Abstract
Severe choline deficiency adversely affects cellular methylation and DNA integrity, with potentially serious implications for disease risk. As part of a 12-wk controlled choline intervention study conducted in folate-compromised Mexican-American men (n = 60; 18–55 y) differing in the methylenetetrahydrofolate reductase (MTHFR) C677T genotype (21 677CC, 29 677TT), this study evaluated the effects of varied choline intakes (300, 550, 1100, and 2200 mg/d) on the change (i.e. wk 12–0) in markers of cellular methylation and DNA integrity. Choline intake affected the change in plasma S-adenosylmethionine (P = 0.044), with decreases tending to be greater (P ≤ 0.08) in the 300 and 550 mg/d groups than in the 2200 mg/d group. Choline intake also interacted with the MTHFR C677T genotype to affect the change in genomic DNA methylation and DNA damage. In men with the MTHFR 677CC genotype, choline intake affected (P = 0.007) the change in DNA methylation, with a greater decrease (P < 0.02) in the 300 mg/d group than in the 1100 and 2200 mg/d groups. In men with the MTHFR 677CC genotype, choline intake also affected (P = 0.047) the change in DNA damage, with the increase tending to be greater (P = 0.07) in the 550 mg/d group than in the 2200 mg/d group. Choline intake did not affect these variables in men with the MTHFR 677TT genotype. Overall, these data suggest that choline intake exceeding current dietary recommendations preserves markers of cellular methylation and attenuates DNA damage in a genetic subgroup of folate-compromised men.
Introduction
The nutritional importance of choline was officially recognized in 1998 with the establishment of choline adequate intakes of 425 and 550 mg/d for women and men, respectively (1). Choline, a trimethylamine, serves as a precursor for several important metabolites, including the phospholipid phosphatidylcholine (PtdCho),6 the methyl donor betaine, and the neurotransmitter acetylcholine (2). Not surprisingly, the metabolic implications of choline deficiency are severe and include fatty liver and steatosis (3–5), cellular membrane fragility (6), DNA damage and apoptosis (7), altered gene expression (8), and cognitive impairments (9,10).
As a major source of labile methyl groups in liver, choline (like folate) is important in the biosynthesis of the universal methyl donor, S-adenosylmethionine (SAM). In this regard, the betaine- (or folate-) derived methyl groups may be used for the conversion of homocysteine (Hcy) to methionine, which can then be activated to SAM. Thus, choline (or folate) insufficiency may diminish the availability of SAM and compromise cellular methylation reactions and genome stability (11). These endpoints may also be compromised by loss-of-function genetic variants in 1-carbon metabolizing genes such as the 677C→T single nucleotide polymorphism in the methylenetetrahydrofolate reductase (MTHFR) gene (12,13).
The requirement for choline can be met in part by de novo biosynthesis of PtdCho through the phosphatidylethanolamine N-methyltransferase (PEMT) pathway. As a main consumer of SAM, PEMT is also a major producer of S-adenosylhomocysteine (SAH), an inhibitor of most methyltransferases (14,15). Thus, suboptimal choline intakes may adversely affect cellular methylation reactions, such as DNA methylation, by decreasing the ratio of SAM:SAH. Methylation of DNA occurs on cytosine residues that precede guanosine (i.e. CpG dinucleotides) and plays a regulatory role in genome expression and stability (16).
While it is well recognized that severe choline deficiency has adverse consequences on cellular methylation and DNA integrity (7,8), the effects of a broad range of choline intake on functional biomarkers of 1-carbon metabolism are less clear. Thus, as an extension of a controlled 12-wk choline intervention trial conducted in Mexican-American men differing in MTHFR C677T genotype (17,18), the present study sought to investigate the effect of varied choline intake (300, 550, 1100, or 2200 mg/d) on biomarkers of cellular methylation and DNA damage.
The results presented herein are within the context of declining folate status [despite consumption of the folate Recommended Dietary Allowance (RDA)] throughout this 12-wk study (17). In this regard, by the end of the study, serum folate had decreased (P < 0.001) by 66% to 7.9 ± 0.7 nmol/L (mean ± SEM) in men with the MTHFR 677TT genotype and by 62% to 11.3 ± 0.3 nmol/L in the 677CC genotype. Moreover, plasma total Hcy had increased (P < 0.0001) by 170% to 31 ± 3 μmol/L in men with the 677TT genotype and by 18% to 11.6 ± 0.3 μmol/L in men with the 677CC genotype.
Study Design and Methods
Study participants.
