Abstract
The intestine exhibits striking diurnal rhythmicity in glucose uptake, mediated by the sodium glucose cotransporter (SGLT1); however, regulatory pathways for these rhythms remain incompletely characterized. We hypothesized that SGLT1 rhythmicity is linked to the circadian clock. To investigate this, we examined rhythmicity of Sglt1 and individual clock genes in rats that consumed food ad libitum (AL). We further compared phase shifts of Sglt1 and clock genes in a second group of rats following restricted feeding to either the dark (DF) or light (LF) phase. Rats fed during the DF were pair-fed to rats fed during the LF. Jejunal mucosa was harvested across the diurnal period to generate expression profiles of Sglt1 and clock genes Clock, Bmal1 (brain-muscle Arnt-like 1), ReverbA/B, Per(Period) 1/2, and Cry (Cryptochrome) 1/2. All clock genes were rhythmic in AL rats (P < 0.05). Sglt1 also exhibited diurnal rhythmicity, with peak expression preceding nutrient arrival (P < 0.05). Light-restricted feeding shifted the expression rhythms of Sglt1 and most clock genes (Bmal1, ReverbA and B, Per1, Per2, and Cry1) compared with dark-restricted feeding (P < 0.05). The Sglt1 rhythm shifted in parallel with rhythms of Per1 and ReverbB. These effects of restricted feeding highlight luminal nutrients as a key Zeitgeber in the intestine, capable of simultaneously shifting the phases of transporter and clock gene expression, and suggest a role for clock genes in regulating Sglt1 and therefore glucose uptake. Understanding the regulatory cues governing rhythms in intestinal function may allow new therapeutic options for conditions of dysregulated absorption such as diabetes and obesity.
Introduction
Circadian rhythmicity in gene and protein expression has been demonstrated in numerous mammalian organs and tissues. These rhythms serve a major physiological role by matching many visceral functions to anticipated environmental demands (1). We and others have documented circadian rhythmicity in intestinal expression of digestive enzymes and transporters for both nutrients and nonnutrients (2–5). Our studies on the intestinal sodium-glucose cotransporter (SGLT1),9 which is responsible for all active intestinal glucose uptake, demonstrate that rhythmicity in intestinal glucose uptake is conferred entirely by rhythmicity in transcription, translation, and function of SGLT1 (3). However, the molecular cues triggering rhythmicity in the Sglt1 gene (Slc5a1) and protein expression remain unknown.
Previous studies have identified a set of genes, referred to as clock genes, involved in the regulation of circadian rhythms, such as hormone secretion, and autonomic functions, including body temperature and blood pressure (6,7). In mammals, the master clock resides in the suprachiasmatic nucleus (SCN) and maintains a 24-h periodicity entrained by light (8) and regulated via opposing positive and negative molecular feedback loops. Mammalian clock components include Per1, Per2, Clock, Bmal1, ReverbA and B, and Cry1 and Cry2. Heterodimers of Clock and Bmal1 positively regulate Per and Cry genes via promoter E-boxes (CAnnTG). Nuclear accumulation of Per and Cry inhibits Clock/Bmal1 activity, which represses Per and Cry, thereby setting up an oscillation in their expression (9,10). Orphan nuclear receptors ReverbA and ReverbB have been identified as key regulators linking the positive and negative limbs of the circadian oscillator, with Reverb transcription driven by Bmal1/Clock and suppressed by Per and Cry (11,12). In addition to the central SCN circadian pacemaker, clock genes are also expressed in many peripheral tissues, including the heart, retina, lung, kidney, peripheral blood cells, and liver (13–16). While several clock genes are known to oscillate in the intestine, the temporal expression patterns of Reverbs have not been characterized.
While light is the predominant Zeitgeber (“time giver” or entraining cue) for the central SCN clock, peripheral clocks can be dissociated from the central clock by various stimuli, including nutrient availability and glucocorticoid exposure (11,17,18; A. T. Stearns, A. Balakrishnan, K. Abolmaali, D. B. Rhoads, S. W. Ashley, A. Tavakkolizadeh, unpublished results). Feeding in particular is a strong Zeitgeber. Restricted feeding can reset the peripheral clocks in the liver, kidney, heart, and pancreas within 1 wk with no change in phase of the SCN clock (19–21) and is a sufficiently potent Zeitgeber to reinstate rhythmicity of the liver clock in otherwise arrhythmic SCN-lesioned mice (22).
We hypothesized that rhythmic expression of SGLT1 in the rat intestine is driven by peripheral circadian clocks to link function to nutrient availability. We surmised that Sglt1 would have a similar phase to 1 or more clock genes and that clock genes and Sglt1 would exhibit parallel phase shifts in response to restricted feeding. Our studies show that nutrient availability acts as a major Zeitgeber in rat intestine, independent of light cycle, and simultaneously phase shifts Sglt1 and clock gene expression. These results provide evidence for regulation of SGLT1 by the peripheral clock.
