Abstract
Recent studies have linked the unfolded protein response (UPR), in particular the inositol-requiring, endoplasmic reticulum-to-nucleus signaling protein 1α (IRE1α)-X-box-binding protein-1 (XBP1) branch of the UPR, to the regulation of lipogenesis and hepatic steatosis. In this study, we examined the hypothesis that the postprandial environment can activate the IRE1α-XBP1 branch of the UPR in the liver via a mammalian target of rapamycin complex 1 (mTORC1)-dependent mechanism. Toward this end, rats were fed a high-carbohydrate diet (68% of energy from corn starch) for 3 h in the absence or presence of rapamycin (intraperitoneal injection of 1 mg/kg) and liver tissue was taken 1 or 7 h following the feeding period. Feeding activated the mTORC1 pathway and IRE1α, induced XBP1 splicing, and increased the expression of XBP1 target genes and lipogenic genes in the liver. The presence of rapamycin prevented the activation of mTORC1 and IRE1α, XBP1 splicing, and the increased expression of XBP1 target genes and lipogenic genes. Rapamycin also prevented the feeding-induced increase in nuclear sterol regulatory element binding protein 1c. These data suggest that the postprandial environment promotes activation of the IRE1-XBP1 branch of the UPR in the liver. This activation appears to be mediated in part by mTORC1.
Introduction
An essential function of the endoplasmic reticulum (ER)4 is the synthesis and processing of secretory and membrane proteins. Disruption of ER homeostasis, collectively termed ER stress, activates the unfolded protein response (UPR), a signaling pathway that links the ER lumen with the cytoplasm and nucleus (1). The UPR is initiated by 3 ER transmembrane proteins, inositol-requiring, ER-to-nucleus signaling protein-1α (IRE1α), RNA-dependent protein kinase-like ER eukaryotic initiation factor-2α ( eIF2α) kinase (PERK), and activating transcription factor (ATF)-6 (2). Activation of IRE1α promotes the splicing of X-box-binding protein-1 (XBP1) mRNA and subsequent transcription of genes involved in protein folding, ER-associated degradation, and translocation (3). PERK activation leads to phosphorylation of eIF2α and subsequent attenuation of translation initiation, as well as increased expression and selective translation of ATF4 (3,4). The ATF6 branch of the UPR is initiated by the release of ATF6 from the ER membrane, ATF6 processing in the Golgi, and the release of its cytoplasmic domain, which acts as a transcriptional activator controlling UPR genes related to ER-associated degradation and folding (3,5).
The ER, in addition to protein processing, is also involved in the production and storage of glycogen and lipids. Recent studies have demonstrated an important link between the IRE1α-XBP1 branch of the UPR and the regulation of the hepatic lipogenic program in mice and adipogenesis in mouse embryonic fibroblasts and 3T3-L1 cells (6,7). These data imply that the postprandial environment may activate the IRE1-XBP1 branch of the UPR in the liver. Hepatic protein synthesis is increased in the postprandial setting (8). The phosphorylation of mammalian target of rapamycin complex-1 (mTORC1) is a principal step in protein translation initiation and is activated in the liver postprandially (8,9). Recent studies have linked mTORC1 to ER stress and activation of the UPR and to the regulation of sterol regulatory element binding proteins (SREBP) (10,11). Therefore, the aim of the present study was to examine the hypothesis that the postprandial environment can activate the IRE1α-XBP1 pathway in the liver via a mTORC1-dependent mechanism.
Methods
Animals and meal-feeding paradigm.
Male Wistar Crl(WI)BR rats (Charles River Laboratories) weighing ∼150 g upon arrival were individually housed in a temperature- and humidity-controlled environment. Rats were maintained on a reverse 12-h-light/-dark cycle and were given free access to water. Rats were provided a purified, high-starch diet (12) consisting of 68% cornstarch, 20% casein, and 12% corn oil (percent of energy, Research Diets) for 3 h/d, 1 h after the beginning of the dark cycle, and food intake and body weight were monitored over a 2-wk adaptation period. All procedures involving rats were reviewed and approved by the Colorado State University Institutional Animal Care and Use Committee.
Experimental procedures.
On the day of the study, rats were either feed-deprived for 24 h (FD; n = 10) or were provided free access to the high-starch diet for 3 h. Rapamycin (RAP; 1 mg/kg; Sigma Chemical, n = 10/time point) or carrier (VEH; dimethylsulfoxide; n = 10/time point) was injected (intraperitoneal) into rats 1 h prior to the start of the feeding period.
Rats were anesthetized with sodium pentobarbital (intraperitoneal; ∼70 mg/kg) 1 or 7 h following the 3-h feeding period. These time points were chosen to examine the early and late response to a defined, uninterrupted interval of feeding. Once deeply anesthetized (absence of response to toe pinch and eye reflex), rats were placed on a heating pad, the abdominal cavity was exposed, and portal vein and inferior vena cava blood samples were obtained. Liver tissue was removed and processed for subsequent analyses.
Processing and analysis of blood samples.