The study participants (n = 60; 18–55 y) were healthy men of Mexican descent preselected for the MTHFR 677CC (n = 31) or TT (n = 29) genotype. All study participants successfully completed a controlled dietary intervention trial that assessed the adequacy of the U.S. folate RDA in men differing in MTHFR genotype (17) as well as the effect of varied choline intake on select biomarkers of choline status (18). Screening and experimental procedures were approved by the Cal Poly Pomona Institutional Review Board for Human Subjects in Pomona, California and informed consent was collected from all participants. Approval to use de-identified samples for the current measurements was granted by the Cornell University Institutional Review Board for Human Subjects in Ithaca, New York.
Study design and diet.
The original study was a 12-wk controlled feeding trial providing 438 μg/d folate as dietary folate equivalents [∼1998 folate RDA; (1)] and choline intakes of 300, 550 (the choline adequate intake), 1100, or 2200 mg/d as previously detailed (17). Throughout the 12 wk of controlled feeding, all meals and snacks were provided on a daily basis by the investigators and study participants were required to consume at least 1 meal per day on-site.
As an extension of the original study, the present study examined the effect of varied choline intake and MTHFR C677T genotype on the change (wk 12 − wk 0) in plasma markers of cellular methylation capacity (SAM and SAH), leukocyte global DNA methylation, and lymphocyte DNA damage indicators.
Sample collection and processing.
Fasting (10 h) venous blood samples were collected from the study participants at baseline (wk 0) and weekly thereafter in EDTA tubes (Vacutainer; Becton Dickinson). Plasma and leukocytes were separated and stored at −80°C as previously detailed (19). Peripheral lymphocytes were separated from blood using the Ficoll-Hypaque gradient in evacuated tubes with sodium citrate (Vacutainer CPT; Becton Dickinson) according to the manufacturer's instructions.
Analytical measurements.
MTHFR C677T genotype determination was achieved in duplicate with PCR and Hinf1 restriction enzyme digestion (20). Plasma SAM and SAH measurements were performed in duplicate using an HPLC method with UV detection (21) and an immunoassay (22), respectively.
Leukocyte percent methylated cytosine (global DNA methylation) was determined in duplicate using the liquid chromatography-MS method of Song et al. (23) with modifications based on our instrumentation. Briefly, DNA was extracted from leukocytes via a commercially available kit (QIAmp DNA Blood Mini kit, Qiagen) and digested with nuclease P1, phosphodiesterase 1, and alkaline phosphatase. The hydrolyzed DNA and calibration curves prepared with 2′-deoxycytidine, 5-methyl-2′-deoxycytidine, and 2′-deoxyguanosine monohydrate were then injected into the liquid chromatography-MS system consisting of a TSQ Quantum mass spectrometer with electrospray ionization source operated in positive ion mode (Thermo), a refrigerated Accela autosampler (Thermo), and an Accela pump with degasser (Thermo). Separation of analytes was achieved by using a 1.9-mm hypersil gold column (Thermo) under a 400-μL/min flow rate with 0.1% formic acid in methanol and 0.1% formic acid in water as the mobile phase. The analytes were detected by monitoring the mass:charge ratio (m/z) transitions of m/z 228 to m/z 112 for 2′-deoxycytidine, m/z 242 to m/z 126 for 5-methyl-2′-deoxycytidine, and m/z 268 to m/z 151 for 2′-deoxyguanosine.
Assessment of lymphocyte DNA damage was evaluated in duplicate using the alkaline version of single-cell gel electrophoresis (COMET assay), which assesses single- and double-strand breaks, incomplete excision repair sites, and cross links (24,25). Lymphocytes were mixed with 1% low-melting point (37°C) agarose (Cambrex) to a final concentration of 1 × 107/L, of which 75 μL was dispensed onto Comet slides (Trevigen) and allowed to solidify at 4°C in the dark for 15 min. The resulting slides were immersed into cold, freshly made lysis solution [10 mmol/L Tris, 2.5 mol/L NaCl, 0.1 mol/L EDTA, 10% dimethylsulfoxide, and 1% Triton X-100 (pH 10)] at 4°C for ≥1 h and then treated for 20 min in cold, freshly made electrophoresis buffer [0.3 mol/L NaOH, 1 mmol/L EDTA (pH 13)]. After electrophoresis was performed at 0.8 volts/cm and 300 mA for 40 min, the slides were submerged for 15 min in neutralization buffer [0.4 mol/L Tris (pH 7.5)], washed for 5 min in 100% ethanol, dried overnight, and stained with SYBR Gold fluorescent dye (Invitrogen) for 15 min at 4°C in the dark. DNA migration from the nucleus was then visualized (typically 100 cells/sample) at 200× magnification using a fluorescent microscope (Olympus BX-61). Percent tail DNA (defined as the proportion of DNA that has migrated from the head) was calculated using computer-based image analysis software (Komet 6.0; Andor Technology). Samples were measured in duplicate on 2 separate occasions in batches. Each batch consisted of a balanced representation of MTHFR C677T genotype and choline intake as well as wk 0 and 12 for each participant and the internal controls (untreated pooled peripheral lymphocytes). The intra-CV for percent tail DNA of the internal controls was 6.7%.