Materials and Methods
Animal studies.
All animal study protocols were prospectively approved by the Harvard Medical Area Standing Committee on Rats.
Sprague-Dawley rats (50 males, 7 wk old) were purchased from Harlan World and acclimatized to a 12-h-light/-dark photoperiod for 5 d with ad libitum access to food (Picolab Rodent Diet 20, LabDiet, containing 21% protein, 9.9% fat, 4.4% fiber, and an energy value of 3.42 kcal/g) and water. Time is designated as H After Light Onset (HALO), with HALO 0 at 0700 h (lights on). In the control arm, rats received food ad libitum (designated AL) and were killed at 3-h intervals beginning at HALO 0 (n = 6–7 per time; Supplemental Fig. 1A). A second group of 50 male rats were similarly acclimatized, then randomly assigned to be fed for 7 d either during only the dark phase (designated DF; HALO 12–24, Supplemental Fig. 1B) or light phase (designated LF; HALO 0–12, Supplemental Fig. 1C). DF rats were pair-fed to LF rats to ensure equal food intake. Rats were housed in pairs in cages. LF animals were given 100 g of food per cage at 0700. The remaining food at 1900 was weighed and subtracted from 100 g to calculate the amount consumed per pair of rats (we assumed that both rats consumed equal amounts of food). The mean daily consumption of LF rats was calculated, multiplied by 2, and provided to each pair of DF rats at 1900. No food remained in the cages of DF rats at 0700 the next day. To minimize disruption during restricted feeding, rats were weighed only 3 times (d 0, 3, and 7). On d 7, rats (n = 6–7) were killed at 6-h intervals beginning at HALO 3.
Tissue harvest.
Rats were anesthetized with sodium pentobarbital (50 mg/kg, Ovation Pharmaceuticals). The small intestine from 2 cm distal to the ligament of Treitz was harvested via midline laparotomy and rinsed with ice-cold saline to remove luminal contents. The 10 cm of jejunum was divided along the antimesenteric border, mucosa scraped from the underlying muscle, snap-frozen in liquid nitrogen, and stored at −80°C for subsequent RNA or protein extraction.
RNA extraction, RT, and real-time PCR.
Total RNA was extracted using the mirVana kit (Ambion). Samples were reverse transcribed simultaneously with Superscript III (Invitrogen) and oligo-dT. Real-time PCR was performed as previously described (3). mRNA levels were expressed as ratios to the stably expressed B-actin. All primers were ordered as custom oligonucleotides from Invitrogen (Supplemental Table 1), except rat Per2, for which mRNA expression was measured using the Taqman primer-probe and gene expression Master mix (Applied Biosystems).
Protein extraction and Western blotting.
SGLT1 protein expression was measured in total lysates from jejunal mucosal scrapings as previously described (3). Diurnal Per1 protein expression in rat jejunum was measured in nuclear extracts of freshly collected mucosal scrapings (Nxtract nuclear extraction kit, Sigma). Western blotting was performed as previously described (3). Nuclear or total protein extracts (75 μg) were resolved on 4–12% Bis-Tris gels, transferred to polyvinylidenefluoride membranes, blocked, then incubated with either rabbit anti-SGLT1 (1:4000; Chemicon International) or rabbit anti-Per1 (1:200; Santa Cruz Biotechnology), respectively. Protein expression was normalized to B-actin (mouse anti-B-actin, 1:1000, Labvision).
Statistical analysis.
Data are presented as means ± SE. Graphical analysis was performed using Graphpad Prism. Circadian rhythmicity was determined as described previously by cross-sectional analysis using the Cosinor procedure, freely available online, and assuming a 24-h period (4,23,24). The acrophase (time of peak expression), mesor (rhythm-adjusted mean), amplitude of rhythmicity, and significance of fit to a 24-h period (as indicated by the P-value) for each gene was abstracted from the program. mRNA levels are expressed as ratios to mean expression of the respective gene at HALO 3 (DF group for all restricted-fed rats). The mesor is an arbitrary value, precluding comparisons between genes. However, amplitudes are independent of scaling and can thus be compared among genes and groups. Two-tailed t tests were used to compare weights of DF and LF groups. The acrophases for AL rats were subtracted from the acrophases of DF and LF rats to identify phase shifts relative to AL rats. A 1-sample t test was used to identify a significant difference in mean phase shift of DF or LF rats from the value 0 (complete absence of phase shift relative to AL). Differences were considered significant at P < 0.05.
Results
Rhythmicity of gene expression in jejunum of AL rats.
