Abstract
The aryl hydrocarbon receptor (AhR) is a period-aryl hydrocarbon receptor nuclear transporter-simple minded domain transcription factor that shares structural similarity with circadian clock genes and readily interacts with components of the molecular clock. Activation of AhR by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) alters behavioral circadian rhythms and represses the Period1 (Per1) gene in murine hematopoietic stem and progenitor cells. Per1 expression is driven by circadian locomotor activity cycles kaput-brain muscle ARNT-like (CLOCK-BMAL1)–dependent activation of Eboxes in the Per1 promoter. We hypothesized that the effects of AhR activation on the circadian clock are mediated by disruption of CLOCK-BMAL1 function and subsequent Per1 gene suppression. Effects of AhR activation on rhythmic Per1 transcripts were examined in livers of mice after treatment with the AhR agonist, TCDD; the molecular mechanisms of Per1 repression by AhR were determined in hepatoma cells using TCDD and β-napthoflavone as AhR activators. This study reports, for the first time, that AhR activation by TCDD alters the Per1 rhythm in the mouse liver and that Per1 gene suppression depends upon the presence of AhR. Furthermore, AhR interaction with BMAL1 attenuates CLOCK-BMAL1 activity and decreases CLOCK binding at Ebox1 and Ebox3 in the Per1 promoter. Taken together, these data suggest that AhR activation represses Per1 through disrupting CLOCK-BMAL1 activity, producing dysregulation of rhythmic Per1 gene expression. These data define alteration of the Per1 rhythm as novel signaling events downstream of AhR activation. Downregulation of Per1 could contribute to metabolic disease, cancer, and other detrimental effects resulting from exposure to certain environmental pollutants.
Keywords: Per1, gene regulation, Ebox, liver, AhR activation, BMAL1, circadian rhythm
Regulation of circadian rhythms is critical for adaptation of organisms to ever-present environmental changes (e.g., light-dark cycle) and coordination of key physiological processes such as the sleep-wake cycle, hormone secretion, and liver metabolism (Bittman et al., 2003; Canaple et al., 2006; Cermakian and Boivin, 2003; Gekakis et al., 1998; Griffin et al., 1999; Ishikawa et al., 2002; Oishi et al., 2003). Coordination of systemic circadian rhythms in mammals is directed by a central clock located in the suprachiasmatic nucleus (SCN) of the hypothalamus (Moore and Eichler, 1972; Stephan and Zucker, 1972). Although the SCN clock operates autonomously, it remains responsive to environmental signals, in particular the light-dark cycle, which acts to precisely regulate the clock phase (Boivin et al., 2003; Matsuo et al., 2003). The SCN transmits timing information to numerous peripheral clocks throughout the organism via neural and humoral signals, assuring the synchrony of multiple physiological processes (Asai et al., 2001; Mendoza et al., 2008; Mizuyachi et al., 2002). However, peripheral clocks may function independent of the SCN under certain circumstances. For example, restricted feeding can shift the rhythmicity of liver metabolism by 10 h within 2 days. Under these same conditions, the rhythmicity of the SCN remains phase locked to the light-dark cycle despite changes in food intake (Mendoza et al., 2008). Disruption of coordinated function between central and peripheral clocks is hypothesized to promote development of diseases (Mishima et al., 2005).
Circadian rhythms are generated by a molecular clock, which has an endogenous period of ∼24 h. The Period1 (Per1) gene is a fundamental component of the circadian timing system. Regulation of Per1 is important both for endogenous rhythmicity and for coupling environmental information to the molecular clock (Hida et al., 2000; Kojima et al., 2003). The Per1 promoter includes five Eboxes, four cyclic adenosine monophosphate-response elements (CRE), and a single SP1-binding site. CLOCK-BMAL1 and phosphorylated CRE-binding protein stimulate Per1 transcription by binding the Eboxes and CRE in the Per1 promoter, respectively. Eboxes are critical for circadian expression of Per1 (Hida et al., 2000). Although CRE elements are vital for light signaling to the central clock, it is not clear whether CRE elements are important for circadian regulation of physiological functions (Hida et al., 2000).
Dioxins and dioxin-like compounds are ubiquitous environmental toxicants that contribute to the development of diseases such as obesity, metabolic syndrome, diabetes, and multisite carcinogenesis (Bock, 1994; Mishima et al., 2005). At the molecular level, dioxins exert their effects through persistent activation of aryl hydrocarbon receptor (AhR; Schmidt and Bradfield, 1996). In the absence of ligand, AhR is bound to heat shock protein-90 (HSP-90) and other proteins to form a cytoplasmic complex. Upon ligand binding, the AhR-HSP-90-dioxin complex translocates into the nucleus, where HSP-90 is displaced by aryl hydrocarbon receptor nuclear translocator (ARNT). AhR and ARNT form a heterodimeric transcription factor that binds and activates specific dioxin-responsive element (DRE)-containing genes (Swanson, 2002). However, this well-described signaling pathway does not account for all the effects of dioxins. Cross talk between AhR-ARNT and other nuclear receptors, their coactivators, and/or corepressors likely also contribute to dioxin-induced disease (Lucier et al., 1991).
As members of the PAS domain family of transcription factors, AhR and the clock genes, CLOCK and BMAL1, share structural similarities; therefore, interactions between AhR signaling and the circadian clock warrants investigation. AhR expression exhibits diurnal changes in multiple tissues comparable with the diurnal variation in expression of the clock genes BMAL1 and Per1 (Mukai et al., 2008; Richardson et al., 1998). AhR-mediated responses stimulated by 2,3,7,8 tetrachloro-dibenzo-p-dioxin (TCDD) treatment, such as induction of Cyp1a1 expression, are enhanced in hepatic cells when Per1 is suppressed using small interfering ribonucleic acid (Qu et al., 2009). In turn, TCDD alters circadian rhythms and the expression of clock genes including Per1 and Per2 in murine hematopoietic stem and progenitor cells (Garrett and Gasiewicz, 2006). Our previous results demonstrate that levels of Per1 are increased in the AhR null mouse liver yet substantially decreased in the TCDD-treated wild-type mouse liver (Mukai et al., 2008). Thus, we tested the hypothesis that AhR directly inhibits Per1 expression through disrupting the activity of CLOCK-BMAL1 at the Per1 promoter. Our data demonstrate that activation of AhR may disrupt CLOCK-BMAL1 activity at Per1 promoter, thus resulting in the downregulation of Per1. These findings suggest that regulation of the circadian clock is likely a novel component in the mechanism by which dioxins may result in metabolic diseases, cancers, and other circadian-related diseases.
MATERIALS AND METHODS
Cell lines and growth conditions.
The mouse Hepa-1c1c7 cell line, a subclone of the Hepa-1 hepatoma cell line, and the c12 cell line derived from Hepa-1c1c7 were purchased from American Type Culture Collection. Cells were cultured in Dulbecco's modified Eagle's medium reduced serum (DMEM-RS; HyClone) with 7.5% bovine growth serum (BGS; HyClone) and penicillin-streptomycin-amphotericin (MP Biomedicals) at 37°C in a humidified 5% CO2 atmosphere. Cultures were treated with β-napthoflavone (β-NF; a gift from J. Flaws, University of Illinois at Urbana-Champaign) and TCDD () in dimethyl sulfoxide (DMSO) vehicle as indicated, and control cultures were treated with equivalent concentration of DMSO.
Animals and tissue collection.
