Abstract
Pseudomonas aeruginosa is an opportunistic Gram-negative pathogen capable of acutely infecting or persistently colonizing susceptible hosts. P. aeruginosa colonizes surfaces in vitro by either biofilm formation or swarming motility. The choice of behaviour is influenced by the physical properties of the surface and specific nutrient availability, and subject to regulatory networks that also govern type 2 and type 3 protein secretion. Biofilm formation by clinical isolates has been well-studied. However, the swarming behaviour of human isolates has not been extensively analysed. We collected isolates from 237 hospitalized patients without cystic fibrosis and analysed motility and secretion phenotypes of each isolate. We found biofilm formation and swarming to be negatively associated, while swarming was positively associated with the secretion of both proteases and type 3 exoenzymes. Most isolates were capable of type 3 secretion and biofilm formation, even though these traits are considered to favour distinct modes of pathogenesis. Our data demonstrate that while clinical isolates display diverse motility, biofilm and secretion phenotypes, many of the predicted relationships between swarming motility and other phenotypes observed in laboratory strains also hold true for bacteria isolated from human patients.
INTRODUCTION
Pseudomonas aeruginosa is a Gram-negative bacterium associated with both acute and chronic infections of humans (Lyczak et al., 2000). Many potential P. aeruginosa virulence factors have been identified using animal models of infection, including those that allow bacteria to move over, attach to, and effectively colonize mammalian tissues, and secreted enzymes and toxins that allow Pseudomonas to cross tissue barriers and evade host immune defences.
Two surface organelles, flagella and type IV pili (tfp), affect virulence in acute and chronic models of Pseudomonas disease. Bacteria lacking flagella caused less inflammation and death than wild-type counterparts in a murine model of acute pneumonia (Feldman et al., 1998), possibly a reflection of flagellin's ability to trigger pro-inflammatory host responses via Toll-like receptor 5 rather than to a loss of motility per se (Balloy et al., 2007). Flagella are required for robust biofilm formation (Klausen et al., 2003), and likely contribute to persistent colonization. tfp, which power twitching motility across solid surfaces (Mattick, 2002), mediate attachment to epithelial cells, contribute to biofilm formation, and are required for full virulence in corneal infection models (Kang et al., 1997; Mattick, 2002; O'Toole & Kolter, 1998).
P. aeruginosa uses flagella and tfp to swarm across semi-solid surfaces. In vitro, swarming occurs on semi-solid surfaces (0.5–0.7 % agar) when specific carbon and nitrogen sources are provided (Kohler et al., 2000; Rashid & Kornberg, 2000). The role of tfp in swarming may be strain-dependent, as some P. aeruginosa isolates require tfp for swarming while others swarm with altered morphology in the absence of tfp (Kohler et al., 2000; Overhage et al., 2007). Overlapping sets of regulators govern swarming and biofilm formation in a reciprocal fashion, leading some investigators to hypothesize that these are alternative behaviours adopted by P. aeruginosa in response to environmental cues (Caiazza et al., 2007; Shrout et al., 2006). For example, strains lacking GacA, required for exopolysaccharide (EPS) production and biofilm formation, display increased swarming (Parkins et al., 2001), while P. aeruginosa with increased EPS production swarms poorly (Caiazza et al., 2007).
A role for swarming motility during in vivo infection or colonization has not been established. However, transposon insertions that attenuate P. aeruginosa virulence in a rat chronic pulmonary infection model map to genes required for swarming (Potvin et al., 2003). Several cues required for swarming in vitro, namely rhamnolipids and elevated glutamate levels, are present in the sputum of cystic fibrosis (CF) patients (Singh et al., 2000). Lastly, microarray data show that genes encoding virulence factors, e.g. type 3 secretion system (T3SS) components and extracellular proteases, are transcribed at higher levels by swarming bacteria (Overhage et al., 2008).
Several studies have examined twitching, swimming, biofilm and T3SS phenotypes of isolates cultured from specific human infection sites, or from specific subsets of patients (Feltman et al., 2001; Fonseca et al., 2007; Head & Yu, 2004). However, no systematic analysis of swarming motility and its relationship to other putative virulence factor phenotypes in clinical isolates has been carried out. We analysed a large set of P. aeruginosa isolates collected prospectively from 237 unique hospitalized individuals without CF. Using this extensive dataset, we examined whether relationships observed in laboratory strains between swarming and the expression of other proposed virulence phenotypes held true in isolates obtained from human patients.