Statistical analysis.
Two-way ANOVA without the interaction term was used to assess differences in the measured variables at the beginning of the 12-wk study between the MTHFR 677CC or TT genotypes and between the choline intake groups. Linear regression analysis was used to examine how various independent variables (i.e. choline intake, MTHFR genotype, baseline measures) affected the change (i.e. wk 12 − wk 0) in the dependent variables of interest (i.e. percent methylated cytosine, plasma SAH, plasma SAM, plasma SAM:SAH ratio, leukocyte DNA damage). Initially, each regression model included the dependent variable of interest, and choline intake, MTHFR genotype, and the baseline measure of the dependent variable, as independent variables. To this basic model, nutritional factors as well as change in plasma total Hcy, age, BMI, and serum creatinine were added as independent variables, as previously described (26). During the construction of the basic models, all possible interactions between variables were investigated. In the final basic models, only significant terms defined as P ≤ 0.10 were preserved. For the models in which the independent variables interacted with the MTHFR C677T genotype to affect the change in the dependent variable, the statistical analysis was repeated separately for each genotype. The Bonferroni method was used for multiple comparison adjustments. The data were analyzed by SPSS software, version 15, and are presented as mean ± SEM unless indicated otherwise. Significance was set at P < 0.05; P ≤ 0.10 was considered to be indicative of trends.
Results
Baseline measures (wk 0).
At baseline, men with the MTHFR 677TT genotype had higher (P = 0.012) plasma SAH concentrations and a lower (P = 0.012) ratio of plasma SAM:SAH (Table 1). Plasma SAH also differed (P = 0.017) among the choline intake groups, with higher (P = 0.011) concentrations in the 2200 mg/d group compared with the 550 mg/d group. No other baseline variables differed between MTHFR C677T genotypes or among the choline intake groups. The interaction term for choline intake and MTHFR C677T genotype was not examined.
TABLE 1.
Baseline plasma SAM and SAH concentrations and markers of cellular methylation and DNA integrity in folate-compromised Mexican-American men with the MTHFR 677CC or 677TT genotype that consumed various levels of choline12
| Choline intake, mg/d |
|||||
|---|---|---|---|---|---|
| Variable | 300 | 550 | 1100 | 2200 | Mean |
| Plasma SAM, nmol/L | |||||
| 677CC | 60.9 ± 3.1 | 58.3 ± 2.6 | 70.8 ± 9.7 | 62.5 ± 2.2 | 63.1 ± 2.3 |
| 677TT | 54.3 ± 2.9 | 61.1 ± 3.0 | 64.0 ± 4.8 | 58.5 ± 4.3 | 59.5 ± 2.5 |
| Mean | 57.6 ± 3.3 | 59.7 ± 3.2 | 67.4 ± 3.3 | 60.5 ± 3.7 | |
| Plasma SAH, nmol/L | |||||
| 677CC | 27.4 ± 1.8 | 25.7 ± 1.1 | 28.0 ± 1.5 | 29.2 ± 1.4 | 27.6 ± 0.8* |
| 677TT | 29.6 ± 1.8 | 27.3 ± 2.0 | 30.7 ± 1.8 | 34.5 ± 1.3 | 30.5 ± 0.8 |
| Mean | 28.5 ± 1.2a,b | 26.5 ± 1.1a | 29.4 ± 1.1a,b | 31.8 ± 1.2b | |
| Plasma SAM:SAH ratio | |||||
| 677CC | 2.3 ± 0.1 | 2.3 ± 0.2 | 2.5 ± 0.3 | 2.2 ± 0.2 | 2.3 ± 0.1* |
| 677TT | 1.8 ± 0.1 | 2.2 ± 0.2 | 2.1 ± 0.2 | 1.7 ± 0.1 | 2.0 ± 0.1 |
| Mean | 2.1 ± 0.1 | 2.2 ± 0.1 | 2.3 ± 0.1 | 1.9 ± 0.2 | |
| Methylated cytosine, % | |||||
| 677CC | 5.3 ± 0.1 | 4.9 ± 0.1 | 5.0 ± 0.1 | 5.1 ± 0.2 | 5.1 ± 0.1 |
| 677TT | 4.9 ± 0.1 | 5.0 ± 0.1 | 5.0 ± 0.1 | 4.8 ± 0.1 | 4.9 ± 0.1 |
| Mean | 5.1 ± 0.1 | 5.0 ± 0.1 | 5.0 ± 0.1 | 4.9 ± 0.1 | |
| Tail DNA, % | |||||
| 677CC | 20.3 | 39.3 ± 7.1 | 30.3 ± 3.2 | 34.3 ± 4.7 | 31.1 ± 2.9 |
| 677TT | 40.7 ± 2.4 | 38.1 ± 2.3 | 35.8 ± 1.7 | 31.8 ± 2.1 | 36.6 ± 1.9 |
| Mean | 30.5 ± 4.5 | 38.7 ± 2.5 | 33.0 ± 2.8 | 33.0 ± 3.5 | |
Values are means ± SEM, n = 5–9/group except groups for tail DNA, n = 1–6. Means in a row with superscripts without a common letter differ, P < 0.05. *Different from MTHFR 677TT genotype, P < 0.05.