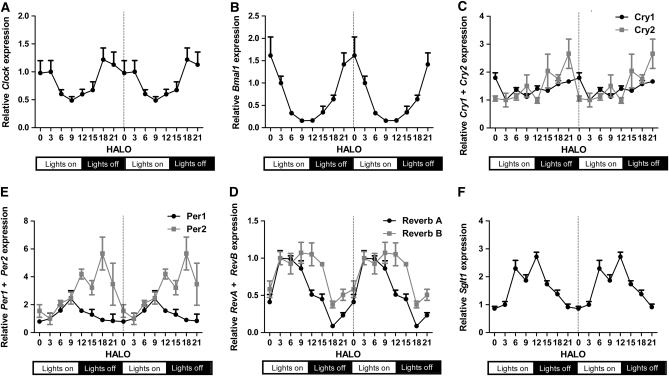
All clock genes examined were expressed in intestinal jejunal mucosa. Cosinor analysis confirmed previously documented rhythmicity in expression of Clock, Bmal1, ReverbA, Per1, Per2, and Cry1 (Table 1; Fig. 1) and demonstrated that rhythmicity for all measured clock genes fit a 24-h periodicity (P < 0.05) (4,23). Periodicities of 24 h were also detected for ReverbB, which exhibited an acrophase (peak) 2 h later than its paralog ReverbA, and for Cry2, which exhibited an acrophase 2 h earlier than Cry1 (Fig. 1C,E). In AL rats, amplitudes were greatest for Bmal1, ReverbA, Per1, and Per2 and more modest for Clock ,Cry1, and Cry2. Positive clock regulators Bmal1 and Clock peaked at late dark phase (P < 0.0001; Table 1; Fig. 1A,B). In contrast, negative regulators Per1, ReverbA, and ReverbB peaked between HALO 6 and 10 and reached a trough at HALO 0 during peak expression of Bmal1 and Clock (P < 0.005; Fig. 1D,E). Per2 expression peaked at HALO 16, a 6-h lag behind Per1 (P < 0.005; Fig. 1D). Sglt1 mRNA exhibited robust 24-h rhythmicity as we have reported previously (3), with peak expression at HALO 11, close to that for Per1 (HALO 9) as well as ReverbB (HALO 9, Table 1; Fig. 1D–F).
TABLE 1.
Rhythmicity, acrophase, mesor, and amplitude of clock gene and Sglt1 mRNA expression in AL rats fed either only during DF or LF periods1
| Gene | Group | P-value2 | Acrophase3 (HALO, hh:mm) | Mesor4 | Amplitude,5% | Phase difference6 |
|---|---|---|---|---|---|---|
| Per1 | AL | 0.0014 | 09:28 | 1.29 | 48 | |
| DF | 0.0004 | 07:38 | 1.09 | 63 | −01:50 | |
| LF | 0.0015 | 01:38 | 0.96 | 57 | −07:50 | |
| Per2 | AL | 0.0008 | 15:59 | 3.02 | 57 | |
| DF | 0.0460 | 13:29 | 4.51 | 57 | −02:30 | |
| LF | 0.0470 | 05:32 | 3.22 | 59 | −10:27 | |
| Bmal1 | AL | <0.0001 | 23:04 | 0.71 | 100 | |
| DF | 0.0003 | 21:35 | 0.83 | 100 | −01:29 | |
| LF | 0.0019 | 12:34 | 1.07 | 78 | −10:30 | |
| Clock | AL | 0.0008 | 21:23 | 0.80 | 38 | |
| DF | 0.5877 | |||||
| LF | 0.5065 | |||||
| ReverbA | AL | <0.0001 | 06:21 | 0.57 | 76 | |
| DF | 0.0001 | 04:34 | 0.37 | 120 | −01:47 | |
| LF | 0.0002 | 21:34 | 0.19 | 60 | −08:47 | |
| ReverbB | AL | 0.0003 | 08:34 | 0.81 | 41 | |
| DF | <0.0001 | 07:50 | 0.85 | 78 | −00:44 | |
| LF | <0.0001 | 01:29 | 0.71 | 67 | −07:05 | |
| Cry1 | AL | 0.0015 | 20:08 | 1.41 | 17 | |
| DF | 0.0145 | 17:24 | 1.57 | 30 | −02:44 | |
| LF | 0.0033 | 08:50 | 1.41 | 33 | −14:18 | |
| Cry2 | AL | 0.0097 | 18:41 | 1.47 | 36 | |
| DF | 0.0790 | |||||
| LF | 0.6250 | |||||
| Sglt1 | AL | <0.0001 | 10:44 | 1.60 | 51 | |
| DF | <0.0001 | 09:27 | 1.86 | 118 | −01:17 | |
| LF | <0.0001 | 02:27 | 1.96 | 88 | −08:17 |
Cosinor analysis was used to determine rhythmicity, acrophase, mesor, and amplitude of clock gene mRNA expression in AL, DF, and LF rats.
The P-values indicate the fit of the data to a 24-h periodicity, with a P-value of 0.05 indicating a 5% probability that the observed 24-h periodicity occurred by chance alone.
The acrophase is expressed as HALO (lights on is at 0700; hence, HALO 0 is 0700).
The mesor is the rhythm-adjusted mean.