METHODS
Specimen collection.
P. aeruginosa isolates cultured from paediatric and adult patients hospitalized at Yale-New Haven Hospital and identified by the clinical microbiology laboratory were collected between September 2005 and April 2007. All patients or their legal guardians provided informed consent, and the study protocol was approved by the Yale University Human Investigation Committee. Patients with CF were excluded from this study. Bacterial isolates were frozen as 15 % glycerol stocks at –80 °C. The first P. aeruginosa isolate identified from each patient after protocol enrolment was analysed in this study.
Motility assays.
Twitching motility was measured by stab-inoculating 1.5 % agar Luria broth (LB) plates with a single bacterial colony. Plates were incubated at 37 °C for 18–20 h, and the diameter of the twitching zone at the plastic–agar interface was noted. Any bacterial strain that had a detectable twitching zone upon visible inspection was scored as positive. Swimming motility was measured as the diameter of zone travelled by bacteria point-inoculated into 0.3 % agar LB plates and incubated for 18–20 h at 30 °C. Swarming motility was assayed as previously described on 0.5 % agar M8 plates supplemented with 0.4 % glucose and 0.05 % glutamate (Kohler et al., 2000). Swarming plates were incubated overnight at 30 °C, and placed at room temperature for an additional 24–48 h prior to measurement of the swarming diameter. Strain PAO1 was included as a control on each twitching, swimming and swarming plate, and the swarming or swimming zone of each isolate was expressed as a ratio relative to PAO1 from the same plate. Any measurable swimming zone was positive in swimming assays and a swarming zone of >10 % of that of PAO1 was considered positive in swarm assays. Swarming assays were repeated in triplicate on swarm-negative strains to confirm that they were truly negative.
Surfactant production.
Swarm-negative isolates were examined for the production of a visible zone of wetting material on swarming plates, characteristic of 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs) and rhamnolipid secretion. These clinical isolates were also inoculated to swarming plates with PAO1, and were scored as positive or negative for their ability to repel PAO1, which is a HAA-dependent phenomenon (Caiazza et al., 2005). Swarming plates were photographed with a Kodak Easyshare DX6340 3.1 megapixel camera.
Static biofilm formation.
Clinical isolates were inoculated to 1.0 ml LB and grown with shaking at 37 °C overnight. This culture was diluted 1 : 100 in LB and grown in triplicate in a polystyrene 96-well plate (Corning) overnight at 37 °C without agitation. PAO1 was inoculated on each plate as a positive control, while wells without bacteria served as blanks. The next day, the OD600 of each well was measured to control for differences in bacterial growth. Plates were subsequently washed three times in standing water to remove unattached bacteria. Wells were stained with 1 % (w/v) crystal violet for 30 min, then washed twice by dipping into standing water baths. Adherent stain was solubilized in 70 % EtOH and the A592 was measured (Head & Yu, 2004). Biofilm staining (A592 sample−A592 blank) was divided by the OD600 of the sample (OD600 sample−OD600 blank) to correct for differences in isolate growth rate. Each value was then normalized to that obtained for PAO1 [(A592 PAO1−A592 blank)/(OD600 PAO1−OD600 blank)] to allow for comparisons between assays.
T3SS ELISA.
Clinical isolates were inoculated from freshly streaked LB agar plates to 1 ml MinS medium [(10× MinS salts l−1=34 g KH2PO4, 50.8 g NH4Cl, 297.1 g C4H4O4Na2.6H2O, 25.7 g C6H6NO6-Na3) 1× complete media l−1=100 ml 10× MinS salts, 100 ml 0.5 M monosodium glutamate, 50 ml 50 % glycerol, 5 ml 1 M MgSO4, 1 ml 18 mM FeSO4 and 744 ml H2O] and grown for 8 h at 37 °C with vigorous aeration. Supernatants were collected after centrifuging cultures at 3000 g for 10 min. The presence of secreted ExoU, ExoS, ExoT, PcrV and PopD in culture supernatants was detected by ELISA as previously described (Li et al., 2005). Values were normalized to those obtained for PAO1 (ExoS) or PA103 (all other secreted proteins), which were included as positive controls in all assays. An isolate that secreted >15 % as much PopD as PA103 was considered positive for type 3 secretion; such isolates also secreted comparable amounts of PcrV and ExoT (data not shown). The effectors ExoU and ExoS are variably encoded and secreted by T3SS-positive P. aeruginosa environmental and clinical isolates and were therefore not used to determine whether a strain was T3SS-positive or -negative.