About 6- to 12 week-old wild-type male c57bl/6J mice (Jackson Labs) were used in these experiments. Animal procedures were approved by the Institutional Animal Care and Use Committee and conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Mice were housed three to four per cage in a light-tight chamber, provided with feed and water ad libitum and entrained for at least 2 weeks under controlled lighting (12:12-h light-dark), temperature (22°C), and humidity (39%). To investigate the effects of long-term AhR activation on the Per1 rhythm, as a marker of circadian clock function, mice were orally dosed with 1 μg/kg of body weight of TCDD or vehicle (corn oil) during their lights-on period. Five days after treatment, animals were released into constant darkness for 48 h prior to initiation of tissue collection. This experimental design allows the assessment of the state of the endogenous clock in the absence of environmental light-dark cues; release into constant darkness prior to tissue collection is a standard protocol for determining endogenous clock state (Hickok and Tischkau, 2010; Mukai et al., 2008). Animals were decapitated and livers obtained at 4-h intervals starting at CT0 (circadian time, where CT0 refers to the time of lights on in the previously entrained light-dark cycle; CT12 is the corresponding time of lights off). Tissues were snap frozen and stored at −80°C until use.
Total RNA isolation, reverse transcription, and quantitative reverse transcription–PCR.
Total cellular RNA was isolated using TRI Reagent according to the manufacturer's protocol (Sigma). Concentration, purity, and quality of the RNA were determined using Nanodrop ND-1000 UV Spectrophotometer at 260 and 280 nm and by gel electrophoresis. For detection of gene transcripts by reverse transcription–PCR, total RNA (1 μg) was used in a 20-μl reaction according to the manufacturer's protocol (Promega). Following complementary DNA (cDNA) synthesis, 5 μl of diluted cDNA (1:5) was used in 20 μl of SYBR green PCR mix (Bio-Rad) containing 300nM Per1-specific primer pairs as previously published (Mukai et al., 2008), and 5 μl of diluted cDNA (1:5) was used in 20 μl of SYBR green PCR mix with 1× Quantitect Primer Assay for Cyp1a1 gene expression analysis (Qiagen). Amplification was performed using a Smart Cycler rapid thermal cycler (Cepheid) according to the following protocol: an initial 10-min denaturation step at 95°C, followed by 40 cycles of denaturation (95°C) for 30 s, annealing (primer-optimized temperature) for 30 s, and extension (72°C) for 30 s. Detection of the fluorescent product was carried out during each 72°C extension period, and emission data were quantified using threshold cycle values. Relative standard curves were created as previously described (Karman and Tischkau, 2006). Resulting standard curves were used to calculate the relative amounts of Per1 and Cyp1a1. Relative amount was converted to fold or percent change compared with control values (DMSO treated) unless otherwise stated. PCR product specificity from each primer pair was confirmed using melting curve analysis and subsequent agarose gel electrophoresis.
Preparation of cell lysates, cytosol, and nuclear lysates and immunoblot analysis.
Following treatment, Hepa-1c1c7 and c12 cells were harvested by washing with cold PBS followed by lysis in Complete Lysis-M (Roche) on ice for 30 min. Liver sections were homogenized with a bead beater using cold glass beads with 1 ml of Complete Lysis-M and shaken twice for 30 s each. The supernatant was collected after centrifuging at 14,000 × g for 5 min. Cytosol and nuclear lysates were isolated using a modified Abcam corporation protocol. In brief, 500 μl of buffer A with protease inhibitor cocktail (Roche; 10mM 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid sodium salt [HEPES], 1.5mM MgCl2, 10mM KCl, 0.5mM dithiothreitol (DTT), and 0.05% NP40, pH 7.9) was added to 100-mm plates on ice after washing with cold PBS; cells were scraped thoroughly and left on ice for 10 min. Lysates were centrifuged at 3000 × g for 10 min at 4°C. Supernatants were removed and kept as cytosol. Pellets were washed once with buffer A without Nonidet P40. Pellets were resuspended in 60 μl buffer B (5mM HEPES, 1.5mM MgCl2, 0.2mM EDTA, 0.5mM DTT, and 26% glycerol, pH 7.9), and 15 μl of 1.5M NaCl was added to give 300mM NaCl. Lysates were vortexed and centrifuged at 10,000 × g for 20 min at 4°C. Supernatants were collected and kept as nuclear lysates. Protein concentrations were determined with BCA kit (Thermo Scientific), and 20–60 μg of protein was heated to 95°C for 5 min in electrophoresis loading buffer. Electrophoresis was conducted at 90 V for 30 min in the stacking gel and 130 V for 60–90 min in the resolving gel. Proteins were transferred onto a nitrocellulose membrane at 260 mA for 160 min. The membrane was blocked with 5% nonfat dry milk in Tris-buffered saline with Tween 20 (TBST; 20mM Tris, pH 7.5, 150mM NaCl, 0.05% Tween 20) for 1 h. The membrane was then incubated overnight at 4°C with specific primary antibodies, AhR (Biomol), CLOCK (Santa Cruz), BMAL1 (ABR), ARNT (ABR), α-tubulin (Sigma), or β-actin (Sigma), prepared in blocking buffer (Thermo Scientific). The membrane was washed for 30 min in TBST, followed by incubation with horseradish peroxidase (HRP)–conjugated secondary antibody (Jackson ImmunoResearch) for 1 h at room temperature. Protein was visualized using the Immun-Star HRP Chemiluminescence Kit (Bio-Rad), followed by phosphor imaging (Fujifilm). To control for equal lane loading, monoclonal anti-α-tubulin or anti-β-actin (nuclear lysates) antibody was used.
Co-immunoprecipitation.
Hepa-1c1c7 cells were cultured in 100-mm plates to 90–100% confluence. Cells were treated with DMSO vehicle or 10μM β-NF for 1.5 h. Cells were lysed in Complete Lysis-M. Lysates were centrifuged at 14,000 × g for 5 min and then quantitated with BCA kit. Soluble proteins (500 μg) were incubated on a rotating platform with 2 μg of goat anti-AhR antibody (Santa Cruz) or rabbit anti-BMAL1 antibody (ABR) at 4°C overnight, followed by incubation with 100 μl of immobilized protein A/G gel slurry (Pierce) at room temperature for 2 h with gentle mixing. Beads were collected by brief centrifugation and washed five times with 0.5 ml of immunoprecipitation (IP) buffer (25mM Tris and 150mM NaCl, pH 7.2). The complex-bound gel was washed with 0.5 ml of water and centrifuged for 3 min at 2500 × g, the supernatant was discarded, and electrophoresis loading buffer was added and boiled for 5 min at 95°C. The gel mixture was centrifuged and supernatants were separated on 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane. The membrane was blocked with 5% nonfat dry milk in TBST (20mM Tris, pH 7.5, 150mM NaCl, 0.05% Tween 20) for 1 h. The membrane containing protein pulled down by anti-AhR antibody was incubated with rabbit anti-BMAL1 antibody, followed by three washes in TBST and then incubation with HRP-conjugated secondary antibody (Jackson ImmunoResearch) for 1 h at room temperature. The same blot was washed three times with TBST and then incubated with rabbit anti-AhR antibody (Biomol), followed by incubation with HRP-conjugated secondary antibody (Jackson ImmunoResearch) for 1 h at room temperature. Using the same procedure, a separate blot was prepared containing protein immunoprecipitated with the anti-BMAL1 antibody and probed with goat anti-CLOCK antibody (Santa Cruz). This blot was then washed three times with TBST and reprobed with rabbit anti-BMAL1 antibody.
Luciferase assay.