The amplitude is expressed as a percentage of the mean to facilitate comparison across means.
The phase difference refers to the shift in peak expression of the gene (acrophase) in DF or LF animals compared with AL animals.
FIGURE 1 .
Circadian rhythmicity of Clock (A), Bmal1 (B), Cry1/Cry2 (C), Per1/Per2 (D), ReverbA/ReverbB (E), and Sglt1 (F) in AL rats. To facilitate comparisons of rhythmicity and amplitude, the x-axis was double-plotted and expression (y-axis) indexed to mean HALO 3 expression for each gene. Values are expressed as mean ± SEM, n = 6 or 7. P-values are shown in Table 1.
Restricted feeding phase-shifts expression rhythms of both SGLT1 and clock genes.
To identify regulatory cues triggering rhythmicity in Sglt1 and clock genes, we sought to define their responses to imposed food availability, thus separating nutrient cues from the light-dark cycle.
Food intake and body weight.
Food consumption by LF rats on d 1 was 16 g (Supplemental Fig. 2A), ∼4 g less than the 20 g/d consumed by AL rats of similar weight (25) but had normalized by d 3. LF rats weighed less than DF rats on d 4 despite equal food intake (244 ± 1.4 g vs. 251 ± 1.4 g; Supplemental Fig. 2B; P < 0.05). This was possibly a catabolic response to the stress of restricted feeding during daytime, or a relatively slower adaptation of intestinal nutrient absorption to the change in period of peak nutrient availability. Weights had equalized by harvest.
Rhythmicity of Sglt1 and clock genes.
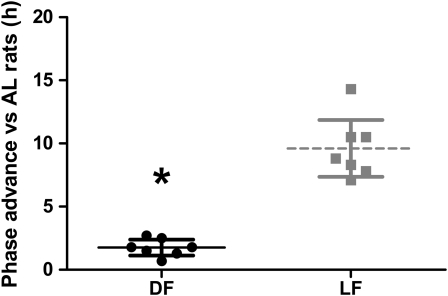
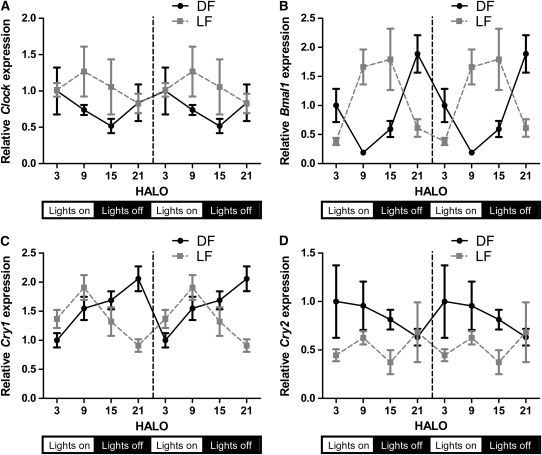
DF rats would be expected to display rhythms similar to AL (Fig. 2). Although this was broadly true, we observed a consistent advance in acrophase for most clock genes and Sglt1 in DF compared with AL rats (mean phase advance of 1.7 h; P = 0.0005) (Table 1; Fig. 2). Periodicity was retained in most clock genes and Sglt1 in LF rats, with a mean phase advance of 9.6 and 7.9 h compared with the AL and DF group, respectively (P < 0.0001) (Table 1; Fig. 2). In contrast, amplitudes for Clock and Cry2 were lower in both restricted groups compared with AL, leading to a loss of rhythmicity (Fig. 3A,D).
FIGURE 2 .
Phase difference in acrophases of clock genes and Sglt1 in DF and LF rats relative to AL rats showing a phase advance of 1.7 and 9.6 h, respectively. Values are individual values (n = 6 or 7) and means ± 95% CI. *Different from DF, P < 0.05.
FIGURE 3 .
Circadian expression of Clock (A), BmalI (B), Cry1 (C), and Cry2 (D) in DF and LF rats. To facilitate comparisons of rhythmicity and amplitude, the x-axis was double-plotted and expression (y-axis) indexed to mean HALO 3 DF expression for each gene. Values are expressed as mean ± SEM, n = 6 or 7. P-values are shown in Table 1.
