Abstract
Background
Fosfomycin is a cell wall inhibitor used efficiently to treat uncomplicated urinary tract and gastrointestinal infections. A very convenient feature of fosfomycin, among others, is that although the expected frequency of resistant mutants is high, the biological cost associated with mutation impedes an effective growth rate, and bacteria cannot offset the obstacles posed by host defenses or compete with sensitive bacteria. Due to the current scarcity of new antibiotics, fosfomycin has been proposed as an alternative treatment for other infections caused by a wide variety of bacteria, particularly Pseudomonas aeruginosa. However, whether fosfomycin resistance in P. aeruginosa provides a fitness cost still remains unknown.
Principal Findings
We herein present experimental evidence to show that fosfomycin resistance cannot only emerge easily during treatment, but that it is also cost-free for P. aeruginosa. We also tested if, as has been reported for other species such as Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis, fosfomycin resistant strains are somewhat compromised in their virulence. As concerns colonization, persistence, lung damage, and lethality, we found no differences between the fosfomycin resistant mutant and its sensitive parental strain. The probability of acquisition in vitro of resistance to the combination of fosfomycin with other antibiotics (tobramycin and imipenem) has also been studied. While the combination of fosfomycin with tobramycin makes improbable the emergence of resistance to both antibiotics when administered together, the combination of fosfomycin plus imipenem does not avoid the appearance of mutants resistant to both antibiotics.
Conclusions
We have reached the conclusion that the use of fosfomycin for P. aeruginosa infections, even in combined therapy, might not be as promising as expected. This study should encourage the scientific community to assess the in vivo cost of resistance for specific antibiotic-bacterial species combinations, and therefore avoid reaching universal conclusions from single model organisms.
Introduction
Amongst a number of old antibiotics being used once more as appealing alternatives for the treatment of different infections, fosfomycin (Fos) displays some features that place it as a very promising compound [1]. Although its use has traditionally been focused on urinary tract infection pathogens, fosfomycin is a broad-spectrum bactericidal antibiotic, that is active against both gram-positive and gram-negative bacteria [2]. It has shown almost no toxicity and allergenicity in humans [3]. It displays little cross-resistance with other antibiotics, probably because it is chemically unrelated to any other known antimicrobial agent [2], [3]. Also, several studies have pointed out the synergistic effect of fosfomycin when used in combination with other antimicrobials [4], [5], [6], [7]. Fosfomycin treatments have shown a relatively low proneness of resistant mutants to persist in vivo, leading to a good effectiveness in the therapy, at least against Escherichia coli [8]. However, recently the acquisition of fosfomycin resistance in a previously circulating CTX-M-15-producing E. coli O25b-ST131-phylogroup B2 strain has been reported. This apparently occurred when the use of fosfomycin was increased by 50% [9].
The emergence of antibiotic resistance in a bacterial population is shaped by several factors, of which the mutation rate to resistance and the fitness cost of the mutants have been considered critical [10]. If resistance implies a considerable cost for the bacteria, their growth rate would not be enough to offset the wash out dynamic imposed by the diverse body fluids, the killing mediated by the immune system, or it might allow the more fit susceptible bacteria to out-compete the resistant ones once the antibiotic is removed [10].
The trend of resistant bacteria to have a competitive disadvantage in a untreated environment is related to the precise mechanism of the resistance in question. Numerous types of resistance achieved by chromosomal mutations are caused by target alterations [11]. In these cases a consistent fitness cost may appear if the resistant target displays a somewhat suboptimal functionality, as has been shown, for example, with some mutations in the gene rpsL conferring streptomycin in bacteria such as E. coli or Salmonella spp. [12]. Another prominent way to attain resistance is determined by accessory elements, where resistance is afforded by enzymes or pumps that inactivate or remove the antimicrobial from the cell [13]. This type of resistance may also give rise to fitness cost, as the replication and activity of the elements themselves could involve some metabolic expenditures [11].
In the specific case of fosfomycin, resistance is mainly acquired by reducing the cell uptake of the drug [14], [15], [16]. This kind of resistance has been widely shown to compromise E. coli at a slower rate of growth when compared to that of the sensitive parental strains [8], [17], [18], probably because most mutations disturb carbon metabolism [8]. In addition, and very conveniently for humans, these mutants also displayed a reduced virulence represented by lower adhesion to epithelial cells [18], [19], [20]. The data for other species are scarce, but it has been reported that Klebsiella pneumoniae and Proteus mirabilis, common bacteria also causing uncomplicated urinary tract infections, follow the same trend as E. coli [18]. Surprisingly, a new resistance mechanism based on amino acid substitutions in MurA, the target protein of fosfomycin, has recently been reported. Unfortunately, the authors have not estimated the cost associated to such a modification [21].
Recently it has been reported that the use of fosfomycin be extended in order to treat pathogens like Pseudomonas aeruginosa [2], where the emergence of antibiotic resistance has led to a unpleasant scarcity of treatments [22]. P. aeruginosa is one of the leading nosocomial pathogens worldwide. The most severe infections produced by P. aeruginosa occur in Intensive Care Unit patients, those suffering from chronic respiratory diseases and immunocompromised individuals [23]. Considering the example of E. coli, it seems reasonable to expect that fosfomycin might serve as a genuine alternative to treat infections caused by multidrug resistant P. aeruginosa. This possibility has been substantiated by published data showing that fosfomycin, furthermore, could reveal a particular effectiveness when used in combined therapy with other well-known antipseudomonal agents [2], [4], [5], [6], [7].
In a previous work, we reported that P. aeruginosa has a high mutation frequency to fosfomycin resistance in vitro [24]. These observations in vitro could undermine the good expectations created about fosfomycin. However, sometimes the data generated in vitro do not agree with the results attained in vivo [10], [25]. Here, we undertook the assessment of this disturbing possibility, monitoring the emergence of fosfomycin resistant (Fos-R) mutants in an in vivo murine model. We then checked the fitness cost caused by the inactivation of glpT in one insertion (null) mutant, as well as its virulence relative to the sensitive parental strain. Our results indicate a deviation from the E. coli model, disproving any connection between fosfomycin resistance, fitness cost or loss of any trait conferring bacterial virulence in P. aeruginosa.