Blood samples were immediately centrifuged at 1000 × g for 2 min. Plasma was collected and frozen at −80°C for later analysis of glucose (Beckman glucose analyzer) and insulin (Linco).
Liver glycogen.
Liver tissue was homogenized in 0.03 mol/L hydrochloric acid and incubated at 100°C for 5 min. Sodium acetate (0.1 mol/L) and amyloglucosidase were added to aliquots of liver homogenates and incubated for 2 h at 30°C. To correct for free glucose, a set of aliquots in which only sodium acetate was added were included. Aliquots were centrifuged at 18,000 × g for 3 min and supernatants were used to analyze glucose concentration using a kit (Sigma).
Tissue preparation and RNA analysis.
Fresh liver was immediately placed into RNALater solution and frozen at −80°C. Total RNA was isolated from liver tissue using TRIzol reagent (Life Technologies) per the manufacturer's instructions. RT and duplex PCR amplification for analysis of XBP1 was performed as described in detail previously (13). Real-time PCR was performed as described in detail previously (13). Primer sets were designed by Beacon designer program version 3.1 (Supplemental Table 1).
Tissue preparation and Western blotting.
Fresh liver was processed as described in detail previously (13). Equivalent amounts of protein (50–100 μg) were subjected to SDS-PAGE and transferred to Hybond-P membranes (Amersham Pharmacia Biotech). Membranes were blocked and incubated with antibodies against eIF2α, phosphorylated eIF2α, phosphorylated ribosomal protein S6 kinase (S6K1), phosphorylated ribosomal protein S6 (RPS6), and phosphorylated IRE1α (Cell Signaling Technology); glucose regulated protein (GRP) 78 and GRP94 (Santa Cruz Technology), and/or actin (Sigma). Proteins were detected using horseradish peroxidase-conjugated secondary antibodies and a chemiluminescence reagent (Santa Cruz). Detection and analysis of density was performed on a UVP Bioimaging system with Labworks Software.
Nuclear protein lysates were prepared from liver samples as described previously (13,14). Western blot analysis was performed as described above using antibodies against XBP1, SREBP1, and Lamin A/C (Santa Cruz).
Data analysis and statistics.
Primary statistical comparisons were made based on a 2 × 2 factorial design (vehicle, rapamycin × 1 h, 7 h) using 2-way ANOVA with Bonferroni's post hoc test. Data that failed Bartlett's test for homogeneous variance were analyzed with the Kruskal-Wallis test. Secondary comparisons between feed-deprived and fed rats were made using 1-way ANOVA. Post hoc comparisons were made using least significant difference test. The level of significance was P < 0.05. Data are reported as means ± SEM.
Results
Food intake, body weight, plasma analytes, and liver glycogen.
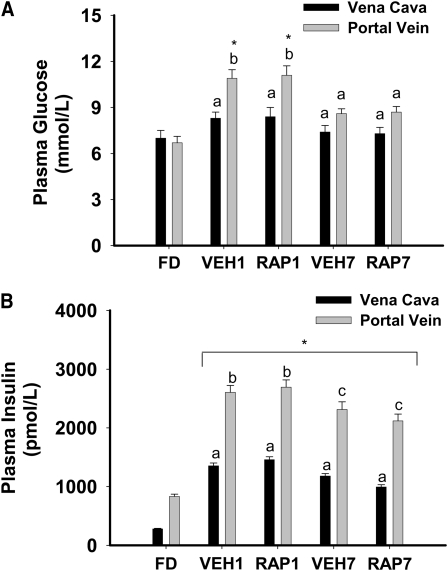
Food intake over the 16-d feeding period, food intake on the day of study, and initial and final body weights are in Supplemental Figure 1. Postprandial glucose and insulin responses were not significantly different between vehicle- and rapamycin-treated rats at 1 or 7 h (Fig. 1A,B). The liver glycogen concentration was 28.7 ± 3.9 μmol/g in FD rats and was not significantly different between VEH- and RAP-treated rats at 1 h (91.7 ± 6.6 and 104.9 ± 2.9, respectively) or 7 h (141.6 ± 1.8 and 150.8 ± 4.9, respectively).
FIGURE 1 .
Plasma glucose (A) and insulin (B) in FD, VEH, or RAP rats. Meal-fed rats were killed 1 h (VEH1, RAP1) or 7 h (VEH7, RAP7) after the 3-h meal-feeding period. Values are the mean ± SE, n = 10. Means without a common letter differ, P < 0.05. *Different from FD, P < 0.05.