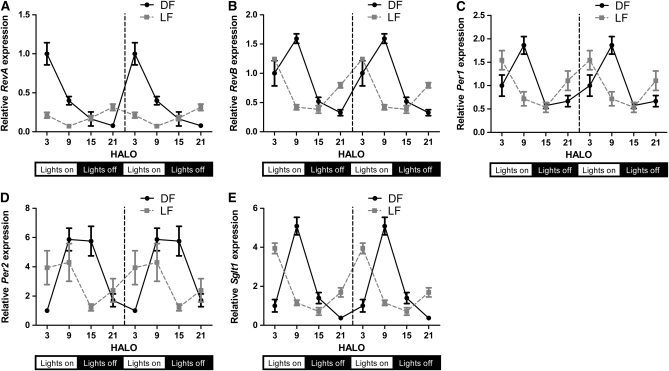
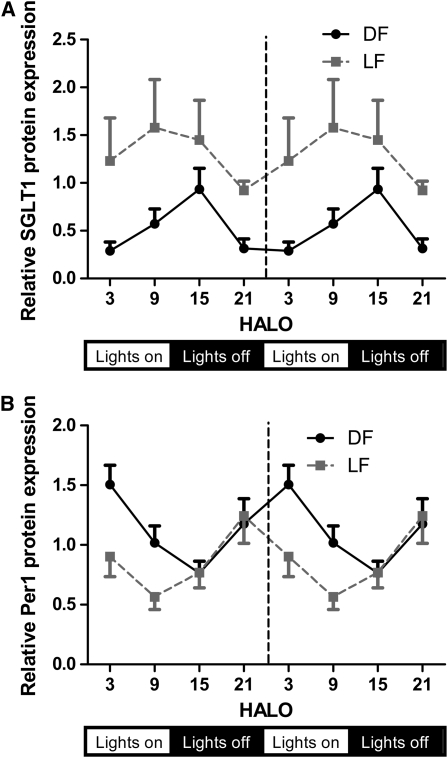
The phase of the Sglt1 mRNA rhythm was advanced in both DF and LF rats (1 and 8 h, respectively). Sglt1 mRNA remained rhythmic in LF rats, but with a lower peak, blunted amplitude, and a 7-h phase difference from DF rats (Fig. 4E; P < 0.05). SGLT1 protein expression in DF rats peaked 4 h later than mRNA expression (HALO 14), 3-fold higher than the trough (Fig. 5A; P < 0.05). SGLT1 protein expression was highest during the day in LF rats, with levels 70% higher at HALO 9 than HALO 21, but did not attain 24-h rhythmicity (Fig. 5A; P = 0.24). We also observed higher trough SGLT1 protein expression in LF than in DF rats (P = 0.001 at HALO 21; Fig. 5A).
FIGURE 4 .
Circadian expression of ReverbA (A), ReverbB (B), Per1 (C), Per2 (D), and Sglt1 (E) in DF and LF rats. To facilitate comparisons of rhythmicity and amplitude, the x-axis was double-plotted and expression (y-axis) indexed to mean HALO 3 DF expression for each gene. Values are expressed as mean ± SEM, n = 6 or 7. P-values are shown in Table 1.
FIGURE 5 .
Protein expression of SGLT1 (A) and Per1 (B) in DF and LF rats. Values are expressed as mean ± SEM, n = 6 or 7.
Clock genes Per1 and ReverbB both displayed similar phase shifts to Sglt1 under the 2 restricted-feeding regimens. The concordance between SGLT1 and Per1 was also observed at the protein level in jejunal nuclear extracts (Fig. 5A,B). A 24-h periodicity was observed in both DF and LF rats, with acrophases of HALO 2 and HALO 22, respectively (Fig. 5B). The period of increasing Per1 protein coincided with the nadir of Sglt1 mRNA and presumably its transcription (Figs. 4E and 5B). ReverbA also exhibited similar phase shifts on restricted feeding, but peak ReverbA expression preceded Sglt1 expression by 4–5 h in all 3 groups.
Discussion
All clock genes exhibited robust circadian rhythmicity in jejunal mucosa of AL rats. Restricting food to the LF dissociated these rhythms from the light cycle. Notably, rhythmicity of both Sglt1 mRNA and protein were shifted. Previous studies from our group and others have shown that rhythmicity of SGLT1 protein expression correlates with rhythmicity at a functional level in the intestine (2,3). These results establish nutrient availability as a key Zeitgeber for the peripheral intestinal clock(s) as well as the expression rhythm of the glucose transporter SGLT1.
Determining the relative shifts in glucose transporter and clock gene rhythms in response to restricted feeding was a major study aim. After 7 d of restricted feeding to either the light or dark period, phase differences of 6–11.5 h were observed for Sglt1 and 5 of the 8 clock genes examined. The lack of a complete 12-h phase shift difference between dark- and light-fed rats may reflect the influence of other factors such as glucocorticoids, which can also partially phase shift gene expression (18,19; A. T. Stearns, A. Balakrishnan, K. Abolmaali, D. B. Rhoads, S. W. Ashley, A. Tavakkolizadeh, unpublished results). The duration of nutrient availability may have also affected the degree of phase shift, because a shorter period of food availability in other studies (HALO 2–8) (26) produced a greater phase shift than we observed. Moreover, peripheral clocks adapt to restricted feeding at different rates; liver shifts much more quickly than lung (19). Although it is possible that the small intestine would achieve a complete phase shift following longer restricted feeding, the sufficiency of 4-d adaptation previously reported (27), the equal weights between rats in the 2 restricted groups, and the plateau in food intake in the LF rats [matching that expected for rats of that weight (25)] all suggest that the partial phase shift was due to factors other than incomplete adaptation. We note that the rapid adaptation observed in liver may result from more direct (i.e. local) stimulus-response pathways. Adaptation in the intestine, particularly for diurnally rhythmic functions, is indirect [as shown by isolated loops (28)] and may entail cephalic and other inputs. The apparently longer period required for adaptation by intestine compared with liver may reflect a tissue-specific feature necessary to stabilize the rhythms in intestinal functions despite moderately varying nutrient intake patterns. Possibly, linkage of phases of critical intestinal functions such as proliferation and absorption to extra-luminal inputs could serve to coordinate these rhythms, thereby assuring that DNA synthesis and peak absorption do not coincide.