Methods
Ethics Statement
All animal experiments were approved by the animal ethics committee of the University of the Balearic Islands, Spain. All animals were handled and housed in strict accordance with guidelines from the University of the Balearic Islands.
Bacteria and media
The P. aeruginosa PA14 and its derivative mutants mutS::MAR2xT7 and glpT::MAR2xT7 were kindly provided by Dr. N. T. Liberati [26]. All the P. aeruginosa strains were cultured in Luria-Bertani (LB) or Mueller-Hilton at 37°C, and gentamycin (10 µg/ml) was added when appropriate. For the biofilm assay, LB and FBA minimal media were used [27].
P. aeruginosa lung infection mouse model
The murine model of P. aeruginosa lung infection was established by intranasal inoculation of bacteria [28], [29], [30]. Briefly, for the preparation of the inocula, bacteria (P. aeruginosa strains PA14, PA14mutS::MAR2xT7, PA14glpT::MAR2xT7 or a 1∶1 mix of PA14/PA14glpT::MAR2xT7) were grown in LB to 0.5 OD 600 nm, centrifuged at 10,000 g for two minutes and diluted 10-fold in saline solution (1 and 5-fold dilutions in saline solution were also tested during the standardization of the model). Finally, serial 1/10 dilutions of bacterial suspensions were plated in MHA to estimate bacterial viability counts.
Female C57BL/6J mice were used. All animals weighed 20 to 25 g and were provided by Harlan Ibérica, S. L. The animals were specific pathogen free and nourished ad libitum with sterile water and food. Before inoculation, the mice were anesthetized by intraperitoneal injection of ketamine and xylazine (100 mg and 10 mg per kg of body weight respectively). Forty microliters of the corresponding bacterial suspension, containing the indicated bacterial counts, were inoculated intranasally by pippeting directly in nasal orifices.
In addition to the evaluation of associated mortality and lung bacterial load, lung histopathology studies were performed. Briefly, lungs were fixed in 10% formalin, embedded in paraffin, sectioned (4 to 6 µm), and stained with hematoxylin and eosin. Lung inflammation was then blindly scored by an expert pathologist using the criteria of Johansen et al. [31] (1, normal; 2, mild focal inflammation; 3, moderate to severe focal inflammation with areas of normal tissue; 4, severe inflammation to necrosis and severe inflammation throughout the lung).
Pulmonary infection survival model
Three groups of 10 mice each per strain were inoculated with two different bacterial inocula of 1×106 and 5×105 of bacteria per animal of strains PA14 and PA14glpT::MAR2xT7 approximately, and mortality was recorded for 7 days. A Kaplan-Meier test was performed to determine the difference between mortality curves in the survival experiments. P values less than 0.05 were considered to be statistically significant.
Estimation of spontaneous Fos-R mutation rates
For an estimation of the in vitro spontaneous mutation rate of PA14 and its derivative mutant mutS::MAR2xT7 strains, approximately 103 cells from overnight cultures were inoculated in 8 tubes, each containing 1 ml of LB, and subsequently incubated at 37°C with strong shaking for 16 h. Aliquots from successive dilutions or concentrated cultures were plated onto Mueller-Hinton agar (MHA) plates with fosfomycin (128 µg/ml). Additionally, appropriate dilutions with no antibiotic were plated onto MHA to estimate viability. The number of colonies growing after 24 h of incubation were determined. The mutation rate was then estimated using the Jones median estimator [32], and 95% confidence intervals were calculated using JONATOR (v0.1), a software developed in gfortran which implements an algorithm described elsewhere [33]. This software is free and available upon request. The same procedure was followed for an estimation of the mutation rate in vivo, but in this case, 8 mice were inoculated intranasally with approximately 5×105 bacteria per animal from log-phase cultures of PA14 and the mutS-deficient strain. Lung homogenates were then obtained 48 hours postinfection, as described elsewhere. Proper dilutions of the lung homogenate were plated, as described for the procedure in vitro.
Estimation of the frequencies of spontaneous mutants resistant to fosfomycin, tobramycin and imipenem
An estimation of the frequencies of spontaneous mutants resistant to fosfomycin (128 µg/ml), tobramycin (4 µg/ml), imipenem (4 µg/ml) and the combinations of tobramycin plus fosfomycin (4 and 128 µg/ml, respectively) and imipenem plus fosfomycin (4 and 128 µg/ml, respectively) was performed for the PA14 and mutS::MAR2xT strains, as previously described [24]. In the case of the glpT::MAR2xT strain, the mutant frequency was determined for tobramycin (4 µg/ml) and imipenem (4 µg/ml) to study the possible epistatic effects of resistance to fosfomycin and these two antibiotics.
Evaluation of the fitness and in vivo selection of fosfomycin resistant mutants
Mice were inoculated with approximately 5×105 cells of strains PA14, PA14mutS::MAR2xT7 and PA14glpT::MAR2xT7 in saline solution. Twenty-four hours after inoculation, two groups of 8 mice per strain were either treated with fosfomycin by receiving two intraperitoneal injections in saline solution with an interval of 8 hours (200 mg/kg/8 h) [34], [35], [36] or untreated. After 48 hours of inoculation (16 h after the end of the treatment in the case of fosfomycin-treated mice), the animals were sacrificed and their lungs aseptically extracted. The left lung was homogenized in 2 ml of saline using the Ultra-Turrax T-25 disperser (IKA, Staufen, Germany). Serial 1/10 dilutions were plated in MHA, and the total bacterial load was determined. To quantify Fos-R mutants, lung homogenates or serial 1/10 dilutions were placed in MHA plates containing Fos 128 µg/ml. The established lower limit of detection was 4 cfu of mutants per lung. The statistical differences among groups were estimated by a Kruskal-Wallis test for lung bacterial counts, and the Mann-Whitney U test was used to compare the proportion of Fos-R mutants between the group receiving fosfomycin and the untreated one. P values of less than 0.05 were considered to be statistically significant.