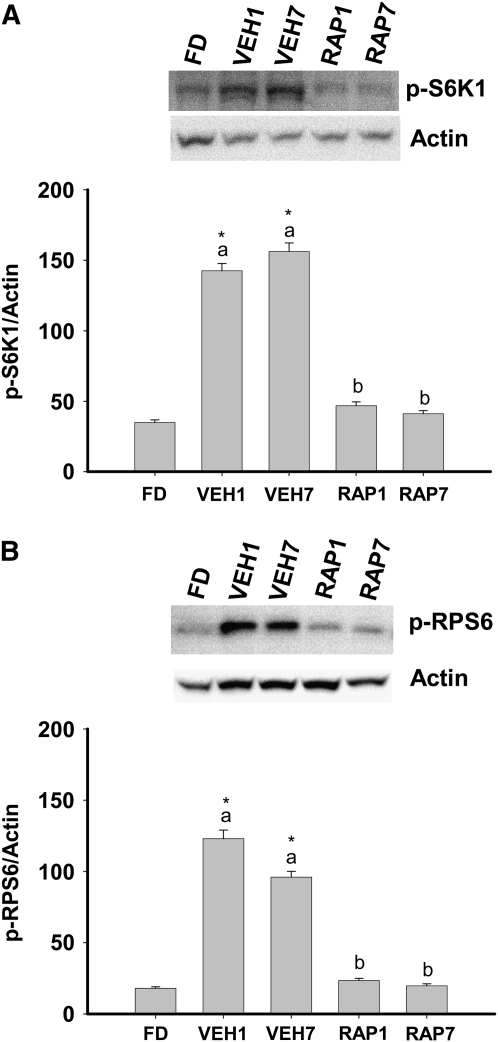
mTORC1 activation in the liver.
mTORC1 transmits growth signals to the translational machinery via phosphorylation and activation of S6K1 and the S6K1 substrate, RPS6 (15). In vehicle-treated fed rats, hepatic S6K1 and RPS6 phosphorylation were increased at 1 and 7 h and the presence of rapamycin prevented this increase (Fig. 2A,B).
FIGURE 2 .
Phosphorylation of S6K1 (A) and RPS6 (B) in the liver of FD, VEH, or RAP rats. Meal-fed rats were killed 1 h (VEH1, RAP1) or 7 h (VEH7, RAP7) after the 3-h meal-feeding period. Representative western blots indicate the proteins detected with antibodies against p-S6K1, p-RPS6, and actin. Values are the mean ± SE, n = 10. Means without a common letter differ, P < 0.05. *Different from FD, P < 0.05.
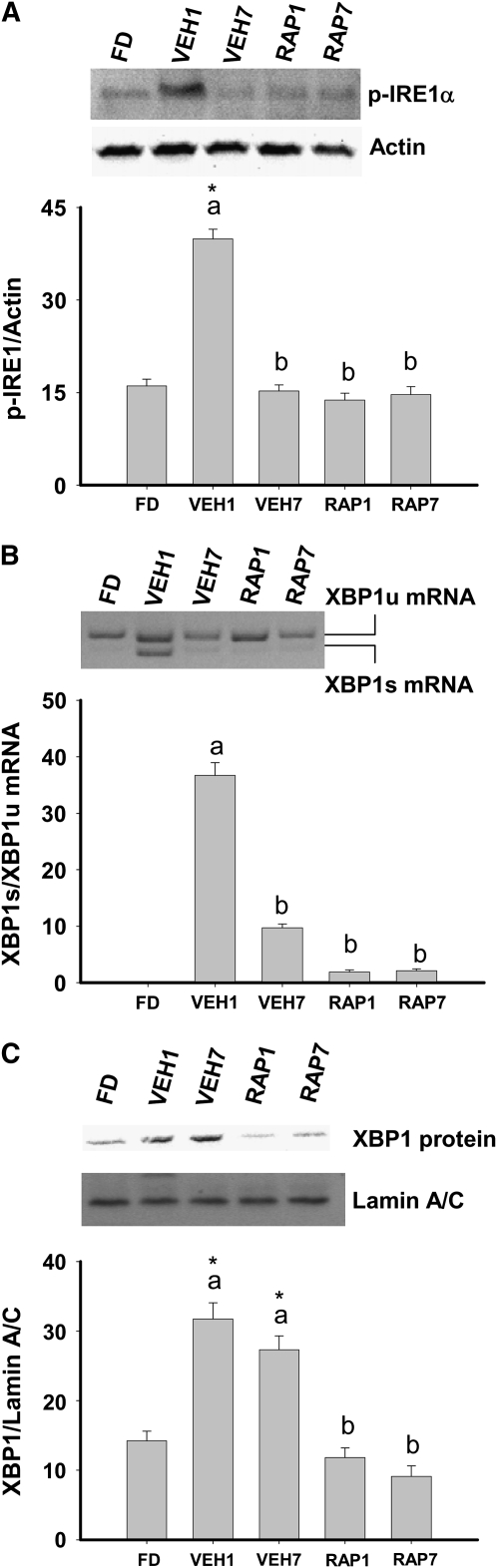
IRE1α and XBP1 in the liver.