Caseinolytic assay.
The caseinolytic activity of secreted LasB (elastase), alkaline protease and protease IV was determined using a microplate assay. Briefly, 1 ml LB was inoculated with clinical isolates and grown for 24 h at 37 °C with shaking (250 r.p.m.). Bacterial cells were removed by centrifugation at 4000 g for 10 min. One microlitre of the supernatant was diluted fivefold into 0.2 M Tris/HCl (pH 8.0), then mixed with 25 μl reaction buffer (0.2 M Tris/HCl, pH 8.0; 20 mM CaCl2) and 25 μl FITC-labelled casein (5 mg ml−1 in 50 mM Tris/HCl, pH 7.5; Sigma C3777). After 75 min at 37 °C, 120 μl 5 % trichloroacetic acid was added to each well. After 20 min further incubation at 37 °C, insoluble material was removed by centrifugation (500 g for 20 min at 4 °C). Forty microlitres of the supernatant was diluted into 160 μl reading buffer (0.5 M Tris/HCl, pH 8.8) and the A492 was measured. Values observed ranged from −0.03 to 0.75 with a median value of 0.34. Repeated measurements of caseinolytic activity present in PA14 supernatants (positive control) yielded values of 0.4–0.5.
Statistical analysis.
Data were compiled in Excel (Microsoft) and analysed using Minitab 15 software. The Kruskal–Wallis test was used as a non-parametric test to compare medians among multiple groups of clinical strains. The Mann–Whitney test was used to compare medians between two groups. For binomial variables, chi-square test analysis was used to assess for significance in comparisons of multiple groups of isolates while the Fisher's exact test was used for comparison of two groups.
RESULTS AND DISCUSSION
Swarming motility of clinical isolates
We collected P. aeruginosa strains from 237 unique patients without CF hospitalized at Yale-New Haven Hospital over a period of 20 months. Bacteria were isolated from blood, the respiratory tract (sputum and bronchoalveolar lavage), urine and deep wound cultures (Table 1). Medical record review documented the presence of signs and symptoms of clinical infection at these tissue sites in a subset of study participants. All isolates were assayed for motility, adherence and protein secretion phenotypes by investigators blinded to the clinical presentation of the patients from whom isolates were derived. Only the first isolate collected from each individual at the time of enrolment was included in our analysis, and no conclusions were drawn about the acuity or chronicity of the isolate's association with any given subject. Host and bacterial variables potentially associated with clinical signs and symptoms of infection were analysed using both unadjusted (bivariate) and multivariate logistic regression models; no association between any form of motility, including swarming, was demonstrated by this analysis (M. Ledizet and others, unpublished). The bacterial phenotype data, however, allowed us to test whether relationships between swarming motility and other potential bacterial virulence phenotypes proposed from studies of laboratory strains were observed in a large set of patient-derived isolates.
Table 1.
Clinical isolate phenotypes as a function of isolate culture site
| Site of culture (n) | Swim-positive [n (%)] | Twitch-positive [n (%)] | Swarm-positive [n (%)]* | Biofilm [median (range)]† | T3SS-positive [n (%)]‡ |
|---|---|---|---|---|---|
| Respir. (136) | 121 (89) | 100 (74 ) | 75 (57 ) | 0.98 (0.17–10) | 77 (57 ) |
| Urine (65) | 54 (83 ) | 49 (75 ) | 41 (63 ) | 0.96 (0.14–5.4) | 31 (48 ) |
| Wound (22) | 19 (86 ) | 18 (81 ) | 16 (76 ) | 0.74 (0.26–1.7) | 9 (41 ) |
| Blood (14) | 14 (100) | 13 (93 ) | 11 (85 ) | 1.2 (0.4–2.2) | 11 (80 ) |
| Total (237) | 208 (88 ) | 180 (76 ) | 143 (63 ) | 0.96 (0.14–10) | 128 (54 ) |
*Isolates were considered positive for swarming if the measured swarming zone diameter was >10 % of that measured for the PAO1 control. Ten isolates failed to grow on swarming medium so n=227 for this column.