Competition experiments
Sixteen mice were inoculated with a 1∶1 mix of PA14 and PA14glpT::MAR2xT7 for a final inoculum size approximately of 5×105. Twenty-four hours after inoculation, the mice were divided into two groups (8 mice each) and treated with Fos (200 mg/Kg/8 h) or untreated. After 48 hours of inoculation (16 h after the end of the treatment in the case of Fos-treated mice), the animals were sacrificed and their lungs aseptically extracted. The left lung was homogenized in 2 ml of saline using the Ultra-Turrax T-25 disperser (IKA, Staufen, Germany). Serial 1/10 dilutions were plated onto MHA and MHA with 10 µg/ml of gentamycin to determine the total bacterial load and the PA14glpT::MAR2xT7 bacterial load, respectively. In a similar way, eight tubes were inoculated with approximately 103 bacteria of each bacterial strain in 5 ml of LB and incubated at 37°C with agitation for 24 h. The bacterial counts were determined following the procedure described above for lung homogenate. The competition index (CI) was defined as the glpT::MAR2xT7 mutant/wild-type ratio. The statistical analysis of the the CI values distribution was performed by a Mann-Whitney U test. P values of less than 0.05 were considered to be statistically significant.
Analysis of Fos-R mutants generated in vivo
For the complementation studies 10 Fos-R mutants isolated from 10 different non-treated mouse lungs inoculated with the wild type strain, were complemented with the plasmid pBBR-glpT, as previously described [37]. The plasmid pBBR-glpT and its parental empty vector pBBR1MCS-3 [38] were introduced by electroporation into all the isolated Fos-R mutants. Transformants were selected on LB-agar supplemented with 150 µg/ml of tetracycline. The minimal inhibitory concentration (MIC) of fosfomycin was determined as described [39] for three colonies of every transformed mutant. The glpT gene of all mutants was sequenced, and compared with the wild type gene as described [37].
Biofilms
An abiotic solid surface biofilm formation assay was performed in 96-well polystyrene microtiter plates in LB and minimal FBA [27] media in a humid chamber at 37°C, as described previously [40]. Biofilms were quantified after seven days of incubation. Forty independent replicas were carried out for each strain. Data were compared by a Student's t-test.
Antibiotic susceptibility testing
Minimal inhibitory concentrations (MICs) of fosfomycin, tobramycin and imipenem were determined for the PA14 strain and its derivative mutants mutS::MAR2xT7 and glpT::MAR2xT7 by the broth microdilution method, as recommended by the CLSI [39].
Results
glpT null mutant is as lethal as its parental strain
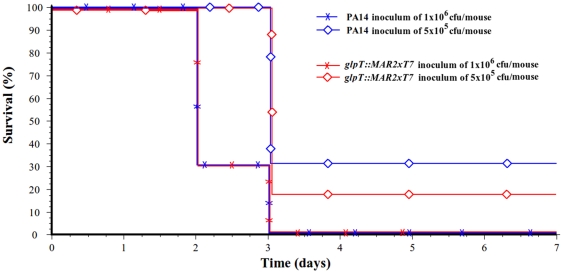
We did not detect any statistically significant difference in mortality between the PA14 wild type strain and the null mutant glpT::MAR2xT7 (Fos-R), independently of the size of the inoculum (Figure 1). The Kaplan-Meier analysis revealed no discrepancy in the mortality curves between strains receiving the same inoculum (p = 0.81, p = 0.21, for approximately 1×106 and 5×105 bacteria, respectively). These results indicate that the disruption of the glpT gene does not interfere with the infective capacity and mortality caused by P. aeruginosa in this model.
Figure 1. Mortality curves.
Mice were infected with two different inocula of P. aeruginosa PA14 (blue lines) and its mutant glpT::MAR2xT7 derivative (red lines) strains. The Kaplan-Meier analysis failed to detect any significant difference between curves from the same bacterial inoculum (1×106 cell (crosses) and 5×105 cells (diamonds)) (p = 0.81 and p = 0.21, respectively).
Effect of fosfomycin treatment in bacterial load of the wild type, Fos-R, and mutS strains
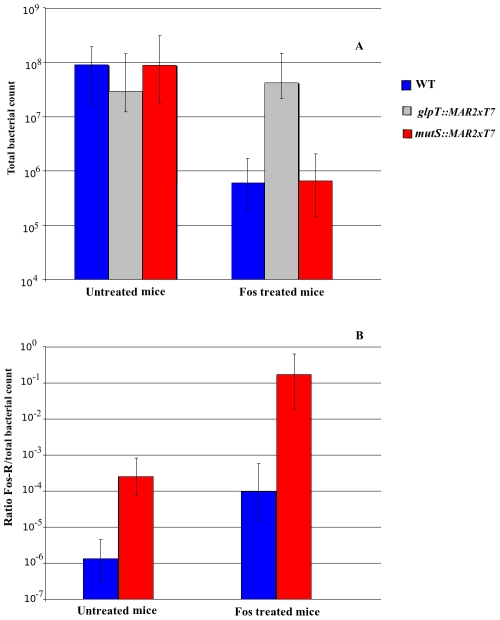
The colonization of mice lungs by P. aeruginosa PA14 (median 9.16×107, interquartile range from 1.57×107 to 1.73×108) forty eight hours post infection did not show statistical differences in the number of colony-forming units (cfu) per mouse, in comparison with the other two strains, mutS::MAR2xT7 (median 8.86×107, interquartile range from 2.15×107 to 2.55×108) and glpT::MAR2xT (median 2.95×107, interquartile range from 1.12×107 to 1.30×108) (p = 0.44). On the other hand, when mice were treated with fosfomycin, bacterial counts of the wild type strain (median 6.02×105, interquartile range from 2.65×105 to 1.66×106) and the mutS-deficient mutant (median 6.60×105, interquartile range from 1.18×105 to 2.06×106) were dramatically decreased by two orders of magnitude (p = 0.005 and p = 0.009, respectively). However, the glpT::MAR2xT mutant did not suffer any drop in bacterial counts (median 4.19×107, interquartile range from 2.01×107 to 1.06×108) with no differences with the non-treated experiment (p = 0.91) (Figure 2A).
Figure 2. Recovery of bacteria from lungs 48 hours post infection.
Mice were infected with approximately 5×105 cfu/animal and treated with two doses of fosfomycin (200 mg/kg) 24 hours after inoculation or with no antibiotic. PA14 (blue columns) and its mutant derivatives mutS::MAR2xT7 (red columns) and glpT::MAR2xT7 (gray columns) (A). Ratio of Fos-R mutants/total bacteria in lungs from treated and non-treated mice (B). Values are medians and error bars represent interquartile ranges.