Data were analyzed by 2-factor ANOVA (choline intake and MTHFR C677T genotype) without the interaction term.
Changes in plasma SAM.
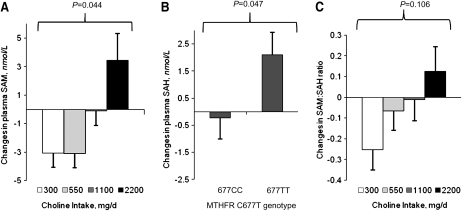
In a statistical model that included baseline plasma SAM concentrations and changes in plasma total Hcy concentrations, choline intake affected the change in plasma SAM (P = 0.044; Fig. 1A). Specifically, plasma SAM concentrations decreased in the 300 and 550 mg/d choline intake groups and the decreases tended to differ (P ≤ 0.08) from the increase in the 2200 mg/d choline intake group. The MTHFR C677T genotype did not predict changes in plasma SAM concentration, nor did the genotype or choline intake interact with other variables included in this model to affect changes in plasma SAM.
FIGURE 1 .
Changes in plasma SAM (A), SAH (B), and the SAM:SAH ratio (C) in folate-compromised Mexican-American men grouped by choline intake or MTHFR C677T genotype. Data are presented as estimated marginal means ± SEM (n = 11–15/choline intake group or n = 25–28 for MTHFR C677T genotypes) and were analyzed using linear regression.
Changes in plasma SAH.
In a statistical model that included baseline plasma SAH concentrations and serum creatinine, the MTHFR C677T genotype affected the change in plasma SAH concentrations (P = 0.047; Fig. 1B). Specifically, the plasma SAH concentration increased only in men with the MTHFR 677TT genotype and the increase differed (P = 0.047) from the modest decline in men with the 677CC genotype. Choline intake did not affect plasma SAH concentrations, nor did choline intake or the MTHFR C677T genotype interact with other variables included in this model to affect changes in plasma SAH.
Changes in the plasma SAM:SAH ratio.
In a statistical model that included the baseline plasma SAM:SAH ratio and changes in plasma total Hcy, choline intake interacted (P = 0.025) with the baseline SAM:SAH ratio to affect the change in the SAM:SAH ratio. Specifically, with the baseline SAM:SAH ratio set at the mean, the SAM:SAH ratio decreased in the 300 mg/d choline intake group and the decrease tended to differ (P = 0.106) from the increase in the 2200 mg/d intake group (Fig. 1C). No significant effect of MTHFR C677T genotype was detected in this model, nor did it interact with other variables to affect changes in the plasma SAM:SAH ratio.
Changes in global DNA methylation.