†Biofilm formation on an abiotic surface was measured in a static assay and normalized to a PAO1 control.
‡A strain was considered positive for T3SS if it secreted >15 % as much PopD as the PA103 control, as measured by ELISA.
Swarming motility allows P. aeruginosa to move across semi-solid surfaces in the presence of specific nutritional cues. The defined medium used in these assays (M8 containing glucose and glutamate as carbon and nitrogen sources) failed to support the growth of 10 clinical isolates. Of the remaining 227 isolates, 143 (63 %) displayed motility of varying morphologies on swarming plates (Fig. 1). Table 1 reports the distribution of swarm-positive strains (defined as swarm diameter ≥10 % of that of PAO1), which did not show significant variation as a function of the clinical site of culture (P=0.23, chi-square).
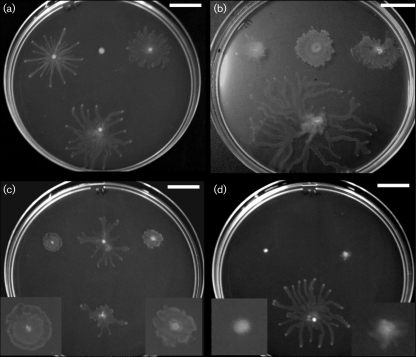
Fig. 1.
Clinical isolates display variable swarming phenotypes. Swarming plates (0.5 % agar M8) were incubated overnight at 30 °C and then at room temperature for 24–48 h. The colony morphology of clinical isolates varied, even among strains with identical twitching and swimming phenotypes. The laboratory strain PAO1 is at the bottom of each plate. (a) Swim-positive/twitch-negative; (b) swim-negative/twitch-positive; (c) swim-positive/twitch-positive; (d) swim-negative/twitch-negative. Enlarged views of colonies with small swarming diameters are provided in insets. Bars, 2 cm.
Swarming motility occurs in the absence of either twitching or swimming motility
Since both flagella and tfp likely influence swarming behaviours, each isolate was tested for swimming and twitching motility under defined laboratory conditions. No significant differences in motility phenotypes were observed when comparing isolates by the site of recovery (Table 1).
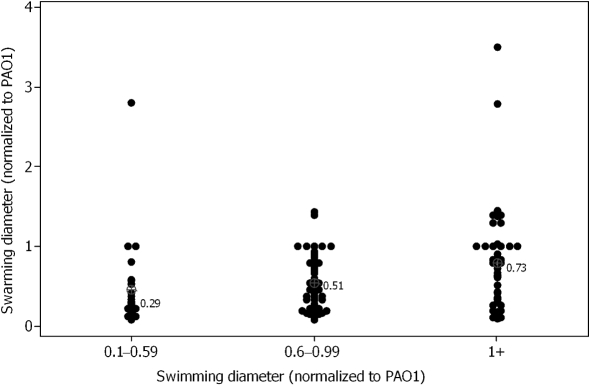
Swarming motility appears to require motile flagella (Kohler et al., 2000; Overhage et al., 2007). Within our series of clinical isolates, we observed that 10 of 20 swim-negative/twitch-positive strains capable of growth on 0.5 % agar M8 plates displayed swarming motility. However, swim-negative isolates swarmed less than swim-positive isolates (Table 2). This difference did not reach statistical significance (P=0.13, Mann–Whitney). Among isolates that were both swim-positive and swarm-positive, isolates with the largest swimming diameters tended to form larger swarming colonies than those with smaller swim diameters (Fig. 2; P=0.008, Kruskal–Wallis). This confirms the importance of functional flagella for swarming motility.
Table 2.