The proportion of resistant mutants is greatly increased in the wild type strain after fosfomycin treatment (Figure 2B), especially if we consider that this increase appeared only after 24 hours from the time the antibiotic was administered. The untreated group has the expected proportion of Fos-R mutants according to its mutation frequency (median 1.58×10−6, interquartile range from 3.62×10−7 to 5.69×10−6). However, the proportion of wild type Fos-R cells increased by two orders of magnitude (median 9.82×10−5, interquartile range from 1.56×10−5 to 5.76×10−4), which represents a more than 60 fold increase and is statistically significant (p = 0.009) (Table 1). Furthermore, for the PA14 mutS::MAR2xT7 strain this phenomenon was agravated. While the proportion of resistant mutants in untreated mice was as expected (median 2.54×10−4, interquartile range from 8.27×10−5 to 7.71×10−4), in treated mice the proportion increased up to two fosfomycin resistant mutants for every ten bacteria (median 1.70×10−1, interquartile range from 2.71×10−2 to 7.15×10−1), which corresponds to a greater than 600 fold increase and is statistically significant (p = 0.001) (Table 1).
Table 1. Amplification of fosfomycin resistant mutants in mice treated with the antibiotic.
| Strains | FosR mutant/total cfu (treated mice) | FosR mutant/total cfu (untreated mice) | Fold increase in Fos-R |
| PA14 WT | 9.8×105 | 1.58×106 | 61.98 |
| mutS::MAR2xT7 | 1.70×101 | 2.54×104 | 666.76 |
Fold increase values were calculated as the ratio between the median of the proportion of Fos-R mutants/total bacterial counts (figure 2B) of treated and untreated mice.
One interesting point is that the median of the absolute number of mutants actually remains constant if we compare the Fos-R mutant load in mice receiving fosfomycin or not, although the total bacterial count decreased by two orders of magnitude in the dosed mice. This indicates that from the start of fosfomycin dosing, probably because the bacterial load was very high at that point, there is a multiplicity problem that limits the amount of mutants. However, when mice are dosed, the number of fosfomycin-susceptible bacteria diminish, thus the resistant ones can prevail at a higher rate in comparison with the non-treated group.
Mutation rates to fosfomycin resistance in vivo are similar to those in vitro
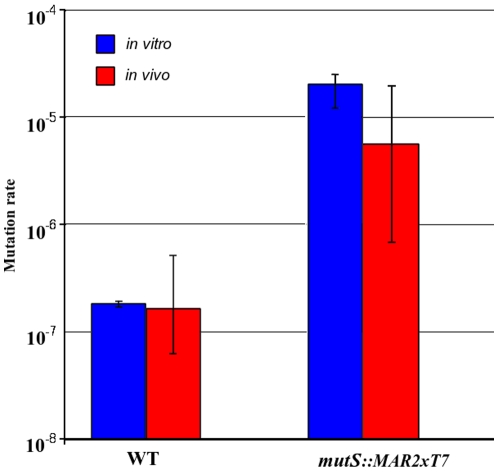
Results shown in figure 3 indicate that the mutation rates obtained in vitro and in vivo are very similar (95% confidence interval). As expected, mutation rates to fosfomycin resistance are in agreement with the previously described values for these P. aeruginosa strains [24]. The values were around 10−5 for the mutS-deficient mutant, while the parental strain showed values two orders of magnitude lower for both the in vitro and the in vivo experiments. The high dispersion in the data corresponding to the in vivo experiment is notable and probably due to the innate variation of the animal models compared to the controlled laboratory conditions of the in vitro experiments. These results indicate that there is no limitation in the emergence of in vivo Fos-R mutants in P. aeruginosa.
Figure 3. Mutation rates.
in vivo (acute mice lung infection model, red columns) and in vitro (blue columns) of P. aeruginosa PA14 and its mutant derivative mutS::MAR2xT7 strains are shown. Bars represent confidence intervals at a signification level of 0.05.
Wild type strain did not outcompete the Fos-R mutant in vivo or in vitro
The in vitro competition experiment rendered no statistical differences between the WT strain and glpT mutant (CI = 1.04; p = 0.80). Similarly, in the in vivo competition experiment in mice which did not receive fosfomycin treatment, the competition index between the wild type strain and glpT-deficient mutant, was 0.78, indicating no fitness advantage in any strain (p = 0.22). As expected, the scenario changed totally when the mice were treated with fosfomycin. In this case the CI rose to 9.70 in favour of the Fos-R mutant (p = 0.02), showing that Fos-R mutants will prevail in the population when pressure is exerted by fosfomycin.
All Fos-R mutants generated in vivo contain mutations in the glpT gene
Complementation studies showed that glpT mutations were the cause of fosfomycin resistance in vivo in all cases. Ten randomly chosen mutants from the wild type strain recovered fosfomycin sensitivity upon complementation with the glpT wild type gene. These results coincide with those from our previous work [37]. The sequence analysis revealed different mutations in the glpT gene (Table 2). The mutations consisted mainly of single-base deletions (5 mutants), which caused inactivation of the gene by frameshifts. There were also deletions of 2, 6 and 12 pb causing frameshifts or loss of 3 or 4 aminoacids respectively. Additionally, a G to A transition was detected at position 900 of the ORF.
Table 2. Mutations in glpT of ten randomly chosen spontaneous mutants of P. aeruginosa PA14 selected from in vitro and in vivo experiments.
| Inactivating mutations of glpT in vitro and in vivo | |||
| in vitro [37] | in vivo (this work) | ||
| Mutant No. | Mutation position | Mutant No. | Mutation position |
| FosR-7 | A59 deletion, 1 bp (frameshift) | FosR-5 | T215CGCCA220 deletion 6 bp |
| FosR-9 | C219ATCGC225 deletion, 6 bp | FosR-6 | T221CGCCT227 deletion 6 bp |
| FosR-1 | A220TCGCC226 deletion 6 bp | FosR-4 | T226A227 deletion, 2 bp (frameshift) |
| FosR-2 | T365CATGTT371 deletion, 7 bp (frameshift) | FosR-9 | T377 deletion, 1 bp (frameshift) |
| FosR-3 | G410 to A transition | FosR-10 | G619 deletion, 1 bp (frameshift) |
| FosR-5 | G596 deletion, 1 bp (frameshift) | FosR-3 | A869 deletion, 1 bp (frameshift) |
| FosR-10 | C975GG insertion, 3 bp | FosR-2 | T887 deletion, 1 bp (frameshift) |
| FosR-4 | A1006 to C transition | FosR-7 | G900 to A transition |
| FosR-8 | C1086 to G transition | FosR-8 | C1032GGCAACCCGGC1043 deletion 12 bp |
| FosR-6 | G1098 to A transition | FosR-10 | T1231 deletion, 1 bp (frameshift) |
Mutants were arranged according to the nucleotide position using the A of the ATG of the ORF as reference.