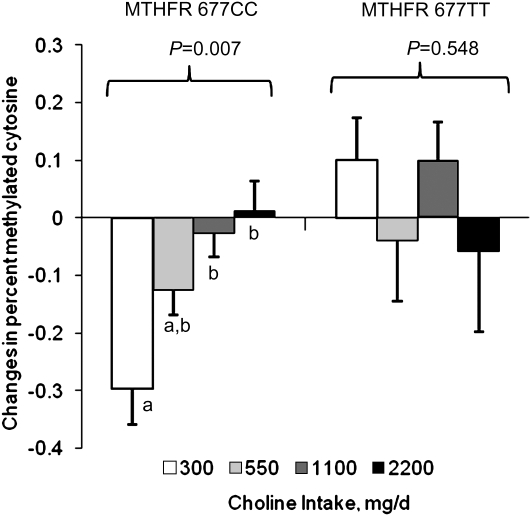
In a statistical model that included baseline percent methylated cytosine and baseline plasma SAH, choline intake interacted with the MTHFR genotype and the baseline measures to affect the change in percent methylated cytosine (P = 0.020). Because of the interaction between choline intake and MTHFR genotype (P = 0.021), the regression analysis was repeated separately for each genotype (Fig. 2). Within the MTHFR 677CC genotype, baseline percent methylated cytosine and baseline plasma SAH interacted with choline intake to affect the change in percent methylated cytosine (P = 0.049). With baseline percent methylated cytosine and baseline plasma SAH at their respective mean levels, choline intake affected the change in percent methylated cytosine (P = 0.007) in this genotype. Specifically, the percent methylated cytosine decreased in the 300 mg/d intake group and the decrease differed from the changes in the 1100 (P = 0.015) and 2200 (P = 0.010) mg/d groups. Within the MTHFR 677TT genotype, no significant main effects or interaction terms were detected.
FIGURE 2 .
Changes in the percent methylated cytosine in folate-compromised Mexican-American men grouped by choline intake and MTHFR C677T genotype. Data are presented as estimated marginal means ± SEM (n = 6–7/group) and were analyzed by linear regression. Because choline intake interacted with the MTHFR genotype (P = 0.021), the regression analysis was repeated separately for each genotype. Labeled means without a common letter differ, P < 0.05.
Changes in DNA damage (percent tail DNA).
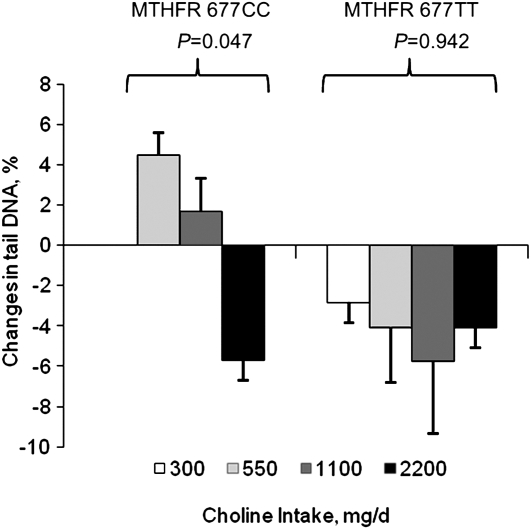
In a subgroup of participants from the main study, choline intake interacted with the MTHFR genotype, baseline DNA damage measures, and the change in plasma tHcy to affect DNA damage (P = 0.006). To assess the effect of choline intake within the MTHFR genotypes, the statistical analysis was repeated separately for each genotype (Fig. 3). Within the MTHFR 677CC genotype, baseline measures of DNA damage interacted with choline intake to affect the change in DNA damage (P = 0.04). In addition, baseline measures of DNA damage interacted with the change in plasma Hcy. With baseline measures of DNA damage and the change in plasma tHcy set at their respective mean levels, choline intake affected the change in DNA damage (P = 0.047) in men with this genotype. Specifically, DNA damage decreased only in the 2200 mg/d choline intake group and the decrease tended to differ (P = 0.07) from the increase in the 550 mg/d intake group. Data from the 300 mg/d choline intake group were excluded from the statistical analysis due to a sample size of one. Within the MTHFR 677TT genotype, no significant main effects or interaction terms were detected.
FIGURE 3 .
Changes in DNA damage (percent tail DNA, comet assay) in a subgroup of folate-compromised Mexican-American men grouped by choline intake and MTHFR C677T genotype. Data are presented as estimated marginal means ± SEM (n = 2–6/group) and were analyzed using linear regression. Because MTHFR interacted with the other independent variables (P = 0.006), the regression analysis was repeated separately for each genotype (excluding the MTHFR 677CC 300 mg/d choline intake group due to its sample size of one).
Discussion
To the best of our knowledge, this is the first human study to demonstrate a potentially beneficial effect of choline intakes exceeding current dietary recommendations (i.e. 550 mg/d) on biomarkers of cellular methylation and DNA damage in men that were not clinically choline deficient (i.e. elevations in serum alanine aminotransferase). Importantly the majority of these effects were confined to men with the MTHFR 677CC genotype and transpired against a background of declining folate status, as previously detailed (17).