Expression and activation of the UPR transcription factor XBP1 are essential to liver development and hepatic lipogenesis (6,16). Activation of XBP1 results from IRE1α-mediated splicing, which leads to a shift in the codon reading frame and subsequent expression of an active transcription factor, spliced XBP1 (XBP1s) (5). The contribution of XBP1s to the regulation of hepatic lipogenesis implies that activation of IRE1α and XBP1 splicing occurs in the postprandial state (17). Therefore, we examined IRE1α phosphorylation, XBP1 mRNA splicing, and nuclear XBP1 protein in the liver of feed-deprived and refed rats. Increased phosphorylation of IRE1α was observed in vehicle-treated fed rats at 1 h and the presence of rapamycin prevented this increase (Fig. 3A). XBP1s mRNA was present in the liver of all vehicle-treated fed rats at 1 h and in 5 of 10 vehicle-treated fed rats at 7 h (Fig. 3B). In contrast, XBP1s mRNA was not observed in the liver of FD rats or in rapamycin-treated, fed rats (Fig. 3B). Feeding increased nuclear XBP1 protein and the presence of rapamycin prevented this increase (Fig. 3C). These data provide evidence that feeding can activate the IRE1α-XBP1 branch of the UPR in the liver via a rapamycin-dependent mechanism.
FIGURE 3 .
Phosphorylation of IRE1α (A), ratio of spliced (s) to unspliced (u) XBP1 mRNA (B), and XBP1 protein in nuclear extracts (C) in the liver of FD, VEH, or RAP rats. Meal-fed rats were killed 1 h (VEH1, RAP1) or 7 h (VEH7, RAP7) after the 3-h meal-feeding period. Representative western blots indicate the proteins detected with antibodies against p-IRE1α, XBP1, actin, or Lamin A/C. Representative gel illustrates unspliced XBP1 and XBP1s mRNA. Values are the mean ± SE, n = 10. Means without a common letter differ, P < 0.05. *Different from FD, P < 0.05.
XBP1 target genes in the liver.
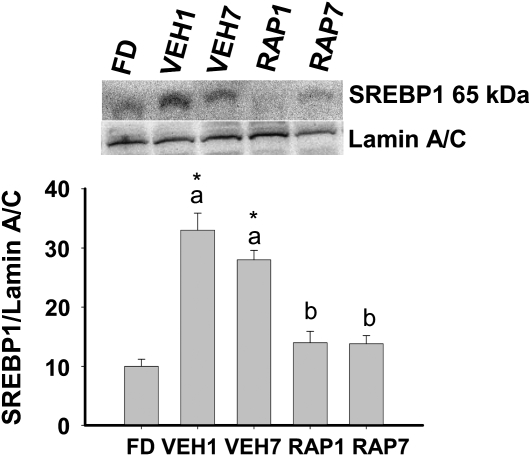
Gene targets of XBP1s include mannosyl (α-1,6-) glycoprotein β-1,2-N-acetylglucosaminiyltransferase (Mgat2) and defender against cell death, which encode proteins involved in N-linked glycosylation (18); signal recognition particle (SRP) 54 and SRP receptor (SRPR), which encode proteins involved in protein translocation into the ER (18); and several lipogenic genes (6). Feeding increased Mgat2, SRPR54, fatty acid synthase, SREBP1c, and sterol-CoA desaturase 1 mRNA and rapamycin reduced or prevented this increase (Table 1). In addition, rapamycin prevented the feeding-induced increase in nuclear SREBP1 protein (Fig. 4). These data demonstrate that mTORC1 signaling participates in postprandial regulation of the lipogenic program in the liver in vivo.
TABLE 1.
| FD | VEH1 | VEH7 | RAP1 | RAP7 | |
|---|---|---|---|---|---|
| Fold change | |||||
| XBP1s target genes | |||||
| Mgat2 | 1.1 ± 0.1 | 2.2 ± 0.3a* | 2.8 ± 0.3a* | 1.3 ± 0.2b | 1.2 ± 0.2b |
| Defender against cell death | 1.0 ± 0.2 | 1.4 ± 0.2 | 1.8 ± 0.2 | 1.1 ± 0.2 | 1.2 ± 0.2 |
| SRP54 | 1.1 ± 0.1 | 2.6 ± 0.2a* | 3.3 ± 0.3a* | 1.4 ± 0.2b | 1.3 ± 0.2b |
| SRPR | 1.0 ± 0.1 | 1.5 ± 0.3 | 1.6 ± 0.3 | 1.3 ± 0.2 | 1.2 ± 0.2 |
| Lipogenic genes | |||||
| Fatty acid synthase | 1.1 ± 0.2 | 23.5 ± 3.1a* | 8.2 ± 0.7b* | 9.2 ± 0.7b* | 2.9 ± 0.2c* |
| SREBP1c | 1.0 ± 0.1 | 19.4 ± 1.3a* | 5.8 ± 0.4b* | 7.7 ± 0.5b* | 2.1 ± 0.2c* |
| Sterol-CoA desaturase 1 | 1.1 ± 0.1 | 7.1 ± 0.4a* | 5.1 ± 0.3a* | 2.3 ± 0.3b* | 1.8 ± 0.2b |
| Diacylglycerol acyl-transferase-2 | 1.2 ± 0.1 | 1.9 ± 0.2 | 2.0 ± 0.3 | 1.7 ± 0.3 | 1.6 ± 0.2 |
| Acetyl-CoA carboxylase-2 | 1.1 ± 0.2 | 1.4 ± 0.2 | 1.3 ± 0.3 | 1.4 ± 0.3 | 1.5 ± 0.3 |
Values are the mean ± SE, n = 10. Means in a row with superscripts without a common letter differ, P< 0.05. *Different from FD, P < 0.05.