Inactivation of glpT does not affect biofilm formation in vitro
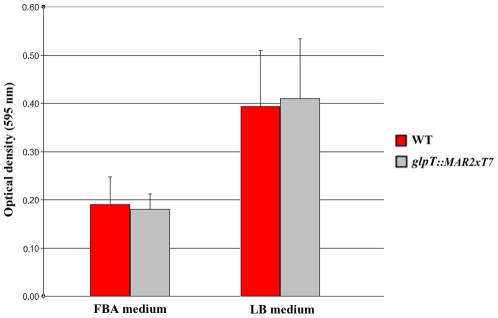
We could not detect any statistically significant difference between the wild type strain and its glpT::MAR2xT derivative in biofilm formation. Figure 4 shows the ability of the two strains to form biofilm in FBA (mean ± standard deviation: 0.19±0.07 for the wild type and 0.17±0.05 for the glpT-deficient mutant, p = 0.31) or LB (mean ± standard deviation: 0.39±0.12 for the wild type and 0.41±0.13 for the glpT-deficient mutant, p = 0.59).
Figure 4. Seven-day biofilm formation.
Biofilm formation of P. aeruginosa PA14 (red columns) and its mutant derivative glpT::MAR2xT7 (gray columns) is shown. The values are represented as the mean ± SD. Forty replicas were performed per strain and medium. No statistically significant differences were detected between strains for each medium (FBA, p = 0.31; LB, p = 0.59).
Histopathology studies
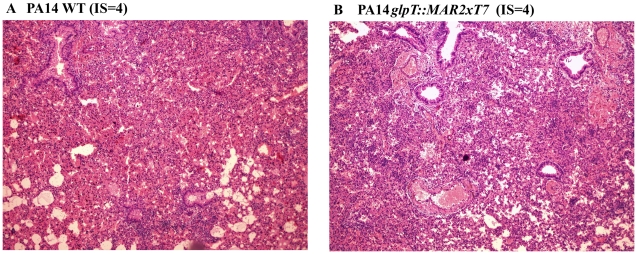
Inflammation was scored in 10 mice per strain by using the criteria of Johansen et al. [31]. In summary, both strains (PA14 and PA14glpT::MAR2xT7) induced a very high level of inflammation (3.9±0.32 and 3.6±0.97; mean and standard deviation of scores for PA14 and PA14glpT::MAR2xT7, respectively) with no significant differences between them (p = 0.40). Most of the infected mice (90% and 80% for PA14 and PA14glpT::MAR2xT7, respectively) had a score of 4 (from severe inflammation to necrosis and severe inflammation throughout the lung) (Figure 5).
Figure 5. Histology of murine lungs after infection with P. aeruginosa.
Shown are representative examples of the histology results obtained for murine lungs 48 hours after infection with the P. aeruginosa strain PA14 (A) or PA14glpT::MAR2xT7 (B). IS, inflammatory score determined as described by Johansen et al. [31].
Frequency of emergence of resistant mutants to combined antibiotics
To investigate the possibility of appearance of double mutants with resistance to the combinations of fosfomycin with different antibiotics, we first determined the MICs of these antibiotics for the PA14 strain and its derivatives mutS::MAR2xT7 and glpT::MAR2xT7 (Table 3). The mutant frequencies of resistance to the combinations of tobramycin plus fosfomycin (4 and 128 µg/ml, respectively) and imipenem plus fosfomycin (4 and 128 µg/ml, respectively) were studied. Obviously, the frequencies of mutants resistant to the single antibiotics fosfomycin (128 µg/ml), tobramycin (4 µg/ml) and imipenem (4 µg/ml) were also calculated. Table 4 shows that the frequency of mutants resistant to the combination of fosfomycin with tobramycin is below our limit of detection (≤10−11), making improbable the emergence of resistance to both antibiotics when administered together, even in a hypermutable background. However, the frequency of mutants resistant to the combination of imipenem plus fosfomycin is relatively high in the wild type (2.1×10−9) with a 2-log increase in the hypermutator mutant (1.1×10−7). These results suggest that the generation of mutants resistant to the combination of fosfomycin with some antibiotics may not be very difficult for P. aeruginosa. Interestingly, the frequencies of mutants resistant to either tobramycin or imipenem of the glpT-deficient strain were similar to those of the wild type. Thus, there is apparently no epistatic effect between the resistance determinants of the pairs tobramycin/fosfomycin and imipenem/fosfomycin.
Table 3. Minimal inhibitory concentrations of the antibiotics fosfomycin, tobramycin and imipenem for PA14 and its derivative mutants mutS::MAR2xT7 and glpT::MAR2xT7.
| Antibiotic | MIC (µg/ml) | ||
| wt | mutS::MAR2xT7 | glpT::MAR2xT7 | |
| Fosfomycin | 8 | 8 | 1024 |
| Tobramycin | 0.25 | 0.25 | 0.25 |
| Imipenem | 0.5 | 0.5 | 0.5 |
Table 4. Frequency of mutants resistant to single antibiotics and the combinations of fosfomycin with tobramycin and imipenem.
| Antibiotic | Concentration a | Mutant frequency | |
| wt | mutS::MAR2xT7 | ||
| Tobramycin | 4 | 2.2×10−9 | 1.4×10−7 |
| Imipenem | 4 | 2.3×10−6 | 9.1×10−4 |
| Fosfomycin | 128 | 1.5×10−6 | 1.1×10−4 |
| Combination | |||
| Tobramycin +Fosfomycin | 4+128 | <10−11 | <10−11 |
| Imipenem +Fosfomycin | 4+128 | 2.1×10−9 | 1.1×10−7 |
a: antibiotic concentation in µg/ml.