The mPer1-luciferase reporter construct (mPer-luc) cloned into pGL3-basic vector, the mCLOCK ORF cloned into pSG5 vector, and mBMAL1 cloned into pCS2-MTK vector were gifts from Dr Paolo Sassone-Corsi at the University of California at Irvine (Travnickova-Bendova et al., 2002). The hAhR cloned in pcDNA3 vector was a gift from Dr Gary Perdew at Pennsylvania State University (Ramadoss and Perdew, 2005). Cotransfection experiments were performed with c12 cells. Cells were plated in 24-well plates in DMEM-RS medium, supplemented with 7.5% bovine growth serum. Transfection experiments were performed using TurboFect (Fermentas) according to the manufacturer's protocol. Transfection mixtures generally contained mPer1-luc (10 ng), CLOCK-BMAL1 (250 ng), and/or AhR (250 ng), and the internal control β-galacatosidase (β-gal) vector (10 ng). Empty vector was used to equalize the amount of plasmid DNA for each transfection. Transfected cells were cultured for an additional 24 h, washed once with cold PBS, and resuspended in passive lysis buffer (Promega). As a reporter gene, firefly luciferase activity was initiated by mixing an aliquot of lysates (20 μl) with Luciferase Assay Reagent II (Promega). Then, β-gal activity was detected as follows: lysis buffer (150 μl) and 150 μl 2× β-gal buffer was added to 50-μl lysates. Reactions were incubated at 37°C until yellow color appeared and then 500 μl of 1M sodium carbonate was added to stop the reaction; optical density was measured at 420 nm. Luciferase was normalized to the β-gal signal.
Chromatin IP assay.
The chromatin IP (ChIP) protocol was performed with minor modifications as described previously (Weinmann and Farnham, 2002). Hepa-1c1c7 cells were grown in 150-cm2 flasks to 90–100% confluence (2 × 107 cells). Cells were treated with DMSO vehicle or 10μM β-NF for 1.5 h. Cross-linking was performed by adding formaldehyde directly to the culture medium to a final concentration of 1%, followed by 10-min incubation. The reaction was quenched using glycine at a final concentration of 0.125M. Cells were rinsed twice with cold PBS, scraped from the dishes, pelleted, and washed again with PBS containing 1.0mM PMSF. Cell pellets were resuspended in cell lysis buffer (5mM piperazine-1,4-bis(2-ethanesulfonic acid), pH 8.0; 85mM KCl; 0.5% Nonidet P-40; 1.0mM PMSF; 5 μg/ml leupeptin; and 5 μg/ml aprotinin) and incubated on ice for 10 min. Cells were then ground with a Dounce homogenizer 10–20 times on ice to aid in nuclei release. Nuclei were pelleted and resuspended in nuclei lysis buffer (50mM Tris-HCl, pH 8.1; 10 mM EDTA; 1% SDS; 0.5mM PMSF; 5 μg/ml leupeptin; and 5 μg/ml aprotinin) and incubated on ice for 10 min. Chromatin was sonicated to an average length of 600 bp with four 10-s pulses of 30 watts, maintaining samples on ice for 30 s between pulses. Sonicated chromatin was then pre-cleared for 60 min at 4°C with bovine serum albumin– and salmon sperm DNA–saturated protein A/G agarose (Thermo Scientific) in preparation for IP. The supernatant was divided equally among all samples and incubated overnight on a rotating platform at 4°C with 2 μg of rabbit anti-AhR (Biomol), rabbit anti-CLOCK (NOVUS), or rabbit anti-BMAL1 (ABR) or mouse anti-p300 (Millipore) antibody. A protein A/G gel slurry (50 μl) was added and incubated for 2 h at room temperature to allow for antibody binding. The gel was pelleted and washed twice with 1× dialysis buffer (50mM Tris-HCl, pH 8.0; 2mM EDTA; and 0.2% sarkosyl) and four times with IP wash buffer (100mM Tris-HCl, pH 9.0; 500mM LiCl; 1% Nonidet P-40; and 1% deoxycholic acid). Immune complexes were eluted from the beads with elution buffer (100mM NaHCO3 and 1% SDS). Reversal of cross-links was carried out by heating the eluates at 65°C for 4–5 h. The elutes were then digested with proteinase K at 56°C for 2 h. DNA was extracted with phenol-chloroform, precipitated with ethanol, and dissolved in Tris-EDTA buffer. Samples were subjected to PCR amplification with promoter-specific primer. mPer1 Ebox1–specific primers (Naruse et al., 2004) were as follows (205 bp): forward, 5′-ATCCTCCCTGAAAAGGGGTA-3′; reverse, 5′-GGATCTCTTCCTGGCATCTG-3’′. mPer1 Ebox2–specific primers (Naruse et al., 2004) were as follows (351 bp): forward, 5′-CTTTCACAGTAGCCATTGGC-3′; reverse, 5′-ACAAGACACCTGTCCTGGTG-3′. mPer1 Ebox3 (Naruse et al., 2004)–specific primers were as follows (257 bp): forward, 5′-AACAGGTCTGTGTCCCAGCA-3′; reverse, 5′-GGACAACATGCCAGTCTGGG-3′. For the positive control, the promoter region of Cyp1a1 containing the proximal DRE enhancer was amplified (357 bp) with the following primers (Marlowe et al., 2004): forward 5′-CTATCTCTTAAACCCCACCCCAA-3′ and reverse 5′-CTAAGTATGGTGGAGGAAAGGGTG-3′. PCR products were separated on 2% agarose gels. Imaging and densitometry of bands were performed using Quantity One software for the VersaDoc gel documentation system (Bio-Rad).
Statistical analyses.
Statistical analysis was performed by one-way ANOVA, followed by Tukey's test for pairwise comparisons. The p values were two sided and considered statistically significant at p < 0.05. All data are shown as mean ± SEM.
RESULTS
Circadian Profiles of Per1 in Vehicle- and TCDD-Treated Mouse Liver during the Light-Dark Cycle
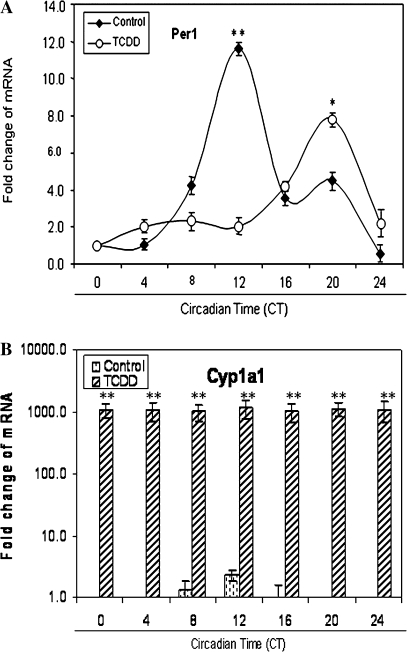
To investigate the effects of long-term AhR activation on the endogenous circadian rhythm of Per1 transcripts, as an indicator of molecular clock function, we determined the circadian profile of Per1 messenger RNA (mRNA) in the liver of vehicle-treated and TCDD-treated mice. Per1 was characterized by a rhythm with peak expression at CT12 in vehicle-treated controls (p < 0.001). After TCDD exposure, Per1 peak expression occurred at CT20 and peak transcript levels were attenuated by 33.3% (p < 0.05; Fig. 1A). As an indicator of AhR activation, we measured Cyp1a1, a primary target of the AhR signaling. Cyp1a1 was increased up to ∼1000-fold relative to vehicle controls (p < 0.001; Fig. 1B).
FIG. 1.
Circadian profiles of Per1 transcripts in the mouse liver during the light-dark cycle. After 2 weeks of entrainment in 12:12-h ligh-dark, mice were orally dosed with 1 μg/kg body weight of vehicle (corn oil) or TCDD during their lights-on period. Five days later, animals were placed in constant darkness to allow expression of their endogenous circadian rhythm. After 48 h in constant darkness, vehicle- and TCDD-treated mice were sacrificed and livers were collected every 4 h. Quantitative PCR was used to measure Per1 (A) and Cyp1a1 (B) mRNA. Results are shown as mean fold change relative to levels at CT0 ± SEM. n = 4–6 per group, *p < 0.05 TCDD-treated peak versus vehicle-treated peak; **p < 0.001 peak versus vehicle control.