The biomarkers of cellular methylation measured in this study included plasma concentrations of SAM, SAH, and the SAM:SAH ratio. Both SAM and the SAM:SAH ratio were influenced (P ≤ 0.10) by choline intake, with decreases in the 300 and/or 550 mg/d group compared with the 2200 mg/d intake group. The preservation of cellular methylation capacity in the higher choline intake was apparent in both MTHFR genotypes. SAM is produced from methionine, which in turn can be derived from the diet or from Hcy remethylation via a folate- or betaine-dependent route. Under the conditions of this study, the higher choline intake may have compensated for the diminished folate availability by increasing the supply of the choline-derived methyl donor, betaine. In this regard, we previously reported higher circulating concentrations of betaine in the 2200 mg/d intake group compared with the 300 and 550 mg/d intake groups (18). The lack of an effect of the MTHFR 677TT genotype on plasma SAM concentrations is consistent with the working hypothesis that the increase in plasma total Hcy (relative to those with the MTHFR 677CC genotype) is due to increased production of Hcy through transmethylation rather than impaired Hcy remethylation (27).
The increase in plasma SAH among men with the MTHFR 677TT genotype (compared with those with the 677CC genotype) is also consistent with enhanced Hcy production by the 677TT genotype. However, given that plasma total Hcy tripled in men with the MTHFR 677TT genotype (17), a greater increase in plasma SAH in this genotype was expected. The hydrolysis of SAH to Hcy is reversible and SAH production is favored if Hcy is not efficiently removed via remethylation to methionine or transsulfuration to cysteine. It is thus likely that Hcy, which is readily transported across plasma membranes, serves as an exportable form of SAH, thereby preserving cellular methylation (28). In turn, this implies that circulating concentrations of Hcy and SAH may not be highly correlated as is the case in the present communication. In men with the MTHFR 677TT genotype, we also expected to detect an adverse effect of high choline intake on plasma SAH based on our previous finding that consumption of 1100 or 2200 mg/d resulted in a doubling in plasma tHcy compared with the 300 or 550 mg/d choline (i.e. ∼20 vs. 40 μmol/L) (26). Again, the lack of effect of choline intake on plasma SAH can be explained by the preferential export of Hcy from tissue (in lieu of SAH).
In the present study, we detected a modest but significant decline in global DNA methylation in the 300 mg/d group that did not occur in the 1100 or 2200 mg/d group (Fig. 2). This effect, however, was genotype specific and confined to men with the MTHFR 677CC genotype. DNA methyltransferases utilize SAM as a substrate, which, in this study, did not differ between the MTHFR C677T genotypes. However, we did detect a modest but significant rise in plasma SAH in participants with the 677TT genotype, suggesting that tissue levels may also be increased. Perhaps this small rise was enough to slow the activity of methyltransferases affected by the inhibitory properties of SAH. Notably, men with the MTHFR 677TT genotype also had lower (P = 0.035) circulating concentrations of PtdCho regardless of their choline intake. Thus, like DNA methyltransferases, PEMT may also have been adversely affected by the modest rise in SAH.
It has been proposed that damage to DNA is the earliest functional effect of choline inadequacy (13). In men and women consuming a choline-deficient diet (<50 mg/d) for up to 42 d, DNA damage (as measured by the comet assay) doubled even in those who did not develop organ dysfunction (13). In the subsample of men included in the present analysis, we detected a decrease in DNA damage in those consuming 2200 vs. 550 mg/d choline. Like DNA methylation, the seemingly beneficial effect of choline intake on this biomarker was confined to those with the MTHFR 677CC genotype. The mechanism by which choline intake/status affects DNA integrity is not entirely clear but may be through effects on mitochondrial membrane integrity and oxidative stress (7,29,30). Given the very small sample size for this specific biomarker, our results should be viewed as preliminary and require confirmation.
In conclusion, data from this study suggest that choline intake exceeding current dietary recommendations beneficially influences markers of cellular methylation and DNA damage in folate-compromised men. Notably, such effects were modest and largely confined to those with the MTHFR 677CC genotype whose folate status was less compromised than the 677TT genotype. Whether preservation of cellular methylation markers and attenuation of DNA damage would translate to improved health outcomes is unclear. It is also unknown whether choline intake that exceeds current recommendations would benefit individuals with superior folate status, as observed in populations exposed to widespread folic acid fortification. Our results show that the MTHFR 677TT genotype adversely affected plasma SAH concentrations, which may have inhibited DNA methyltransferases and abolished the effects of high choline intakes on global DNA methylation. Although this study was conducted under highly controlled conditions, additional studies with larger sample sizes are required to further examine the potential metabolic, genomic, and physiological implications of consuming choline intakes that exceed current recommendations.