Meal-fed rats were killed 1 h (VEH1, RAP1) or 7 h (VEH7, RAP7) after the 3-h meal-feeding period.
FIGURE 4 .
SREBP1 in nuclear extracts (A) in the liver of FD, VEH, or RAP rats. Meal-fed rats were killed 1 h (VEH1, RAP1) or 7 h (VEH7, RAP7) after the 3-h meal-feeding period. Representative western blot indicates the protein detected with antibodies against SREBP1 or Lamin A/C. Values are the mean ± SE, n = 10. Means without a common letter differ, P < 0.05. *Different from FD, P < 0.05.
Other components of the UPR.
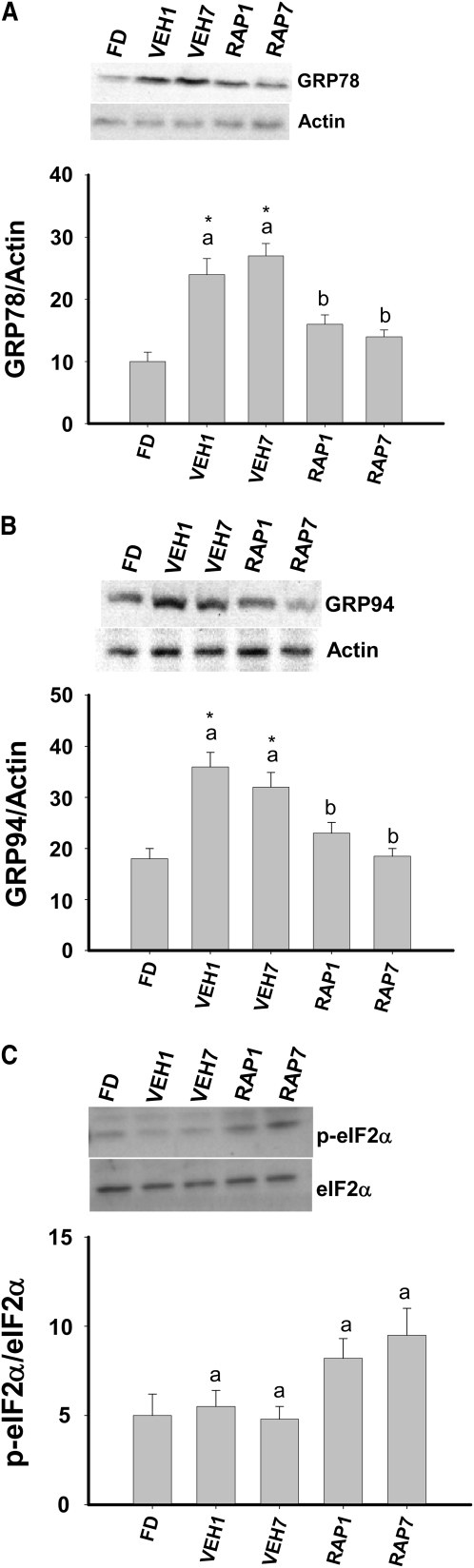
In addition to activation of the IRE1-XBP1 pathway, the UPR is also characterized by activation of PERK and ATF6. PERK activation results in the phosphorylation of eIF2α, translational attenuation, and upregulation of ATF4. ATF6 activation involves Golgi-mediated splicing and entry into the nucleus. Downstream targets of PERK and ATF6 include the glucose regulation proteins, GRP78 and GRP94, the proapoptotic gene CCAAT-enhancer homologous protein (Chop), and ATF4. Feeding increased the expression of GRP78, GRP94, Chop, and ATF4 mRNA at 1 h, and GRP78 and GRP94 mRNA at 7 h. Rapamycin prevented these feeding-mediated increases in mRNA (Table 2). Feeding also increased the expression of GRP78 and GRP94 protein and this increase was prevented in the presence of rapamycin (Fig. 5A,B). In contrast, phosphorylation of eIF2α was not increased by feeding (Fig. 5C).
TABLE 2.
| FD | VEH1 | VEH7 | RAP1 | RAP7 | |
|---|---|---|---|---|---|
| Fold change | |||||
| GRP78 | 1.2 ± 0.1 | 6.7 ± 0.4a* | 3.8 ± 0.2a* | 1.1 ± 0.2b | 1.3 ± 0.3b |
| GRP94 | 1.1 ± 0.1 | 3.3 ± 0.3a* | 3.1 ± 0.3a* | 1.2 ± 0.3b | 1.1 ± 0.2b |
| Chop | 1.0 ± 0.2 | 4.5 ± 0.3a* | 1.9 ± 0.4b | 1.2 ± 0.2b | 1.1 ± 0.3b |
| ATF4 | 1.1 ± 0.1 | 3.4 ± 0.2a* | 1.5 ± 0.2b | 1.1 ± 0.2b | 1.2 ± 0.1b |
Values are the mean ± SE, n = 10. Means in a row with superscripts without a common letter differ, P < 0.05. *Different from FD, P < 0.05.