Activation of AhR Represses Per1 Gene Expression
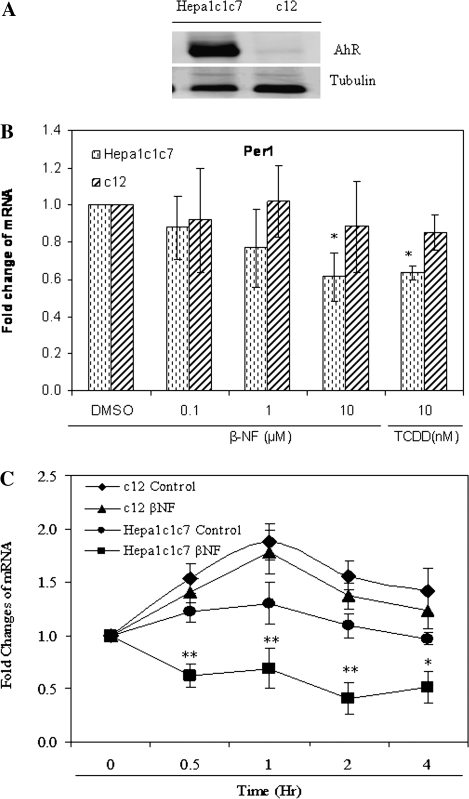
The effects of AhR activation on Per1 levels were examined in cell lines that express various levels of AhR to begin to elucidate potential mechanisms for the observed alteration of the liver's molecular clock in response to TCDD in vivo. Western blot results showed that basal AhR levels were very high in Hepa-1c1c7 but very low in c12 cells (Fig. 2A). Thus, these lines were used for all subsequent experiments.
FIG. 2.
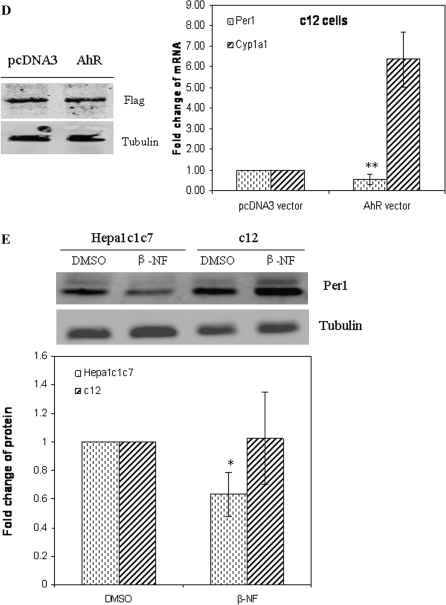
Activation of AhR inhibits Per1 gene expression. (A) Proteins from Hepa-1c1c7 and c12 cell lines were prepared and 30 μg of total cell lysates for each cell line was subjected to Western blot for AhR using α-tubulin as a loading control. (B) One hour after the addition of vehicle (DMSO, 0.01%), increasing concentrations (0.1, 1, and 10μM) of β-NF, or 10nM of TCDD, total RNA was harvested in Hepa-1c1c7 and c12 cells. Quantitative PCR was used to measure Per1 mRNA. (C) RNA was harvested at the various times (0, 0.5, 1, 2, and 4 h) after the addition of DMSO vehicle media or 10μM β-NF in Hepa-1c1c7 and c12 cells. (D) c12 cells were transfected by FLAG-pcDNA3 control vector or pcDNA3-FLAG-AhR vector and incubated for 24 h, and total protein and RNA was extracted. Quantitative PCR was used to measure Per1 and Cyp1a1 mRNA. Western blot was used to detect transfected FLAG-tagged protein. (E) Hepa-1c1c7 and c12 cells were treated with 10μM of β-NF for 4 h. Cell lysates were collected and Per1 protein was analyzed by Western blot using 60 μg of total cell lysates. Quantitative PCR and Western blot was used to measure Per1 mRNA and protein levels, respectively. Results are shown as mean fold change relative to the vehicle or 0-h level ± SEM. n = 3–4 per group, *p < 0.05 and **p < 0.01 versus the vehicle or control. Results are a representative of three to four independent experiments.
To examine the effect of AhR activation on Per1 gene expression, Hepa-1c1c7 and c12 cells were treated with vehicle, increasing concentrations of the AhR ligand β-NF for 1 h, or 10nM of TCDD for 4 h (Fig. 2B). Quantitative PCR results revealed that 10μM β-NF and 10nM TCDD treatment resulted in a 38.5 and 36.0% decrease of Per1 mRNA expression relative to vehicle in Hepa-1c1c7 (p < 0.05), respectively (Fig. 2B). Cyp1a1 transcript levels were significantly increased in the same samples, indicating that the effects of β-NF and TCDD were associated with typical signs of AhR activation (data not shown). In contrast, exposure of AhR-deficient c12 cells to either β-NF or TCDD did not alter levels of Per1 mRNA (Fig. 2B) and caused only a minor increase in Cyp1a1 (data not shown).
Subsequently, we examined the time required for AhR-dependent suppression of the Per1 transcript. Hepa-1c1c7 and c12 cells were treated with DMSO vehicle as control or 10μM of β-NF for 0, 0.5, 1, 2, and 4 h. The Per1 transcript was not significantly changed in Hepa-1c1c7 cells after vehicle control treatment but was significantly decreased at 0.5, 1, and 2 h (p < 0.01) and 4 h (p < 0.05) by β-NF. In AhR-deficient c12 cells, the Per1 transcript was strongly elevated by media change and was not significantly inhibited after β-NF exposure (Fig. 2C). Cyp1a1 levels were dramatically elevated in a time-dependent manner in β-NF-exposed Hepa-1c1c7 cells but only slightly elevated in c12 cells in the same samples (data not shown).
To show that AhR is activated upon transfection of AhR vector, and then inhibits the Per1 transcript without the addition of exogenous AhR agonist, c12 cells were transfected with a FLAG-tagged AhR vector. Western blot results showed that c12 expressed same level of FLAG after transfection with FLAG-pcDNA3 control or pcDNA3-FLAG-AhR vector (Fig. 2D). Transfected AhR vector significantly repressed Per1 transcript (p < 0.05) and dramatically elevated the Cyp1a1 transcript (p < 0.001; Fig. 2D). These data suggest that ectopically expressed AhR is activate, induces its principal target gene Cyp1a1, and represses Per1 transcript in the absence of a known exogenous ligand in c12 cells.
To test whether AhR activation also inhibits protein expression, we investigated Per1 protein levels after vehicle or 10μM β-NF treatment for 4 h in Hepa-1c1c7 and c12 cells (Fig. 2E). Western blot analysis demonstrated that β-NF treatment decreased Per1 protein expression in the Hepa-1c1c7 cells (p < 0.05). However, in AhR-deficient c12 cells, β-NF had no significant effect on the Per1 protein expression. Collectively, these results illustrate that inhibition of Per1 occurs downstream of AhR activation.
β-NF Results in Accumulation of Endogenous AhR in the Nucleus and Produces AhR Degradation
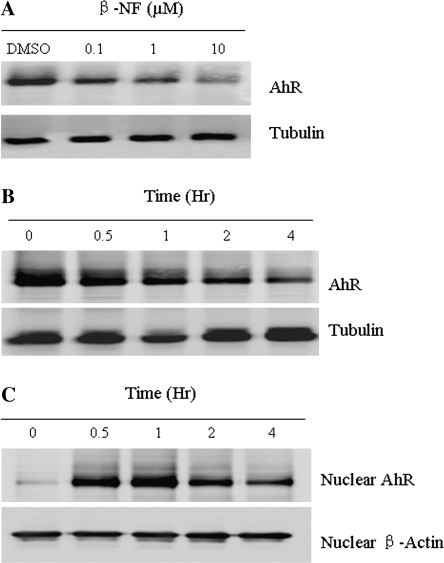
Previous studies have demonstrated that AhR accumulates in the nucleus of several cell lines within 60-90 min of TCDD exposure (Pollenz, 2002). The effect of β-NF on AhR protein levels was evaluated by Western blot analysis of total lysates. Treatment with 10μM of β-NF for 4 h in Hepa-1c1c7 cells resulted in a marked reduction of AhR (Figs. 3A and 3B). The nuclear fraction was analyzed by Western blot to investigate the effect of β-NF on AhR accumulation inside the nucleus. AhR protein became predominantly nuclear following 0.5 h of β-NF exposure (Fig. 3C), but nuclear protein decreased dramatically during continued 4 h of β-NF exposure. These results demonstrate a correlation between the effects of β-NF on AhR accumulation in the nucleus and time-dependent inhibition of Per1 expression.