In AL rats, Per1 and ReverbB mRNA expression peaked in phase with Sglt1, slightly preceding Sglt1 expression by 1–2 h. Restricted feeding produced similar phase shifts for Sglt1, Per1, and ReverbB; all 3 genes were phase shifted by 1–2 h in DF rats and 7–8 h in LF rats compared with AL rats. The presence of 4 canonical E-boxes in the Sglt1 promoter raises the possibility that the Per1 transcription factor is involved in controlling Sglt1 rhythmicity. If so, occurrence of the Sglt1 mRNA nadir when the Per1 protein level is rising suggests that Per1 exerts a negative influence. Lack of Reverb response elements in the Sglt1 promoter argues against ReverbB involvement but does not preclude indirect regulation or use of a noncanonical element.
We were surprised that nocturnal food restriction advanced the phases of Sglt1 and intestinal clock genes by 1–2 h compared with AL feeding. Although only a modest amount of food is usually consumed during the day (10–20% daily intake) (27), the restriction was apparently sufficient to shift gene expression phases. The daytime food deprivation in DF rats effectively prevented “early phase eating,” consumption of food in the late LF, and may have enhanced entrainment signals normally produced by hunger or hormonal responses, thereby sharpening the anticipatory intestinal gene induction and advancing the acrophases in DF rats. In either case, it is clear that restricting food access to 12 h led to detectable alterations in intestinal rhythms.
Overall SGLT1 protein expression was higher in LF rats compared with DF rats (P = 0.010), despite no significant difference in mRNA levels (P = 0.946). This result is consistent with our previous finding that post-transcriptional events are also important in regulating intestinal SGLT1 expression (28). In light of reports that SGLT1 expression is increased in obesity and diabetes (29,30), it would be interesting to assess the functional consequences of our observation by comparing glucose homeostasis in light- and dark-fed rats as well as measuring SGLT1 expression in shift workers who are forced to eat off schedule and have increased risk of developing glucose intolerance (31).
Our findings lend support to the notion that clock genes cue intestinal rhythmicity in response to nutrient availability. Clock genes are clearly important transcriptional regulators (26,32,33). Clock and clock-controlled genes have been implicated in the regulation of other proteins such as the Na+/H+ exchanger Nhe3 in the kidney (32), the oligopeptide transporter Pept1 (26), and the multidrug resistance 1 gene (33). Pan and Hussain (34), using Clock mutant mice, presented evidence for its involvement in intestinal absorptive rhythms. Our studies add to the existing evidence implicating clock genes in absorptive rhythms and provide important information on the role of clock genes in regulating SGLT1 rhythmicity and thereby rhythmicity of glucose uptake in the intestine.
The glucose concentration generated from digestion may be a major stimulus in regulating the expression of clock and Sglt1 genes in the intestine. In an intriguing study, glucose was shown to downregulate Per1 and Per2 mRNA expression in rat-1 fibroblasts (35). The authors hypothesized that glucose itself, which displays a modest circadian rhythm in rodents (36), provides a Zeitgeber for peripheral clocks, acting to downregulate Per1 and Per2 via other transcriptional regulators. This hypothesis is consistent with decreased Per1 mRNA levels during the period of nutrient consumption in both AL and DF rats. Although plasma glucose levels are relatively constant, enterocytes (and probably also hepatocytes) are unique in experiencing abrupt increases in glucose supply and intracellular concentrations following feeding. Thus, glucose suppression of Per expression may be the molecular basis for resetting intestinal (and liver) clocks by nutrient availability. The ability of these 2 “gateway” organs to respond rapidly to nutrient intake patterns via peripheral clocks would have great adaptive value by optimally coordinating absorptive functions with nutrient delivery.
In summary, we have shown that nutrients provide a major Zeitgeber for intestinal clock genes and that shifting the period of availability simultaneously phase shifts expression of clock genes and intestinal transporters. Further studies are required to define the molecular mechanism linking clock genes to Sglt1 rhythmicity. The regulatory mechanisms governing circadian rhythmicity of intestinal function may have a considerable role in obesity and diabetes and a better understanding could lead to new therapies for these worsening epidemics.