Meal-fed rats were killed 1 h (VEH1, RAP1) or 7 h (VEH7, RAP7) after the 3-h meal-feeding period.
FIGURE 5 .
GRP78 (A) and GRP94 (B) and phosphorylation of eIF2α (C) in the liver of FD, VEH, or RAP rats. Meal-fed rats were killed 1 h (VEH1, RAP1) or 7 h (VEH7, RAP7) after the 3-h meal-feeding period. Representative western blots indicate the proteins detected with antibodies against GRP78, GRP94, actin, p-eIF2α, or eIF2α. Values are the mean ± SE, n = 10. Means without a common letter differ, P < 0.05. *Different from FD, P < 0.05.
Discussion
A fundamental function of the UPR is to alleviate ER stress provoked by the accumulation of unfolded proteins through the upregulation of protein folding and degradation pathways in the ER and attenuation of global protein synthesis (1). Although groundbreaking research over the past few years has led to a comprehensive description of the basic pathways involved in UPR activation, signal transduction and transcriptional activation in mammalian systems, much less is known about the role and regulation of the UPR in vivo (5,19–24). Recent studies have suggested that both proximal sensors/initiators of the UPR (e.g. PERK, IRE1α, ATF6) and downstream responses (e.g. XBP1 splicing, phosphorylation of eIF2α, increased chaperone expression) can participate in a diverse array of cellular functions, including differentiation, ER and mitochondrial biogenesis, insulin action, and glucose and lipid metabolism (6,14,25–29).
Selective deletion of XBP1 in the liver of mice reduced plasma cholesterol and triglycerides, secondary to a decreased production of lipids from the liver (6). This study, from Lee et al. (6), identified XBP1 as a potentially important regulator of hepatic lipogenesis. For this to be the case, we hypothesized that a single meal should activate the IRE1α-XBP1 branch of the UPR in the liver. Indeed, a single, high-carbohydrate meal resulted in the phosphorylation of IRE1α, induced XBP1 splicing, and increased the amount of nuclear XBP1. Thus, the postprandial environment elicits a signal that activates IRE1α-mediated XBP1 splicing and XBP1 entry into the nucleus. Chronic exposure to a high-sucrose diet results in hepatic steatosis characterized by increased SFA and activation of the IRE1α-XBP1 branch of the UPR in the liver of feed-deprived rats (13). Thus, the regulation of this branch of the UPR, in particular the ability to turn off postprandial activation of IRE1α and XBP1 splicing, may be linked to the development of dyslipidemia.
Previous studies have linked mTORC1 activity to the regulation of ER homeostasis, SREBP activity, and lipid stores (10,11,30). Therefore, we next examined the role of mTORC1 in postprandial-mediated activation of IRE1α-XBP1 splicing using rapamycin. The presence of rapamycin prevented the activation of mTORC1 and IRE1α, XBP1 splicing, and the enrichment of nuclear fractions with XBP1 protein. Therefore, it is hypothesized that postprandial regulation of IRE1α-XBP1 is mediated via mTORC1. Rapamycin also reduced the meal-induced increase in some lipogenic genes and nuclear SREBP1 protein in the liver. Taken together, these data are consistent with the notion that mTORC1 and XBP1 splicing participate in the regulation of the lipogenic program in the liver (6,10,17,31). However, because rapamycin was the sole intervention in the present study, further work is required to definitively demonstrate whether and how mTORC1 influences IRE1α, XBP1, and lipogenesis in the liver.
TOR functions in the context of 2 complexes, mTORC1 and mTORC2. The mTORC1 complex is composed of mTOR, raptor, and mLST8/GβL (9,32). mTOR-mediated phosphorylation of eIF4E-binding proteins and S6K1 leads to increased protein synthesis via stimulation of cap-dependent translation and increased ribosome biogenesis (11,32). Rapamycin has been shown to reduce liver growth during refeeding via control of ribosomal protein translation but not cap-dependent translation initiation (9). The UPR is activated when the protein load entering the ER exceeds the existing capacity of the ER lumen to fold or degrade these proteins (1). Thus, the postprandial state may provoke activation of the IRE1α-XBP1 branch of the UPR as a result of mTOR-mediated activation of protein synthesis that is, acutely, in excess of the capacity of the ER lumen to fold and degrade entering proteins. This scenario would predict that other components of the UPR, in addition to IRE1α-XBP1, would be activated in the liver following a meal.