FIG. 3.
β-NF induces AhR translocation and degradation. (A) Hepa-1c1c7 cells were treated with vehicle (DMSO, 0.01%) or increasing concentrations (0.1, 1, and 10μM) of β-NF for 4 h. (B) and (C) Hepa-1c1c7 cells were treated with 10μM of β-NF for various times (0, 0.5, 1, 2, and 4 h). Total cell lysates and nuclear lysates were prepared as detailed in the “Materials and Methods” section. Western blot was performed using 20 μg lysates of each sample to measure AhR protein. All blots were stained with α-tubulin or β-actin (nuclear lysates) antibody as a loading control. Results are a representative of three independent experiments.
β-NF Induces AhR-BMAL1 and Decreases CLOCK-BMAL1 Heterodimer Formation
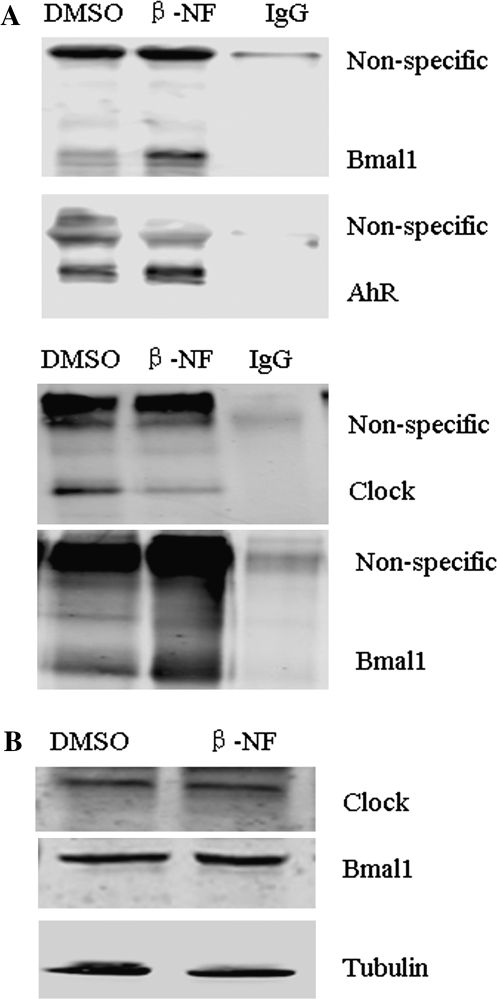
The molecular mechanism for AhR-dependent Per1 repression was investigated. CLOCK-BMAL1 heterodimers bind to Eboxes in the Per1 promoter to induce Per1. Previous studies indicate that AhR interacts with BMAL1 in vitro (Hogenesch et al., 1997). To determine if AhR can interact with CLOCK-BMAL1 in hepatoma cells, co-IP was performed. Hepa-1c1c7 cells were exposed to vehicle or β-NF (10μM) for 1.5 h. Cell lysates were collected and immunoprecipitated by specific anti-AhR or anti-BMAL1 antibody, and then, associated proteins were detected by Western blot. As shown in Figure 4A top two blots, BMAL1 was present in protein complexes pulled down with the AhR-specific antibody after β-NF exposure. Furthermore, the bottom two blots in Figure 4A show that CLOCK immunoreactivity was decreased in protein complexes immunoprecipitated with the BMAL1 antibody after β-NF exposure. Western blot (Fig. 4B) indicated that the amounts of CLOCK and BMAL1 proteins were not changed after β-NF (10μM) exposure for 1.5 h in Hepa-1c1c7 cells. These results clearly demonstrate that β-NF promotes the formation of complexes that include both AhR and BMAL1 in Hepa-1c1c7 cells; furthermore, β-NF decreases the amount of CLOCK-BMAL1 complexes without altering the expression of CLOCK and BMAL1 proteins. AhR also interacts with BMAL1 in TCDD-treated mouse ovaries (data not shown).
FIG. 4.
β-NF induces AhR-BMAL1 and decreases CLOCK-BMAL1 associations. Hepa-1c1c7 cells were treated with vehicle (DMSO, 0.01%) or 10μM of β-NF for 1.5 h. (A) A total of 500 μg of vehicle or β-NF-treated cell lysates was used for co-IP. Two micrograms of AhR (goat) antibody or BMAL1 (rabbit) was used for precipitation of protein. IgG was a negative control. Protein complexes were collected using protein A/G subjected to Western blot. Lysates in the top two blots were precipitated with anti-AhR (goat) primary antibody and then probed with anti-BMAL1 primary antibody (rabbit, 1:200, top blot) or with anti-AhR (rabbit, 1 μg/ml, second blot) primary antibody, respectively. Lysates shown in the bottom two blots were precipitated with anti-BMAL1 (rabbit) primary antibody and probed with anti-CLOCK (goat, 1:500, third blot from top) primary antibody, or with anti-BMAL1 (rabbit, 2 μg/ml, fourth blot from top) primary antibody, respectively. (B) Cell lysates of vehicle or β-NF treated for 1.5 h were subjected to Western blot analysis for CLOCK, BMAL1, and α-tubulin (loading control). Results are representative of three independent experiments.
AhR Transfection Represses CLOCK-BMAL1–Induced Activation of Per1 Promoter
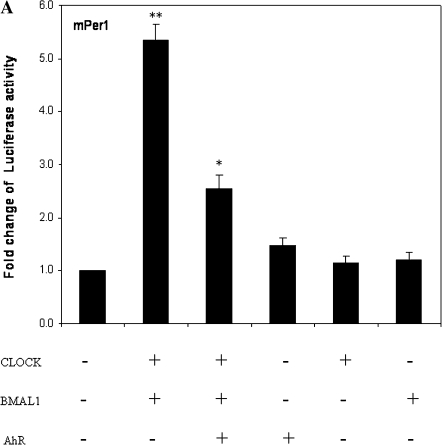
Because activated AhR can form complexes that include BMAL1, we explored whether this interaction affects CLOCK-BMAL1–regulated Per1 promoter activity. Cotransfection experiments were used to test the hypothesis that transfection of AhR may repress CLOCK-BMAL1 activity and thus inhibit the Per1 promoter. AhR-deficient c12 cells were cotransfected with vectors containing Per1-luc, CLOCK-BMAL1, CLOCK, BMAL1, and/or AhR. As expected, luciferase assay results show that CLOCK and BMAL1 together induced 5.6-fold increase in transcriptional activity of Per1 (p < 0.001; Fig. 5). Cotransfection of AhR protein inhibited the CLOCK-BMAL1–induced increase in transcriptional activity by ∼49% (p < 0.05), whereas the addition of AhR without CLOCK and BMAL1, or transfection of either CLOCK or BMAL1 alone did not alter the basal levels of Per1 (Fig. 5).
FIG. 5.