Discussion
Among the different factors contributing to the ascent of antibiotic resistance, fitness cost is now recognized as prominent and one that ought to be evaluated when assessing the suitability of any drug treatment [8], [10]. Here we have shown that in the absence of fosfomycin, the P. aeruginosa PA14 wild type strain (sensitive) was unable to outcompete the null glpT mutant (Fos-R strain) after two days of in vivo or 24 hours of in vitro competition. Similar results were obtained previously in an in vitro experiment comparing growth rate in minimal media [37].
On the other hand, after two days of acute lung infection, the bacterial lung counts and the ability of the glpT mutant to cause death did not differ from the parental strain. These results clearly indicate that fosfomycin resistance does not exert a significant burden on fitness or virulence for P. aeruginosa.
This divergence from the E. coli patterns could be explained to some extent by the fact that, although both species share the reduction in uptake of fosfomycin on the basis of resistance mechanism [8], [14], [15], [16], [37], they apparently differ in the specific target genes for mutation and their metabolic implications. It is well established that, in E. coli and other gram negative species fosfomycin access primarily through the L-alpha-glycerol-3-phosphate transport system (glpT), and to a lesser extent via the hexose phosphate transport system (uhpT) [15]. Moreover, these two genes are known to be positively regulated by cAMP, and so mutations that lower the cAMP level in the cell (i.e. in the cyaA and ptsI genes) can induce resistance as well [41]. This reduction in cAMP levels can produce profound alterations to the bacterial metabolism, and can probably explain the significant reduction in fitness and virulence shown for many resistant isolates in E. coli [8]. Aldoght, Fos-R clinical isolates of E. coli are now being found and suggesting that this is a viable threat [9], [21].
In contrast, the uptake of fosfomycin in P. aeruginosa depends exclusively on GlpT permease [37], which does not seem to be regulated by cAMP levels. Indeed, the glpT gene is isolated in the P. aeruginosa genome (www.pseudomonas.com), while the E. coli ortholog is included inside the well-known glg operon [42]. This may be related to another radical difference between these two species, as has been stated in the case of P. aeruginosa which displays a higher preference for glycerol as carbon source than for glycerol-3-phosphate [43]. These observations could be understood considering the different ecology of both species.
We have also performed an analysis of the glpT sequences from Fos-R mutants obtained in vivo and complementation studies, which show that fosfomycin resistance is acquired in P. aeruginosa by any mutation capable of inactivating GlpT activity. From a comparative point of view with in vitro generated mutants, mutational pathways appear to be very similar in both conditions.
Our results have at least two relevant implications from a medical stand point. First, our data demonstrate that the fitness cost of antibiotic resistance needs to be measured for each particular species of interest, and that the observations made in just one species cannot be extrapolated to another, since the genetic particularities might modulate the effects of bacterial fitness. Moreover, fosfomycin resistant clinical isolates of E. coli are now being found, suggesting that this is a viable threat [9], [21]. Second, our results curb the good prospects that fosfomycin has recently promised, at least for its use on P. aeruginosa infections. Similarly, it has been reported that there is no repercussion for fitness in any other resistance mechanism mediated by gene inactivation in P. aeruginosa. A partial deregulation of ampC expression by mutation of its repressor ampD confers efficient resistance to cephalosporins, without biological cost, due to two other copies of ampD [44]. However, contrarily to what occurs with glpT mutations, where a very high level of resistance is conferred, the ampD-based resistance is far from being complete.
The high proportion of fosfomycin mutants generated by the hypermutable mutS-deficient strain indicates a real danger of rapid emergence of resistant mutants during a treatment. This is especially true in chronic respiratory infections, where one of the most prevalent and dangerous bacterial pathogens is P. aeruginosa [45]. In these infections the prevalence of hypermutable strains is abnormally high [46] and have been linked to antibiotic resistance development and persistence in respiratory airways [47], [48]. Thus, one can expect that in such infections fosfomycin resistance would develop as soon as patients begin the treatment.
As fosfomycin has been proposed as potentially useful in combined therapy [49], we studied in vitro the probability of acquisition of resistance to the combination of fosfomycin with two different antibiotics, tobramycin and imipenem. While the combination of fosfomycin with tobramycin makes improbable the emergence of resistance to both antibiotics when administered together, even in an hypermutable background, the results from the combination of fosfomycin plus imipenem suggest that the generation of mutants resistant to both antibiotics is not very difficult for P. aeruginosa. However, additional studies are still needed to determine the real effectiveness of fosfomycin when administered together with these and other antibiotics. Recently Ward et al [50] have reported that the biological cost of resistance to a second antibiotic in P. aeruginosa could be greater than that of previously acquired resistance. Finally, the total cost depends on the genetic background in which resistance evolve and its interaction with the environment. Well conducted studies are needed to find a good combination of antibiotics where double resistance, if available, has a sufficiently high cost to impede the persistence of infection.
We were unable to identify any condition where the inactivation of glpT has some significant repercussion on P. aeruginosa fitness. Biofilm formation of the defective mutant did not show any alteration when compared with the wild type strain, which probably indicates that this gene does not influence biofilm development. These results are consistent with the fact that some metabolic genes are down regulated during biofilm formation [51], [52].
In conclusion, apart from the absence of in vivo fitness cost of fosfomycin resistance in P. aeruginosa, our findings reveal that we need a deeper understanding of the interplay between antibiotic resistance, biological fitness and virulence. This knowledge will be invaluable if we aspire to improve the development of drug antibiotherapy for the different pathogens nowadays threatening humankind.