PERK is 1 of 4 proteins capable of mediating the phosphorylation of eIF2α, a cellular event that typically is associated with a reduction in general translation (1,33). However, recent studies suggest that PERK-dependent regulation of lipogenesis occurs during mouse mammary gland development and that a high-fat meal induces phosphorylation of eIF2α in the liver (29,34). In the present study, phosphorylation of eIF2α was not increased in the liver of fed rats. That we did not observe any increase in phosphorylation of eIF2α in the present study may be due to the experimental protocol, which involved the sampling of liver 1 and 7 h after cessation of a 3-h feeding period, and/or to differential regulation of the phosphorylation state of eIF2α by dietary fat. We certainly cannot rule out the possibility that phosphorylation of eIF2α increased during the 3-h feeding period, given that ATF4 and Chop mRNA were increased and both are downstream targets of phosphorylated eIF2α (33).
Dhahbi et al. (35) demonstrated that several ER-associated protein chaperones (e.g. GRP78, GRP94, calreticulin) were increased in the liver of mice refed following 48 h of food deprivation. In the present study, GRP78 and 94 mRNA and protein increased in the liver of fed rats and rapamycin prevented this increase. Thus, the postprandial regulation of ER-associated protein chaperones, similar to the IRE1α-XBP1 branch of the UPR, appears to be mediated by mTORC1.
It is possible that the IRE1α-XBP1 branch of the UPR may be regulated by signals that do not involve the accumulation of unfolded proteins or ER stress. Although PERK and IRE1α share functionally similar ER-luminal sensing domains and both are simultaneously activated by chemically induced ER stress in vitro, they can be selectively engaged in vivo (5). In particular, recent studies have identified several proteins that directly interact with and/or regulate the activity of IRE1α (36–38). In addition, the transcriptional response to XBP1 may be regulated by protein interactions with XBP1. Future studies will investigate how postprandial signals activate the IRE1α-XBP1 branch of the UPR in the liver in vivo.
In summary, the results from this study demonstrate that the postprandial environment activated the IRE1/XBP1 branch of the UPR. Rapamycin prevented these responses, suggesting that mTORC1 links the postprandial environment to the UPR and regulation of the lipogenic program in the liver.
Supplementary Material
Acknowledgments
K.T.P. and M.J.P. designed research; K.T.P., A.M.N., and L.R. conducted research; K.T.P., A.M.N., L.R., F.E., D.W., and Y.W. analyzed data; K.T.P. and M.J.P. wrote the paper. K.T.P. and M.J.P. had primary responsibility for final content. All authors read and approved the final manuscript.
Supported in part by NIH grants DK072017 and DK47416 and the Lillian Fountain Smith Endowment.
Author disclosures: K. T. Pfaffenbach, L. Reese, A. M. Nivala, F. Ellis, D. Wang, Y. Wei, and M. J. Pagliassotti, no conflicts of interest.
Supplemental Table 1 and Figure 1 are available with the online posting of this paper at jn.nutrition.org.
Abbreviations used: ATF, activating transcription factor; Chop, CCAAT-enhancer homologous protein; ER, endoplasmic reticulum; eIF2α, eukaryotic initiation factor 2α; FD, feed-deprived group; GRP, glucose regulated protein; IRE1α, inositol-requiring, endoplasmic reticulum-to-nucleus signaling protein; Mgat2, mannosyl (α-1,6-) glycoprotein β-1,2-N-acetylglucosaminiyltransferase; mTORC1, mammalian target of rapamycin complex 1; PERK, RNA-dependent protein kinase-like endoplasmic reticulum eukaryotic initiation factor-2α kinase; RAP, rapamycin-treated group; RPS6, ribosomal protein S6; S6K1, ribosomal protein S6 kinase; SREBP, sterol regulatory element binding protein; SRP, signal recognition particle; SRPR, signal recognition particle receptor; UPR, unfolded protein response; VEH, vehicle-treated group; XBP1, X-box-binding protein-1; XBP1s, spliced XBP1 mRNA.