Activation of AhR represses CLOCK-BMAL1–induced transactivation. c12 cells were transfected with a reporter construct mPer1-luc (10 ng), CLOCK-BMAL1 (250 ng each), AhR construct (250 ng), CLOCK (250 ng), BMAL1 (250 ng), β-gal (10 ng), or the empty vectors. After 24 h, cells were harvested for luciferase assay. The relative firefly luciferase of mPer1-luc from analysis for individual groups was normalized against β-gal luciferase. Results are shown as mean fold change relative to the untransfected CLOCK-BMAL1 and AhR relative firefly luciferase level ± SEM. n = 3–4 per group, *p < 0.05 versus cotransfected CLOCK and BMAL1 vector group; **p < 0.001 versus cotransfected empty vector group.
Activation of AhR Displaces CLOCK from the Per1 Promoter and Results in Per1 Inhibition
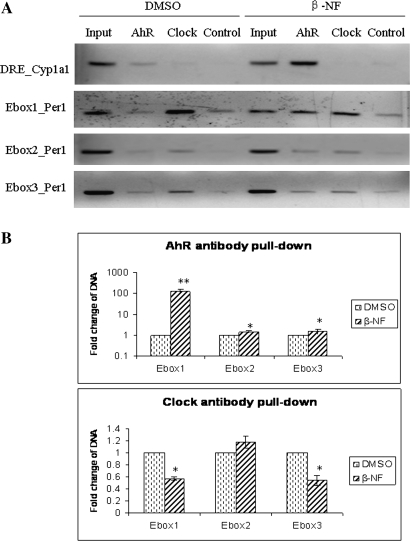
The transient transfection experiment reported above suggests the possibility that AhR protein may repress Per1 gene expression by blocking CLOCK-BMAL1 heterodimer binding to Eboxes. ChIP was used to analyze the interaction between CLOCK-BMAL1, AhR, and Per1 promoter to test this hypothesis. Hepa-1c1c7 cells were treated with the vehicle DMSO or 10μM of β-NF for 1.5 h, and native chromatin was extracted from nuclear lysates. IPs were carried out using AhR, BMAL1, and CLOCK antibodies. Controls containing no antibody were included to verify the specificity of precipitation. Figure 6 demonstrates that β-NF treatment increased AhR binding to Ebox1, Ebox2, and Ebox3 in the Per1 promoter by 127.4- (p < 0.001), 1.5- (p < 0.05), and 1.6-fold (p < 0.05), respectively. β-NF treatment reduced CLOCK binding to Ebox1 (p < 0.001) and Ebox3 (p < 0.05) of the Per1 promoter by 50% but had no significant effect on CLOCK binding to Ebox2. BMAL1 binding to Eboxes (Ebox1, Ebox2, and Ebox3) in the Per1 promoter was unaffected by β-NF exposure (data not shown). Cyp1a1 was used as a positive control; AhR binding to the DRE in the Cyp1a1 promoter was increased in the β-NF-treated group.
FIG. 6.
Recruitment of AhR to the Per1 promoter results in the inhibition of Per1. Hepa-1c1c7 cells were treated with either vehicle (DMSO, 0.01%) or 10μM β-NF for 1.5 h. Cells were harvested and native chromatin from nuclear lysates was prepared. IPs were carried out using AhR and CLOCK antibodies. A control (no antibody) was included to confirm the specificity of the precipitation. Purified genomic DNA was subjected to PCR amplifications with gene-specific primers. Input lane represents promoter-specific amplification of 0.2% of the total chromatin sample. (A) Products were separated and analyzed using a 2% agarose gel. (B) Densitometric analysis of AhR and CLOCK. Results are shown as mean fold change relative to vehicle level ± SEM. n = 3–4 group, *p < 0.05 and **p < 0.001 versus vehicle. Results are a representative of three independent experiments.
DISCUSSION
In the present study, we report that TCDD treatment markedly decreases the peak expression levels and causes an altered circadian expression pattern of Per1 transcripts in mouse liver (Fig. 1). Thus, TCDD alters Per1 gene expression in peripheral tissues that may lead to dysfunction of peripheral circadian rhythms. We used Hepa-1c1c7 hepatoma cells, which express high levels of AhR, and their c12 derivative, which expresses low levels of AhR, to test the hypothesis that the repression of Per1 observed in vivo after exposure to dioxin is dependent on AhR. The AhR agonist, β-NF, strongly inhibits Per1 mRNA and protein in Hepa-1c1c7 but not in c12 cells (Figs. 2B and 2E). These results suggest that activation of AhR strongly inhibits Per1 gene expression, which is a key component of the molecular circadian clockworks. These data represent the first evidence of this novel mechanism of action for the AhR.
The time required for Per1 repression after treatment with AhR agonists is consistent with the proposed model of AhR activation, nuclear translocation, and degradation of AhR. Upon ligand binding, AhR translocates into the nucleus and ultimately undergoes degradation (Pollenz, 2002). Serum can increase the Per1 transcript, as indicated by use of 7.5% BGS media as control in the time course for β-NF treatment. The Per1 transcript was moderately increased in control due to the presence of serum, but β-NF treatment significantly repressed the Per1 transcript at 0.5, 1, and 2 h after treatment; Per1 transcript levels recovered partly at 4 h in Hepa-1c1c7 cells (Fig. 2C). In the AhR-deficient c12 cells, Per1 was significantly elevated by 7.5% BGS media change and was not significantly inhibited after β-NF exposure (Fig. 2C). These results suggest that Per1 can be easily induced in AhR-deficient cells and that AhR is responsible for the inhibitory effects of β-NF. Evident at 0.5 h after β-NF treatment, continuous repression of the Per1 transcript was observed until 2 h of treatment, but the repression was attenuated at 4 h of treatment. Thus, the effects of β-NF on Per1 likely result from transient nuclear accumulation followed by degradation of nuclear AhR. AhR protein rapidly increased in nuclear extracts at 0.5 h of exposure but markedly declined at 4 h of exposure (Fig. 3C), consistent with a previous study that demonstrated similar changes in AhR after treatment of cultured cells with TCDD (Pollenz, 2002). These results provide strong evidence that β-NF represses Per1 through the AhR signaling pathway.
Previous co-IP studies have reported that AhR associates with BMAL1 in vitro, but yeast two-hybrid assays did not detect a direct interaction between AhR and BMAL1 in vivo (Hogenesch et al., 1997 1998). In the present study, co-IP experiments demonstrated the presence of BMAL1 in protein complexes pulled down with an AhR-specific antibody in Hepa-1c1c7 cells after β-NF treatment (Fig. 4). AhR co-immunoprecipitated BMAL1 in the mouse ovary (data not shown) and the TCDD treatment increases this interaction. There are several possible explanations for these data. First, BMAL1 co-immunoprecipitates with AhR in vitro, indicating that the two proteins have suitable domain structures to allow such interaction. Second, BMAL1 is similar in amino acid sequence to the known AhR dimerization partner, ARNT, suggesting that interaction might be possible. Third, the lack of interaction in the yeast two-hybrid system may not accurately reflect interactions in the mammalian system.
The AhR principal target gene Cyp1a1 was dramatically induced after transfection with AhR vector in c12 cells. Per1 also was repressed after transfection with the AhR vector in c12 cells (Fig. 2D). Ectopic expression of AhR demonstrated that AhR can inhibit CLOCK-BMAL1 transcriptional activity in c12 cells. However, transfection of AhR alone did not change basal levels of Per1-luc (Fig. 5). These results suggest that ectopically expressed AhR can be activated and can repress the Per1 transcript in the absence of an exogenous ligand, which is in accordance with the report that ectopically expressed AhR activates and localizes to the nucleus in the absence of an exogenous ligand (Chang and Puga, 1998). Together with the co-IP data, these results suggest that AhR may repress Per1 through its interaction with BMAL1 and, consequently, may disrupt activity of CLOCK-BMAL1 and/or other coactivators.