Supplementary Material
Acknowledgments
We thank John Young for excellent technical assistance and Jan Rounds for invaluable managerial assistance. A.B., S.W.A., A.T., and D.B.R. designed the studies; A.B. and A.T.S. conducted the research; A.B. analyzed the data; A.B., A.T., and D.B.R. wrote the paper; and A.B. and D.B.R. had primary responsibility for final content. All authors read and approved the final manuscript.
Supported by the NIH grant 5 R01 DK047326 (S.W.A.), ADA grant 7-05-RA-121 (D.B.R.), the Harvard Clinical Nutrition Research Center grant (A.T.) P30-DK040561, the Nutricia Research Foundation (A.B.), and the Berkeley Fellowship (A.T.S.).
Author disclosures: A. Balakrishnan, A. T. Stearns, S. W. Ashley, A. Tavakkolizadeh, and D. B. Rhoads, no conflicts of interest.
Supplemental Figures 1 and 2 and Supplemental Table 1 are available with the online posting of this paper at jn.nutrition.org.
This manuscript was presented in poster form at Digestive Diseases Week 2007 and 2008, and published in abstract form only in the supplementary issue of Gastroenterology (less than 400 words).
Abbreviations used: AL, ad libitum; DF, dark fed; HALO, hours after light onset; LF, light fed; SCN: suprachiasmatic nucleus.
References
- 1.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–90. [DOI] [PubMed] [Google Scholar]
- 2.Tavakkolizadeh A, Berger UV, Shen KR, Levitsky LL, Zinner MJ, Hediger MA, Ashley SW, Whang EE, Rhoads DB. Diurnal rhythmicity in intestinal SGLT-1 function, V (max), and mRNA expression topography. Am J Physiol Gastrointest Liver Physiol. 2001;280:G209–15. [DOI] [PubMed] [Google Scholar]
- 3.Balakrishnan A, Stearns AT, Rounds J, Irani J, Giuffrida M, Rhoads DB, Ashley SW, Tavakkolizadeh A. Diurnal rhythmicity in glucose uptake is mediated by temporal periodicity in the expression of the sodium-glucose cotransporter (SGLT1). Surgery. 2008;143:813–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stearns AT, Balakrishnan A, Rhoads DB, Ashley SW, Tavakkolizadeh A. Diurnal rhythmicity in the transcription of jejunal drug transporters. J Pharmacol Sci. 2008;108:144–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan X, Terada T, Irie M, Saito H, Inui K. Diurnal rhythm of H+-peptide cotransporter in rat small intestine. Am J Physiol Gastrointest Liver Physiol. 2002;283:G57–64. [DOI] [PubMed] [Google Scholar]
- 6.Richards AM, Nicholls MG, Espiner EA, Ikram H, Cullens M, Hinton D. Diurnal patterns of blood pressure, heart rate and vasoactive hormones in normal man. Clin Exp Hypertens A. 1986;8:153–66. [DOI] [PubMed] [Google Scholar]
- 7.Selmaoui B, Touitou Y. Reproducibility of the circadian rhythms of serum cortisol and melatonin in healthy subjects: a study of three different 24-h cycles over six weeks. Life Sci. 2003;73:3339–49. [DOI] [PubMed] [Google Scholar]
- 8.Hastings MH. Circadian clocks. Curr Biol. 1997;7:R670–2. [DOI] [PubMed] [Google Scholar]
- 9.Panda S, Hogenesch JB. It's all in the timing: many clocks, many outputs. J Biol Rhythms. 2004;19:374–87. [DOI] [PubMed] [Google Scholar]
- 10.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–41. [DOI] [PubMed] [Google Scholar]
- 11.Torra IP, Tsibulsky V, Delaunay F, Saladin R, Laudet V, Fruchart JC, Kosykh V, Staels B. Circadian and glucocorticoid regulation of Rev-erbalpha expression in liver. Endocrinology. 2000;141:3799–806. [DOI] [PubMed] [Google Scholar]
- 12.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–60. [DOI] [PubMed] [Google Scholar]
- 13.Oishi K, Sakamoto K, Okada T, Nagase T, Ishida N. Humoral signals mediate the circadian expression of rat period homologue (rPer2) mRNA in peripheral tissues. Neurosci Lett. 1998;256:117–9. [DOI] [PubMed] [Google Scholar]
- 14.Oishi K, Sakamoto K, Okada T, Nagase T, Ishida N. Antiphase circadian expression between BMAL1 and period homologue mRNA in the suprachiasmatic nucleus and peripheral tissues of rats. Biochem Biophys Res Commun. 1998;253:199–203. [DOI] [PubMed] [Google Scholar]
- 15.Takata M, Burioka N, Ohdo S, Takane H, Terazono H, Miyata M, Sako T, Suyama H, Fukuoka Y, et al. Daily expression of mRNAs for the mammalian Clock genes Per2 and clock in mouse suprachiasmatic nuclei and liver and human peripheral blood mononuclear cells. Jpn J Pharmacol. 2002;90:263–9. [DOI] [PubMed] [Google Scholar]
- 16.Sakamoto K, Oishi K, Nagase T, Miyazaki K, Ishida N. Circadian expression of clock genes during ontogeny in the rat heart. Neuroreport. 2002;13:1239–42. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto T, Nakahata Y, Tanaka M, Yoshida M, Soma H, Shinohara K, Yasuda A, Mamine T, Takumi T. Acute physical stress elevates mouse period1 mRNA expression in mouse peripheral tissues via a glucocorticoid-responsive element. J Biol Chem. 2005;280:42036–43. [DOI] [PubMed] [Google Scholar]
- 18.Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–7. [DOI] [PubMed] [Google Scholar]
- 19.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–3. [DOI] [PubMed] [Google Scholar]
- 20.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. [DOI] [PubMed] [Google Scholar]
- 22.Hara R, Wan K, Wakamatsu H, Aida R, Moriya T, Akiyama M, Shibata S. Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells. 2001;6:269–78. [DOI] [PubMed] [Google Scholar]
- 23.Nelson W, Tong YL, Lee JK, Halberg F. Methods for cosinor-rhythmometry. Chronobiologia. 1979;6:305–23. [PubMed] [Google Scholar]
- 24.Circadian Rhythm Laboratory. University of South Carolina. [cited 2010 Feb 16]. Available from: http://www.circadian.org.