Acknowledgments
We thank Conrad Wagner for measurements of plasma SAM and SAH. M.A.C. designed the research; W.S. and C.M.A. conducted the research; M.A.C., F.V., and J.Y. analyzed the data; M.A.C. and J.Y. wrote the paper with significant input from F.V. M.A.C. had primary responsibility for final content. All authors read and approved the final manuscript.
Supported by NIH grant no. S06GM053933, the California State University Agricultural Research Initiative with master grant funding from the Federal Agricultural Appropriations Bill allocation, grant no. 2006-38908-17681, and the USDA/Cooperative State Research, Education and Extension Service (CSREES) Hatch program.
Author disclosures: W. Shin, J. Yan, C. M. Abratte, F. Vermeylen, and M. A. Caudill, no conflicts of interest.
Abbreviations used: Hcy, homocysteine; MTHFR, 5,10-methylenetetrahydrofolate reductase; PEMT, phosphatidylethanolamine N-methyltransferase; PtdCho, phosphatidylcholine; RDA, Recommended Dietary Allowance; SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine.
References
- 1.Institute of Medicine. National Academy of Sciences USA. Dietary reference intakes for thiamin, riboflavin, niacin, vitamin B6, folate, vitamin B12, pantothenic acid, biotin and choline. Washington, DC: National Academy Press; 1998. [PubMed]
- 2.Zeisel SH, Blusztajn JK. Choline and human nutrition. Annu Rev Nutr. 1994;14:269–96. [DOI] [PubMed] [Google Scholar]
- 3.Zeisel SH, Da Costa KA, Franklin PD, Alexander EA, Lamont JT, Sheard NF, Beiser A. Choline, an essential nutrient for humans. FASEB J. 1991;5:2093–8. [PubMed] [Google Scholar]
- 4.Buchman AL, Ament ME, Sohel M, Dubin M, Jenden DJ, Roch M, Pownall H, Farley W, Awal M, et al. Choline deficiency causes reversible hepatic abnormalities in patients receiving parenteral nutrition: proof of a human choline requirement: a placebo-controlled trial. JPEN. 2001;25:260–8. [DOI] [PubMed] [Google Scholar]
- 5.Yao ZM, Vance DE. The active synthesis of phosphatidylcholine is required for very low density lipoprotein secretion from rat hepatocytes. J Biol Chem. 1988;263:2998–3004. [PubMed] [Google Scholar]
- 6.Li Z, Agellon LB, Allen TM, Umeda M, Jewell L, Mason A, Vance DE. The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab. 2006;3:321–31. [DOI] [PubMed] [Google Scholar]
- 7.da Costa KA, Niculescu MD, Craciunescu CN, Fischer LM, Zeisel SH. Choline deficiency increases lymphocyte apoptosis and DNA damage in humans. Am J Clin Nutr. 2006;84:88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niculescu MD, da Costa KA, Fischer LM, Zeisel SH. Lymphocyte gene expression in subjects fed a low-choline diet differs between those who develop organ dysfunction and those who do not. Am J Clin Nutr. 2007;86:230–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meck WH, Williams CL. Simultaneous temporal processing is sensitive to prenatal choline availability in mature and aged rats. Neuroreport. 1997;8:3045–51. [DOI] [PubMed] [Google Scholar]
- 10.Niculescu MD, Yamamuro Y, Zeisel SH. Choline availability modulates human neuroblastoma cell proliferation and alters the methylation of the promoter region of the cyclin-dependent kinase inhibitor 3 gene. J Neurochem. 2004;89:1252–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caudill M. Folate and choline interrelationships: metabolic and potential health implications. In: Bailey L, editor. Folate in health and disease. Boca Raton (FL): CRC Press; 2009. p. 449–65.