Acknowledgments
The authors would like to thank an anonymous referee for his/her constructive comments and N.T. Liberati and F. Ausubel for the P. aeruginosa strains PA14 and its glpT::MAR2xT7 and mutS::MAR2xT7 derivatives.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Grants PI070215 and the Spanish Network for Research in Infectious Diseases (REIPI RD06/0008), both from the Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Falagas ME, Grammatikos AP, Michalopoulos A. Potential of old-generation antibiotics to address current need for new antibiotics. Expert Rev Anti Infect Ther. 2008;6:593–600. doi: 10.1586/14787210.6.5.593. [DOI] [PubMed] [Google Scholar]
- 2.Falagas ME, Giannopoulou KP, Kokolakis GN, Rafailidis PI. Fosfomycin: use beyond urinary tract and gastrointestinal infections. Clin Infect Dis. 2008;46:1069–1077. doi: 10.1086/527442. [DOI] [PubMed] [Google Scholar]
- 3.Suarez JE, Mendoza MC. Plasmid-encoded fosfomycin resistance. Antimicrob Agents Chemother. 1991;35:791–795. doi: 10.1128/aac.35.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chin NX, Neu NM, Neu HC. Synergy of fosfomycin with beta-lactam antibiotics against staphylococci and aerobic gram-negative bacilli. Drugs Exp Clin Res. 1986;12:943–947. [PubMed] [Google Scholar]
- 5.Olay T, Rodriguez A, Oliver LE, Vicente MV, Quecedo MC. Interaction of fosfomycin with other antimicrobial agents: in vitro and in vivo studies. J Antimicrob Chemother. 1978;4:569–576. doi: 10.1093/jac/4.6.569. [DOI] [PubMed] [Google Scholar]
- 6.Tessier F, Quentin C. In vitro activity of fosfomycin combined with ceftazidime, imipenem, amikacin, and ciprofloxacin against Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis. 1997;16:159–162. doi: 10.1007/BF01709477. [DOI] [PubMed] [Google Scholar]
- 7.Visalli MA, Jacobs MR, Appelbaum PC. Determination of activities of levofloxacin, alone and combined with gentamicin, ceftazidime, cefpirome, and meropenem, against 124 strains of Pseudomonas aeruginosa by checkerboard and time-kill methodology. Antimicrob Agents Chemother. 1998;42:953–955. doi: 10.1128/aac.42.4.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nilsson AI, Berg OG, Aspevall O, Kahlmeter G, Andersson DI. Biological costs and mechanisms of fosfomycin resistance in Escherichia coli. Antimicrob Agents Chemother. 2003;47:2850–2858. doi: 10.1128/AAC.47.9.2850-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oteo J, Orden B, Bautista V, Cuevas O, Arroyo M, et al. CTX-M-15-producing urinary Escherichia coli O25b-ST131-phylogroup B2 has acquired resistance to fosfomycin. J Antimicrob Chemother. 2009;64:712–717. doi: 10.1093/jac/dkp288. [DOI] [PubMed] [Google Scholar]
- 10.Andersson DI, Levin BR. The biological cost of antibiotic resistance. Curr Opin Microbiol. 1999;2:489–493. doi: 10.1016/s1369-5274(99)00005-3. [DOI] [PubMed] [Google Scholar]
- 11.Andersson DI. The biological cost of mutational antibiotic resistance: any practical conclusions? Curr Opin Microbiol. 2006;9:461–465. doi: 10.1016/j.mib.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Zengel JM, Young R, Dennis PP, Nomura M. Role of ribosomal protein S12 in peptide chain elongation: analysis of pleiotropic, streptomycin-resistant mutants of Escherichia coli. J Bacteriol. 1977;129:1320–1329. doi: 10.1128/jb.129.3.1320-1329.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poole K. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother. 2005;56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 14.Kadner RJ, Winkler HH. Isolation and characterization of mutations affecting the transport of hexose phosphates in Escherichia coli. J Bacteriol. 1973;113:895–900. doi: 10.1128/jb.113.2.895-900.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kahan FM, Kahan JS, Cassidy PJ, Kropp H. The mechanism of action of fosfomycin (phosphonomycin). Ann N Y Acad Sci. 1974;235:364–386. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- 16.Cordaro JC, Melton T, Stratis JP, Atagun M, Gladding C, et al. Fosfomycin resistance: selection method for internal and extended deletions of the phosphoenolpyruvate: sugar phosphotransferase genes of Salmonella typhimurium. J Bacteriol. 1976;128:785–793. doi: 10.1128/jb.128.3.785-793.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alos JI, Garcia-Pena P, Tamayo J. [Biological cost associated with fosfomycin resistance in Escherichia coli isolates from urinary tract infections]. Rev Esp Quimioter. 2007;20:211–215. [PubMed] [Google Scholar]
- 18.Marchese A, Gualco L, Debbia EA, Schito GC, Schito AM. In vitro activity of fosfomycin against gram-negative urinary pathogens and the biological cost of fosfomycin resistance. Int J Antimicrob Agents. 2003;22(Suppl 2):53–59. doi: 10.1016/s0924-8579(03)00230-9. [DOI] [PubMed] [Google Scholar]
- 19.Gismondo MR, Drago L, Fassina C, Garlaschi ML, Rosina M, et al. Escherichia coli: effect of fosfomycin trometamol on some urovirulence factors. J Chemother. 1994;6:167–172. doi: 10.1080/1120009x.1994.11741147. [DOI] [PubMed] [Google Scholar]
- 20.Klein U, Pawelzik M, Opferkuch W. Influence of beta-lactam antibiotics, fosfomycin and vancomycin on the adherence (hemagglutination) of Escherichia coli-containing different adhesins. Chemotherapy. 1985;31:138–145. doi: 10.1159/000238326. [DOI] [PubMed] [Google Scholar]
- 21.Takahata S, Ida T, Hiraishi T, Sakakibara S, Maebashi K, et al. Molecular mechanisms of fosfomycin resistance in clinical isolates of Escherichia coli. Int J Antimicrob Agents. 2010;35:333–337. doi: 10.1016/j.ijantimicag.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 22.Shorr AF. Review of studies of the impact on Gram-negative bacterial resistance on outcomes in the intensive care unit. Crit Care Med. 2009;37:1463–1469. doi: 10.1097/CCM.0b013e31819ced02. [DOI] [PubMed] [Google Scholar]
- 23.Blanc DS, Petignat C, Janin B, Bille J, Francioli P. Frequency and molecular diversity of Pseudomonas aeruginosa upon admission and during hospitalization: a prospective epidemiologic study. Clin Microbiol Infect. 1998;4:242–247. doi: 10.1111/j.1469-0691.1998.tb00051.x. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez-Rojas A, Blazquez J. The Pseudomonas aeruginosa pfpI gene plays an antimutator role and provides general stress protection. J Bacteriol. 2009;191:844–850. doi: 10.1128/JB.01081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bjorkman J, Nagaev I, Berg OG, Hughes D, Andersson DI. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science. 2000;287:1479–1482. doi: 10.1126/science.287.5457.1479. [DOI] [PubMed] [Google Scholar]