References
- 1.Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–33. [DOI] [PubMed] [Google Scholar]
- 2.Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14:20–8. [DOI] [PubMed] [Google Scholar]
- 3.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–89. [DOI] [PubMed] [Google Scholar]
- 4.Kaufman RJ. Orchestrating the unfolded protein response in health and disease. J Clin Invest. 2002;110:1389–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hetz C, Glimcher LH. Fine-tuning of the unfolded protein response: assembling the IRE1alpha interactome. Mol Cell. 2009;35:551–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee A-H, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sha H, He Y, Chen H, Wang C, Zenno A, Shi H, Yang X, Zhang X, Qi L. The IRE1alpha-XBP1 pathway of the unfolded protein response is required for adipogenesis. Cell Metab. 2009;9:556–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reiter AK, Crozier SJ, Kimball SR, Jefferson LS. Meal feeding alters translational control of gene expression in rat liver. J Nutr. 2005;135:367–75. [DOI] [PubMed] [Google Scholar]
- 9.Anand P, Gruppuso PA. Rapamycin inhibits liver growth during refeeding in rats via control of ribosomal protein translation but not cap-dependent translation initiation. J Nutr. 2006;136:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozcan U, Ozcan L, Yilmaz E, Duvel K, Sahin M, Manning BD, Hotamisligil GS. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol Cell. 2008;29:541–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pagliassotti MJ, Wei Y, Bizeau ME. Glucose-6-phosphatase activity is not suppressed but the mRNA level is increased by a sucrose-enriched meal in rats. J Nutr. 2003;133:32–7. [DOI] [PubMed] [Google Scholar]
- 13.Wang D, Wei Y, Pagliassotti MJ. Saturated fatty acids promote endoplasmic reticulum stress and liver injury in rats with hepatic steatosis. Endocrinology. 2006;147:943–51. [DOI] [PubMed] [Google Scholar]
- 14.Wang D, Wei Y, Schmoll D, Maclean KN, Pagliassotti MJ. Endoplasmic reticulum stress increases glucose-6-phosphatase and glucose cycling in liver cells. Endocrinology. 2006;147:350–8. [DOI] [PubMed] [Google Scholar]
- 15.Wouters BG, Koritzinsky M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat Rev Cancer. 2008;8:851–64. [DOI] [PubMed] [Google Scholar]
- 16.Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, et al. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14:152–7. [PMC free article] [PubMed] [Google Scholar]
- 17.Glimcher LH, Lee AH. From sugar to fat: how the transcription factor XBP1 regulates hepatic lipogenesis. Ann N Y Acad Sci. 2009;1173 Suppl 1:E2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sriburi R, Bommiasamy H, Buldak GL, Robbins GR, Frank M, Jackowski S, Brewer JW. Coordinate regulation of phospholipid biosynthesis and secretory pathway gene expression in XBP-1 (S)-induced endoplasmic reticulum biogenesis. J Biol Chem. 2007;282:7024–34. [DOI] [PubMed] [Google Scholar]
- 19.Zhang K, Kaufman RJ. Signaling the unfolded protein response from the endoplasmic reticulum. J Biol Chem. 2004;279:25935–8. [DOI] [PubMed] [Google Scholar]
- 20.Okada T, Yoshida H, Akazawa R, Negishi M, Mori K. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem J. 2002;366:585–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J, Kaufman RJ. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 2006;13:374–84. [DOI] [PubMed] [Google Scholar]
- 22.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–29. [DOI] [PubMed] [Google Scholar]
- 23.Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, Miao H, Hassler JR, et al. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spear E, Ng DTW. The unfolded protein response: no longer just a special teams player. Traffic. 2001;2:515–23. [DOI] [PubMed] [Google Scholar]
- 25.Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 2007;27:53–66. [DOI] [PubMed] [Google Scholar]
- 27.Ozcan U, Cao Q, Yilmaz E, Lee A-H, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun CZ, Glimcher LH, et al. Endoplasmic reticulum stress links obesity, insulin action and type 2 diabetes. Science. 2004;306:457–61. [DOI] [PubMed] [Google Scholar]
- 28.Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–76. [DOI] [PubMed] [Google Scholar]
- 29.Bobrovnikova-Marjon E, Hatzivassiliou G, Grigoriadou C, Romero M, Cavener DR, Thompson CB, Diehl JA. PERK-dependent regulation of lipogenesis during mouse mammary gland development and adipocyte differentiation. Proc Natl Acad Sci USA. 2008;105:16314–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luong N, Davies CR, Wessells RJ, Graham SM, King MT, Veech R, Bodmer R, Oldham SM. Activated FOXO-mediated insulin resistance is blocked by reduction of TOR activity. Cell Metab. 2006;4:133–42. [DOI] [PubMed] [Google Scholar]
- 31.Porstmann T, Santos CR, Lewis C, Griffiths B, Schulze A. A new player in the orchestra of cell growth: SREBP activity is regulated by mTORC1 and contributes to the regulation of cell and organ size. Biochem Soc Trans. 2009;37:278–83. [DOI] [PubMed] [Google Scholar]
- 32.Yang Q, Guan KL. Expanding mTOR signaling. Cell Res. 2007;17:666–81. [DOI] [PubMed] [Google Scholar]
- 33.Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. [DOI] [PubMed] [Google Scholar]
- 34.Oyadomari S, Harding H, Zhang Y, Oyadomari M, Ron D. Dephosphorylation of translation initiation factor 2α enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7:520–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dhahbi JM, Cao SX, Mote PL, Rowley BC, Wingo J, Spindler SR. Postprandial induction of chaperone gene expression is rapid in mice. J Nutr. 2002;132:31–7. [DOI] [PubMed] [Google Scholar]
- 36.Hetz C, Bernasconi P, Fisher J, Lee AH, Bassik MC, Antonsson B, Brandt GS, Iwakoshi NN, Schinzel A, et al. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science. 2006;312:572–6. [DOI] [PubMed] [Google Scholar]
- 37.Lisbona F, Rojas-Rivera D, Thielen P, Zamorano S, Todd D, Martinon F, Glavic A, Kress C, Lin JH, et al. BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1alpha. Mol Cell. 2009;33:679–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu F, Nguyen DT, Stuible M, Dube N, Tremblay ML, Chevet E. Protein-tyrosine phosphatase 1B potentiates IRE1 signaling during endoplasmic reticulum stress. J Biol Chem. 2004;279:49689–93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.