- 12.Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, Olivieri O, Jacques PF, Rosenberg IH, et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci USA. 2002;99:5606–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Axume J, Smith SS, Pogribny IP, Moriarty DJ, Caudill MA. The MTHFR 677TT genotype and folate intake interact to lower global leukocyte DNA methylation in young Mexican American women. Nutr Res. 2007;27:13–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stead LM, Brosnan JT, Brosnan ME, Vance DE, Jacobs RL. Is it time to reevaluate methyl balance in humans? Am J Clin Nutr. 2006;83:5–10. [DOI] [PubMed] [Google Scholar]
- 15.Mudd SH, Brosnan JT, Brosnan ME, Jacobs RL, Stabler SP, Allen RH, Vance DE, Wagner C. Methyl balance and transmethylation fluxes in humans. Am J Clin Nutr. 2007;85:19–25. [DOI] [PubMed] [Google Scholar]
- 16.McCabe DC, Caudill MA. DNA methylation, genomic silencing, and links to nutrition and cancer. Nutr Rev. 2005;63:183–95. [DOI] [PubMed] [Google Scholar]
- 17.Solis C, Veenema K, Ivanov AA, Tran S, Li R, Wang W, Moriarty DJ, Maletz CV, Caudill MA. Folate intake at RDA levels is inadequate for Mexican American men with the methylenetetrahydrofolate reductase 677TT genotype. J Nutr. 2008;138:67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veenema K, Solis C, Li R, Wang W, Maletz CV, Abratte CM, Caudill MA. Adequate Intake levels of choline are sufficient for preventing elevations in serum markers of liver dysfunction in Mexican American men but are not optimal for minimizing plasma total homocysteine increases after a methionine load. Am J Clin Nutr. 2008;88:685–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guinotte CL, Burns MG, Axume JA, Hata H, Urrutia TF, Alamilla A, McCabe D, Singgih A, Cogger EA, et al. Methylenetetrahydrofolate reductase 677C→T variant modulates folate status response to controlled folate intakes in young women. J Nutr. 2003;133:1272–80. [DOI] [PubMed] [Google Scholar]
- 20.Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, Boers GJ, den Heijer M, Kluijtmans LA, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–3. [DOI] [PubMed] [Google Scholar]
- 21.Capdevila A, Wagner C. Measurement of plasma S-adenosylmethionine and S-adenosylhomocysteine as their fluorescent isoindoles. Anal Biochem. 1998;264:180–4. [DOI] [PubMed] [Google Scholar]
- 22.Capdevila A, Burk RF, Freedman J, Frantzen F, Alfheim I, Wagner C. A simple rapid immunoassay for S-adenosylhomocysteine in plasma. J Nutr Biochem. 2007;18:827–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song L, James SR, Kazim L, Karpf AR. Specific method for the determination of genomic DNA methylation by liquid chromatography-electrospray ionization tandem mass spectrometry. Anal Chem. 2005;77:504–10. [DOI] [PubMed] [Google Scholar]
- 24.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–91. [DOI] [PubMed] [Google Scholar]
- 25.Tice RR, Agurell E, Anderson D, Burlinson B, Hartmann A, Kobayashi H, Miyamae Y, Rojas E, Ryu JC, et al. Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ Mol Mutagen. 2000;35:206–21. [DOI] [PubMed] [Google Scholar]
- 26.Caudill MA, Dellschaft N, Solis C, Hinkis S, Ivanov AA, Nash-Barboza S, Randall KE, Jackson B, Solomita GN, et al. Choline intake, plasma riboflavin, and the phosphatidylethanolamine N-methyltransferase G5465A genotype predict plasma homocysteine in folate-deplete Mexican-American men with the methylenetetrahydrofolate reductase 677TT genotype. J Nutr. 2009;139:727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis SR, Quinlivan EP, Shelnutt KP, Ghandour H, Capdevila A, Coats BS, Wagner C, Shane B, Selhub J, et al. Homocysteine synthesis is elevated but total remethylation is unchanged by the methylenetetrahydrofolate reductase 677C->T polymorphism and by dietary folate restriction in young women. J Nutr. 2005;135:1045–50. [DOI] [PubMed] [Google Scholar]
- 28.Caudill MA, Wang JC, Melnyk S, Pogribny IP, Jernigan S, Collins MD, Santos-Guzman J, Swendseid ME, Cogger EA, et al. Intracellular S-adenosylhomocysteine concentrations predict global DNA hypomethylation in tissues of methyl-deficient cystathionine beta-synthase heterozygous mice. J Nutr. 2001;131:2811–8. [DOI] [PubMed] [Google Scholar]
- 29.Filipowicz CM, McCauley RB. The effect of choline deficiency on the outer membranes of rat liver mitochondria. Biochim Biophys Acta. 1983;734:373–7. [DOI] [PubMed] [Google Scholar]
- 30.Hensley K, Kotake Y, Sang H, Pye QN, Wallis GL, Kolker LM, Tabatabaie T, Stewart CA, Konishi Y, et al. Dietary choline restriction causes complex I dysfunction and increased H2O2 generation in liver mitochondria. Carcinogenesis. 2000;21:983–9. [DOI] [PubMed] [Google Scholar]