Although the classical signaling pathway attributed to AhR consists of heterodimerization with ARNT and binding to DREs in dioxin-responsive genes, more recent studies highlight alternative strategies for AhR signaling. AhR reportedly binds to E2F-dependent gene promoters through displacement of the coactivator p300 to repress S phase–specific gene expression (Marlowe et al., 2004). Similarly, alternative dimerization partners for BMAL1 have been identified. For example, the coactivator CREB binding protein can interact with BMAL1 (Takahata et al., 2000), whereas p300 can interact with CLOCK to regulate Per1 transactivation (Etchegaray et al., 2003). In this report, β-NF treatment increased AhR binding to Eboxes of the Per1 promoter and decreased CLOCK binding to Ebox1 and Ebox3 (Fig. 6). However, β-NF treatment did not change p300 binding to Eboxes (data not shown). In addition, β-NF treatment decreased CLOCK-BMAL1 interactions. These results suggest at least four possible mechanisms for the repression of Per1 by AhR. First, AhR may displace CLOCK from the CLOCK-BMAL1 heterodimer, thereby prevent binding to Ebox1 and Ebox3. Second, AhR may bind with the CLOCK-BMAL1 heterodimer at Ebox2 and disrupt its activity, without necessarily removing CLOCK from Eboxes. Third, AhR may displace other coactivators (excluding p300) from Eboxes. Fourth, AhR may also recruit a corepressor to the Ebox. Future experiments will explore the interactions of AhR with other potential coactivators and/or corepressors.
Circadian rhythms are generated by molecular “clocks” present in virtually every cell. Peripheral clocks are synchronized to each other and to solar time by a “master clock” in the SCN (Asai et al., 2001; Mendoza et al., 2008; Mizuyachi et al., 2002). Shift work, which likely acts to disrupt the clock in human populations, predisposes the affected people to numerous health risks, including increased risk for breast and prostate cancers, increased risk for cardiovascular disease, and increased incidence of type 2 diabetes (Froy, 2007; Turek et al., 2005). Furthermore, clock disruption in mouse models has suggested a link between disruption of Per genes and cancer; Per gene–deficient mice are more susceptible to cancer, developing tumors earlier and more often than wild-type mice (Fu et al., 2002; Lee, 2005); Per1, Per2, and Per3 are downregulated in human breast cancer cells, and overexpression of Per1 in human cancer cells retards growth and increases sensitivity to ionizing radiation (Chen et al., 2005; Gery et al., 2006, 2007).
Long-term follow-up studies of dioxin exposure have revealed that dioxins can contribute to the development of cancers, metabolic disease, and other related diseases (Arisawa et al., 2005; Bertazzi et al., 1998; Bock, 1994; Henriksen et al., 1997; Larsen, 2006; Mishima et al., 2005; Roybal et al., 2007; Zambon et al., 2007). Although the mechanisms behind these metabolic and tumor-generating effects of dioxins have not been elucidated, circadian clock disruption in the liver by dioxins, as suggested by the current investigation, may be important. The current report clearly shows that AhR activation represses Per1 gene expression and alters circadian expression of Per1. These findings suggest heretofore unexplored mechanisms by which environmental pollutants may contribute to the development of metabolic disease and carcinogenesis through disrupting the circadian rhythm and repression of clock genes.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institute of Environmental Health Sciences grant (ES012948); American Cancer Society grant to S.A.T.
Supplementary Material
Acknowledgments
The authors express their appreciation to K. Bottum and S. Karmarkar for assistance with manuscript writing.
References
- Arisawa K, Takeda H, Mikasa H. Background exposure to PCDDs/PCDFs/PCBs and its potential health effects: a review of epidemiologic studies. J. Med. Invest. 2005;52:10–21. doi: 10.2152/jmi.52.10. [DOI] [PubMed] [Google Scholar]
- Asai M, Yamaguchi S, Isejima H, Jonouchi M, Moriya T, Shibata S, Kobayashi M, Okamura H. Visualization of mPer1 transcription in vitro: NMDA induces a rapid phase shift of mPer1 gene in cultured SCN. Curr. Biol. 2001;11:1524–1527. doi: 10.1016/s0960-9822(01)00445-6. [DOI] [PubMed] [Google Scholar]
- Bertazzi PA, Bernucci I, Brambilla G, Consonni D, Pesatori AC. The Seveso studies on early and long-term effects of dioxin exposure: a review. Environ. Health Perspect. 1998;106(Suppl. 2):625–633. doi: 10.1289/ehp.98106625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittman EL, Doherty L, Huang L, Paroskie A. Period gene expression in mouse endocrine tissues. Am. J. Physiol. 2003;285:R561–R569. doi: 10.1152/ajpregu.00783.2002. [DOI] [PubMed] [Google Scholar]
- Bock KW. Aryl hydrocarbon or dioxin receptor: biologic and toxic responses. Rev. Physiol. Biochem. Pharmacol. 1994;125:1–42. doi: 10.1007/BFb0030908. [DOI] [PubMed] [Google Scholar]
- Boivin DB, James FO, Wu A, Cho-Park PF, Xiong H, Sun ZS. Circadian clock genes oscillate in human peripheral blood mononuclear cells. Blood. 2003;102:4143–4145. doi: 10.1182/blood-2003-03-0779. [DOI] [PubMed] [Google Scholar]
- Canaple L, Rambaud J, Dkhissi-Benyahya O, Rayet B, Tan NS, Michalik L, Delaunay F, Wahli W, Laudet V. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor alpha defines a novel positive feedback loop in the rodent liver circadian clock. Mol. Endocrinol. 2006;20:1715–1727. doi: 10.1210/me.2006-0052. [DOI] [PubMed] [Google Scholar]
- Cermakian N, Boivin DB. A molecular perspective of human circadian rhythm disorders. Brain Res. 2003;42:204–220. doi: 10.1016/s0165-0173(03)00171-1. [DOI] [PubMed] [Google Scholar]
- Chang CY, Puga A. Constitutive activation of the aromatic hydrocarbon receptor. Mol. Cell. Biol. 1998;18:525–535. doi: 10.1128/mcb.18.1.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ, Chang JG. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26:1241–1246. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- Froy O. The relationship between nutrition and circadian rhythms in mammals. Front Neuroendocrinol. 2007;28:61–71. doi: 10.1016/j.yfrne.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- Garrett RW, Gasiewicz TA. The aryl hydrocarbon receptor agonist 2,3,7,8-tetrachlorodibenzo-p-dioxin alters the circadian rhythms, quiescence, and expression of clock genes in murine hematopoietic stem and progenitor cells. Mol. Pharmacol. 2006;69:2076–2083. doi: 10.1124/mol.105.021006. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol. Cell. 2006;22:375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- Gery S, Komatsu N, Kawamata N, Miller CW, Desmond J, Virk RK, Marchevsky A, McKenna R, Taguchi H, Koeffler HP. Epigenetic silencing of the candidate tumor suppressor gene Per1 in non-small cell lung cancer. Clin. Cancer Res. 2007;13:1399–1404. doi: 10.1158/1078-0432.CCR-06-1730. [DOI] [PubMed] [Google Scholar]
- Griffin EA, Jr, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- Henriksen GL, Ketchum NS, Michalek JE, Swaby JA. Serum dioxin and diabetes mellitus in veterans of Operation Ranch Hand. Epidemiology. 1997;8:252–258. doi: 10.1097/00001648-199705000-00005. [DOI] [PubMed] [Google Scholar]