- 26.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heydorn A, Ersboll BK, Hentzer M, Parsek MR, Givskov M, et al. Experimental reproducibility in flow-chamber biofilms. Microbiology. 2000;146 (Pt 10):2409–2415. doi: 10.1099/00221287-146-10-2409. [DOI] [PubMed] [Google Scholar]
- 28.DiGiandomenico A, Rao J, Harcher K, Zaidi TS, Gardner J, et al. Intranasal immunization with heterologously expressed polysaccharide protects against multiple Pseudomonas aeruginosa infections. Proc Natl Acad Sci U S A. 2007;104:4624–4629. doi: 10.1073/pnas.0608657104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mueller-Ortiz SL, Drouin SM, Wetsel RA. The alternative activation pathway and complement component C3 are critical for a protective immune response against Pseudomonas aeruginosa in a murine model of pneumonia. Infect Immun. 2004;72:2899–2906. doi: 10.1128/IAI.72.5.2899-2906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yonezawa M, Sugiyama H, Kizawa K, Hori R, Mitsuyama J, et al. A new model of pulmonary superinfection with Aspergillus fumigatus and Pseudomonas aeruginosa in mice. J Infect Chemother. 2000;6:155–161. doi: 10.1007/s101560070015. [DOI] [PubMed] [Google Scholar]
- 31.Johansen HK, Espersen F, Cryz SJ, Jr, Hougen HP, Fomsgaard A, et al. Immunization with Pseudomonas aeruginosa vaccines and adjuvant can modulate the type of inflammatory response subsequent to infection. Infect Immun. 1994;62:3146–3155. doi: 10.1128/iai.62.8.3146-3155.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones ME, Thomas SM, Rogers A. Luria-Delbruck fluctuation experiments: design and analysis. Genetics. 1994;136:1209–1216. doi: 10.1093/genetics/136.3.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foster PL. Methods for determining spontaneous mutation rates. Methods Enzymol. 2006;409:195–213. doi: 10.1016/S0076-6879(05)09012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawaguchi K, Hasunuma R, Kikuchi S, Ryll R, Morikawa K, et al. Time- and dose-dependent effect of fosfomycin on suppression of infection-induced endotoxin shock in mice. Biol Pharm Bull. 2002;25:1658–1661. doi: 10.1248/bpb.25.1658. [DOI] [PubMed] [Google Scholar]
- 35.Haag R, Vomel W, Schaumann W. [Comparison of in vitro methods for the determination of the activity of fosfomycin with the efficacy determined in experimentally infected mice (author's transl)]. Immun Infekt. 1981;9:177–182. [PubMed] [Google Scholar]
- 36.Schindler J, Langsadl L. Fosfomycin therapy of experimental infections in mice. Zentralbl Bakteriol Orig A. 1979;243:136–139. [PubMed] [Google Scholar]
- 37.Castaneda-Garcia A, Rodriguez-Rojas A, Guelfo JR, Blazquez J. The Glycerol-3-Phosphate Permease GlpT Is the Only Fosfomycin Transporter in Pseudomonas aeruginosa. J Bacteriol. 2009;191:6968–6974. doi: 10.1128/JB.00748-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 39.CLSI. Wayne, PA: Clinical and Laboratory Standards Institute; 2006. CLSI. CLSI Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. Approved Standard - 7th Edition. pp. CLSI M7–A7. [Google Scholar]
- 40.O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 41.Tsuruoka T, Miyata A, Yamada Y. Two kinds of mutants defective in multiple carbohydrate utilization isolated from in vitro fosfomycin-resistant strains of Escherichia coli K–12. J Antibiot (Tokyo) 1978;31:192–201. doi: 10.7164/antibiotics.31.192. [DOI] [PubMed] [Google Scholar]
- 42.Lemieux MJ, Huang Y, Wang DN. Glycerol-3-phosphate transporter of Escherichia coli: structure, function and regulation. Res Microbiol. 2004;155:623–629. doi: 10.1016/j.resmic.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 43.Schweizer HP, Po C. Regulation of glycerol metabolism in Pseudomonas aeruginosa: characterization of the glpR repressor gene. J Bacteriol. 1996;178:5215–5221. doi: 10.1128/jb.178.17.5215-5221.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moya B, Juan C, Alberti S, Perez JL, Oliver A. Benefit of having multiple ampD genes for acquiring beta-lactam resistance without losing fitness and virulence in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2008;52:3694–3700. doi: 10.1128/AAC.00172-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strateva T, Yordanov D. Pseudomonas aeruginosa - a Phenomenon of Bacterial Resistance. J Med Microbiol. 2009 doi: 10.1099/jmm.0.009142-0. [DOI] [PubMed] [Google Scholar]
- 46.Oliver A, Canton R, Campo P, Baquero F, Blazquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 47.Macia MD, Blanquer D, Togores B, Sauleda J, Perez JL, et al. Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrob Agents Chemother. 2005;49:3382–3386. doi: 10.1128/AAC.49.8.3382-3386.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mena A, Smith EE, Burns JL, Speert DP, Moskowitz SM, et al. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J Bacteriol. 2008;190:7910–7917. doi: 10.1128/JB.01147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Falagas ME, Kastoris AC, Karageorgopoulos DE, Rafailidis PI. Fosfomycin for the treatment of infections caused by multidrug-resistant non-fermenting Gram-negative bacilli: a systematic review of microbiological, animal and clinical studies. Int J Antimicrob Agents. 2009 doi: 10.1016/j.ijantimicag.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 50.Ward H, Perron GG, Maclean RC. The cost of multiple drug resistance in Pseudomonas aeruginosa. J Evol Biol. 2009;22:997–1003. doi: 10.1111/j.1420-9101.2009.01712.x. [DOI] [PubMed] [Google Scholar]
- 51.Waite RD, Paccanaro A, Papakonstantinopoulou A, Hurst JM, Saqi M, et al. Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles. BMC Genomics. 2006;7:162. doi: 10.1186/1471-2164-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waite RD, Papakonstantinopoulou A, Littler E, Curtis MA. Transcriptome analysis of Pseudomonas aeruginosa growth: comparison of gene expression in planktonic cultures and developing and mature biofilms. J Bacteriol. 2005;187:6571–6576. doi: 10.1128/JB.187.18.6571-6576.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]