- 25.Stearns AT, Balakrishnan A, Rounds J, Rhoads DB, Ashley SW, Tavakkolizadeh A. Capsaicin-sensitive vagal afferents modulate posttranscriptional regulation of the rat Na+/glucose cotransporter SGLT1. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1078–83. [DOI] [PubMed] [Google Scholar]
- 26.Saito H, Terada T, Shimakura J, Katsura T, Inui K. Regulatory mechanism governing the diurnal rhythm of intestinal H+/peptide cotransporter 1 (PEPT1). Am J Physiol Gastrointest Liver Physiol. 2008;295:G395–402. [DOI] [PubMed] [Google Scholar]
- 27.Pan X, Terada T, Okuda M, Inui K. The diurnal rhythm of the intestinal transporters SGLT1 and PEPT1 is regulated by the feeding conditions in rats. J Nutr. 2004;134:2211–5. [DOI] [PubMed] [Google Scholar]
- 28.Stearns AT, Balakrishnan A, Rhoads DB, Ashley SW, Tavakkolizadeh A. Diurnal expression of the rat intestinal sodium-glucose cotransporter 1 (SGLT1) is independent of local luminal factors. Surgery. 2009;145:294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dyer J, Wood IS, Palejwala A, Ellis A, Shirazi-Beechey SP. Expression of monosaccharide transporters in intestine of diabetic humans. Am J Physiol Gastrointest Liver Physiol. 2002;282:G241–8. [DOI] [PubMed] [Google Scholar]
- 30.Osswald C, Baumgarten K, Stumpel F, Gorboulev V, Akimjanova M, Knobeloch KP, Horak I, Kluge R, Joost HG, Koepsell H. Mice without the regulator gene Rsc1A1 exhibit increased Na+-D-glucose cotransport in small intestine and develop obesity. Mol Cell Biol. 2005;25:78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hampton SM, Morgan LM, Lawrence N, Anastasiadou T, Norris F, Deacon S, Ribeiro D, Arendt J. Postprandial hormone and metabolic responses in simulated shift work. J Endocrinol. 1996;151:259–67. [DOI] [PubMed] [Google Scholar]
- 32.Saifur Rohman M, Emoto N, Nonaka H, Okura R, Nishimura M, Yagita K, van der Horst GT, Matsuo M, Okamura H, Yokoyama M. Circadian clock genes directly regulate expression of the Na (+)/H(+) exchanger NHE3 in the kidney. Kidney Int. 2005;67:1410–9. [DOI] [PubMed] [Google Scholar]
- 33.Murakami Y, Higashi Y, Matsunaga N, Koyanagi S, Ohdo S. Circadian clock-controlled intestinal expression of the multidrug-resistance gene mdr1a in mice. Gastroenterology. 2008;135:1636–44 e3. [DOI] [PubMed] [Google Scholar]
- 34.Pan X, Hussain MM. Clock is important for food and circadian regulation of macronutrient absorption in mice. J Lipid Research. 2009;50:1800–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirota T, Okano T, Kokame K, Shirotani-Ikejima H, Miyata T, Fukada Y. Glucose down-regulates Per1 and Per2 mRNA levels and induces circadian gene expression in cultured Rat-1 fibroblasts. J Biol Chem. 2002;277:44244–51. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto H, Nagai K, Nakagawa H. Role of SCN in daily rhythms of plasma glucose, FFA, insulin and glucagon. Chronobiol Int. 1987;4:483–91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.