- Hickok J, Tischkau S. In vivo circadian rhythms in gonadotropin releasing hormone neurons. Neuroendocrinology. 2010;91:110–120. doi: 10.1159/000243163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hida A, Koike N, Hirose M, Hattori M, Sakaki Y, Tei H. The human and mouse Period1 genes: five well-conserved Eboxes additively contribute to the enhancement of mPer1 transcription. Genomics. 2000;65:224–233. doi: 10.1006/geno.2000.6166. [DOI] [PubMed] [Google Scholar]
- Hogenesch JB, Chan WK, Jackiw VH, Brown RC, Gu Y-Z, Pray-Grant M, Perdew GH, Bradfield CA. Characterization of a subset of the basic-helix-loop-helix-PAS superfamily that interacts with components of the dioxin signaling pathway. J. Biol. Chem. 1997;272:8581–8593. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl Acad. Sci. U.S.A. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Hirayama J, Kobayashi Y, Todo T. Zebrafish CRY represses transcription mediated by CLOCK-BMAL heterodimer without inhibiting its binding to DNA. Genes Cells. 2002;7:1073–1086. doi: 10.1046/j.1365-2443.2002.00579.x. [DOI] [PubMed] [Google Scholar]
- Karman BN, Tischkau SA. Circadian clock gene expression in the ovary: effects of luteinizing hormone. Biol. Reprod. 2006;75:624–632. doi: 10.1095/biolreprod.106.050732. [DOI] [PubMed] [Google Scholar]
- Kojima S, Hirose M, Tokunaga K, Sakaki Y, Tei H. Structural and functional analysis of 3’ untranslated region of mouse Period1 mRNA. Biochem. Biophys. Res. Commun. 2003;301:1–7. doi: 10.1016/s0006-291x(02)02938-8. [DOI] [PubMed] [Google Scholar]
- Larsen JC. Risk assessments of polychlorinated dibenzo-p-dioxins, polychlorinated dibenzofurans, and dioxin-like polychlorinated biphenyls in food. Mol. Nutr. Food Res. 2006;50:885–896. doi: 10.1002/mnfr.200500247. [DOI] [PubMed] [Google Scholar]
- Lee CC. The circadian clock and tumor suppression by mammalian period genes. Methods Enzymol. 2005;393:852–861. doi: 10.1016/S0076-6879(05)93045-0. [DOI] [PubMed] [Google Scholar]
- Lucier GW, Tritscher A, Goldsworthy T, Foley J, Clark G, Goldstein J, Maronpot R. Ovarian hormones enhance 2,3,7,8-tetrachlorodibenzo-p-dioxin-mediated increases in cell proliferation and preneoplastic foci in a two-stage model for rat hepatocarcinogenesis. Cancer Res. 1991;51:1391–1397. [PubMed] [Google Scholar]
- Marlowe JL, Knudsen ES, Schwemberger S, Puga A. The aryl hydrocarbon receptor displaces p300 from E2F-dependent promoters and represses S phase-specific gene expression. J. Biol. Chem. 2004;279:29013–29022. doi: 10.1074/jbc.M404315200. [DOI] [PubMed] [Google Scholar]
- Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- Mendoza J, Pevet P, Challet E. High-fat feeding alters the clock synchronization to light. J. Physiol. 2008;586:5901–5910. doi: 10.1113/jphysiol.2008.159566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima K, Tozawa T, Satoh K, Saitoh H, Mishima Y. The 3111T/C polymorphism of hClock is associated with evening preference and delayed sleep timing in a Japanese population sample. Am J Med. Genet. 2005;133:101–104. doi: 10.1002/ajmg.b.30110. [DOI] [PubMed] [Google Scholar]
- Mizuyachi K, Son D-S, Rozman KK, Terranova PF. Alteration in ovarian gene expression in response to 2,3,7,8-tetrachlorodibenzo-p-dioxin: reduction of cyclooxygenase-2 in the blockage of ovulation. Reprod. Toxicol. 2002;16:299–307. doi: 10.1016/s0890-6238(02)00024-2. [DOI] [PubMed] [Google Scholar]
- Moore RY, Eichler VB. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 1972;42:201–206. doi: 10.1016/0006-8993(72)90054-6. [DOI] [PubMed] [Google Scholar]
- Mukai M, Lin TM, Peterson RE, Cooke PS, Tischkau SA. Behavioral rhythmicity of mice lacking AhR and attenuation of light-induced phase shift by 2,3,7,8-tetrachlorodibenzo-p-dioxin. J. Biol. Rhythms. 2008;23:200–210. doi: 10.1177/0748730408316022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse Y, Oh-hashi K, Iijima N, Naruse M, Yoshioka H, Tanaka M. Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. Mol. Cell. Biol. 2004;24:6278–6287. doi: 10.1128/MCB.24.14.6278-6287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Miyazaki K, Kadota K, Kikuno R, Nagase T, Atsumi G, Ohkura N, Azama T, Mesaki M, Yukimasa S, et al. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J. Biol. Chem. 2003;278:41519–41527. doi: 10.1074/jbc.M304564200. [DOI] [PubMed] [Google Scholar]
- Pollenz RS. The mechanism of AH receptor protein down-regulation (degradation) and its impact on AH receptor-mediated gene regulation. Chem. Biol Interact. 2002;141:41–61. doi: 10.1016/s0009-2797(02)00065-0. [DOI] [PubMed] [Google Scholar]
- Qu X, Metz RP, Porter WW, Cassone VM, Earnest DJ. Disruption of period gene expression alters the inductive effects of dioxin on the AhR signaling pathway in the mouse liver. Toxicol. Applied Pharmacol. 2009;234:370–377. doi: 10.1016/j.taap.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadoss P, Perdew GH. The transactivation domain of the Ah receptor is a key determinant of cellular localization and ligand-independent nucleocytoplasmic shuttling properties. Biochemistry. 2005;44:11148–11159. doi: 10.1021/bi050948b. [DOI] [PubMed] [Google Scholar]
- Richardson V, Santostefano M, LS B. Daily cycle of bHLH-PAS proteins, Ah receptor and Arnt, in multiple tissues of female Sprague-Dawley rats. Biochem. Biophys. Res. Commun. 1998;252:225–231. doi: 10.1006/bbrc.1998.9634. [DOI] [PubMed] [Google Scholar]
- Roybal K, Theobold D, Graham A, Dinieri J, Russo S, Krishnan V, Chakravarty S, Peevey J, Oehrlein N, Birnbaum S, et al. Mania-like behavior induced by disruption of Clock. Proc. Natl Acad. Sci. U.S.A. 2007;104:6406–6411. doi: 10.1073/pnas.0609625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JV, Bradfield CA. Ah receptor signaling pathways. Ann. Rev. Cell Dev. Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomoter activity of rats are eliminated by hypothalamic lesions. Proc. Natl Acad. Sci. U.S.A. 1972;69:1583–1586. doi: 10.1073/pnas.69.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson HI. DNA binding and protein interactions of the AHR/ARNT heterodimer that facilitate gene activation. Chem. Biol. Interact. 2002;141:63–76. doi: 10.1016/s0009-2797(02)00066-2. [DOI] [PubMed] [Google Scholar]
- Takahata S, Ozaki T, Mimura J, Kikuchi Y, Sogawa K, Fujii-Kuriyama Y. Transactivation mechanisms of mouse clock transcription factors, mClock and mArnt3. Genes Cells. 2000;5:739–747. doi: 10.1046/j.1365-2443.2000.00363.x. [DOI] [PubMed] [Google Scholar]
- Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci U S A. 2002;99:7728–7733. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann AS, Farnham PJ. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods. 2002;26:37–47. doi: 10.1016/S1046-2023(02)00006-3. [DOI] [PubMed] [Google Scholar]
- Zambon P, Ricci P, Bovo E, Casula A, Gattolin M, Fiore AR, Chiosi F, Guzzinati S. Sarcoma risk and dioxin emissions from incinerators and industrial plants: a population-based case-control study (Italy) Environ. Health. 2007;6:19. doi: 10.1186/1476-069X-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.