microRNAs in a mouse model of multidrug resistance protein 2 gene silencing. J. Virol. 2006;80:12236–12247. doi: 10.1128/JVI.01205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka K. Pastore L. Kim I.H. Merched A. Nomura S. Lee H.J. Merched-Sauvage M. Arden-Riley C. Lee B. Finegold M. Beaudet A. Chan L. Long-term stable correction of low-density lipoprotein receptor-deficient mice with a helper-dependent adenoviral vector expressing the very low-density lipoprotein receptor. Circulation. 2001;103:1274–1281. doi: 10.1161/01.cir.103.9.1274. [DOI] [PubMed] [Google Scholar]
- Palmer D. Ng P. Improved system for helper-dependent adenoviral vector production. Mol. Ther. 2003;8:846–852. doi: 10.1016/j.ymthe.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Parks R.J. Chen L. Anton M. Sankar U. Rudnicki M.A. Graham F.L. A helper-dependent adenovirus vector system: Removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13565–13570. doi: 10.1073/pnas.93.24.13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkert C.A. Ornitz D.M. Brinster R.L. Palmiter R.D. An albumin enhancer located 10 kb upstream functions along with its promoter to direct efficient, liver-specific expression in transgenic mice. Genes Dev. 1987;1:268–276. doi: 10.1101/gad.1.3.268. [DOI] [PubMed] [Google Scholar]
- Sakhuja K. Reddy P.S. Ganesh S. Cantaniag F. Pattison S. Limbach P. Kayda D.B. Kadan M.J. Kaleko M. Connelly S. Optimization of the generation and propagation of gutless adenoviral vectors. Hum. Gene Ther. 2003;14:243–254. doi: 10.1089/10430340360535797. [DOI] [PubMed] [Google Scholar]
- Sandig V. Youil R. Bett A.J. Franlin L.L. Oshima M. Maione D. Wang F. Metzker M.L. Savino R. Caskey C.T. Optimization of the helper-dependent adenovirus system for production and potency in vivo. Proc. Natl. Acad. Sci. U.S.A. 2000;97:1002–1007. doi: 10.1073/pnas.97.3.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiedner G. Morral N. Parks R. Wu Y. Koopmans S.C. Langston C. Graham F.L. Beaudet A.L. Kochanek S. Genomic DNA transfer with a high-capacity adenovirus vector results in improved in vivo gene expression and decreased toxicity. Nat. Genet. 1998;18:180–183. doi: 10.1038/ng0298-180. [DOI] [PubMed] [Google Scholar]
- Schiedner G. Hertel S. Kochanek S. Efficient transformation of primary human amniocytes by E1 functions of Ad5: Generation of new cell lines for adenoviral vector production. Hum. Gene Ther. 2000;11:2105–2116. doi: 10.1089/104303400750001417. [DOI] [PubMed] [Google Scholar]
- Shimomura I. Bashmakov Y. Horton J.D. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J. Biol. Chem. 1999;274:30028–30032. doi: 10.1074/jbc.274.42.30028. [DOI] [PubMed] [Google Scholar]
- Shimomura I. Matsuda M. Hammer R.E. Bashmakov Y. Brown M.S. Goldstein J.L. Decreased IRS-2 and increased SREBP-1c lead to mixed insulin resistance and sensitivity in livers of lipodystrophic and ob/ob mice. Mol. Cell. 2000;6:77–86. [PubMed] [Google Scholar]
- Toietta G. Mane V.P. Norona W.S. Finegold M.J. Ng P. McDonagh A.F. Beaudet A.L. Lee B. Lifelong elimination of hyperbilirubinemia in the Gunn rat with a single injection of helper-dependent adenoviral vector. Proc. Natl. Acad. Sci. U.S.A. 2005;102:3930–3935. doi: 10.1073/pnas.0500930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witting S.R. Brown M. Saxena R. Nabinger S. Morral N. Helper-dependent adenovirus-mediated short hairpin RNA expression in the liver activates the interferon response. J. Biol. Chem. 2008;283:2120–2128. doi: 10.1074/jbc.M704178200. [DOI] [PubMed] [Google Scholar]
- Yang Y. Ertl H.C.J. Wilson J.M. MHC class I restricted cytotoxic T lymphocytes to viral antigens destroy hepatocytes in mice infected with E1 deleted recombinant adenoviruses. Immunity. 1994;1:433–442. doi: 10.1016/1074-7613(94)90074-4. [DOI] [PubMed] [Google Scholar]
- Youil R. Toner T.J. Su Q. Kaslow D.C. Rapid method for the isolation of full length adenoviral genomes by bacterial intermolecular homologous recombination. J. Virol. Methods. 2001;92:91–97. doi: 10.1016/s0166-0934(00)00280-9. [DOI] [PubMed] [Google Scholar]