Biofilm formation and swarming motility of isolates with specific swimming and twitching phenotypes
| Swim-positive | Swim-negative | |||||
|---|---|---|---|---|---|---|
| Twitch-positive | Twitch-negative | Total | Twitch-positive | Twitch-negative | Total | |
| Swarm-positive | 106/154 (69 %) | 26/40 (65 %) | 132/199 (66 %) | 10/20 (50 %) | 1/8 (13 %) | 11/28 (39 %) |
| Swarm diameter, median* | 0.52 | 0.47 | 0.52 | 0.34 | 0.21 | 0.31 |
| Biofilm, median (range)† | 1.05 (0.4–2.2) | 0.56 (0.2–3.8) | 0.96 (0.2–3.8) | 0.74 (0.1–2.8) | 0.65 (0.2–2.6) | 0.68 (0.1–2.8) |
*The swarming colony diameter is normalized to the value obtained for PAO1. Swarm-negative isolates are excluded from the calculation of the median swarm diameter.
†Biofilm formation is normalized to a PAO1 control.
Fig. 2.
Increased swimming motility correlates with increased swarming motility among swim-positive/swarm-positive clinical isolates. Swarming and swimming diameters are normalized to PAO1. Median normalized swarm diameters are indicated for each group and are significantly different (P=0.008, Kruskal–Wallis).
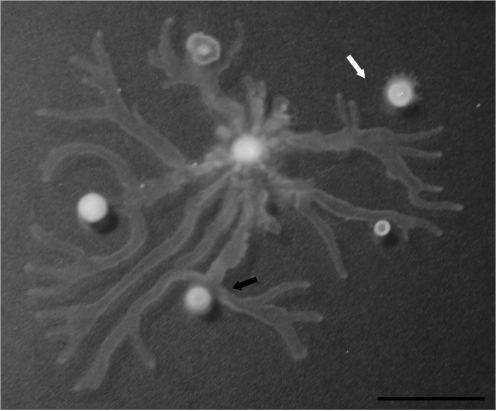
Many swim-positive isolates (n=67) displayed no motility on swarm plates, consistent with observations that factors in addition to flagellar motility are required for swarming (Deziel et al., 2003). We tested whether these isolates were capable of producing wetting agents such as HAAs or rhamnolipids, which appear to be required for swarming motility in most laboratory strains, by examining whether they could repel the leading edge of a PAO1 swarming colony (Fig. 3) (Caiazza et al., 2005; Tremblay et al., 2007). Fully 49 of the 67 isolates were deficient in this assay, and also failed to produce a visible ring of wetting material when grown on swarming plates (Fig. 3). This suggests that production of HAAs and/or rhamnolipids is a major factor regulating the expression of swarming motility in clinical isolates.
Fig. 3.
Most swarm-negative isolates do not produce wetting material. Wild-type PAO1 was inoculated in the centre of the swarming plate. The black arrow shows a swarm-negative clinical isolate that does not produce wetting material and does not repel PAO1, while the white arrow shows PAO1 avoiding a swarm-negative clinical isolate due to the production of wetting material. Bar, 2 cm.
The role of tfp in swarming is still subject to debate, as conflicting data have been published indicating that tfp are required versus dispensable for swarming in vitro (Overhage et al., 2007; Shrout et al., 2006). Swarming colony morphology is affected by the presence or absence of tfp in some laboratory strains, suggesting that tfp are expressed and alter the adhesive properties of swarming bacteria (Murray & Kazmierczak, 2008). We found that twitching motility did not affect swarming frequency or swarm diameter among swim-positive isolates (Table 2). We observed a number of distinct swarming colony morphologies among our clinical isolates (Fig. 1a–d). However, morphology was not related to bacterial swimming or twitching phenotypes in any consistent fashion, as illustrated by Fig. 1.
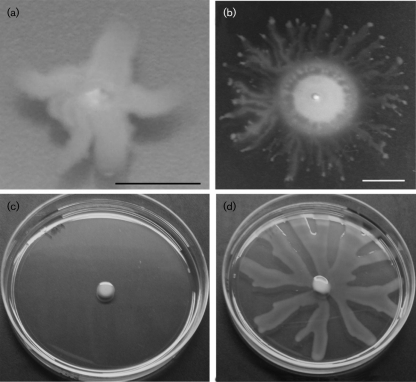
Swarming motility occurs when particular amino acids serve as sole nitrogen sources (Rashid & Kornberg, 2000) and is not usually observed on rich (undefined) media. However, two isolates of 237 displayed the unusual phenotype of swarming on the 0.3 % agar LB plates used to assay swimming motility (Fig. 4); these isolates also produced a visible zone of surfactant under these conditions. We grew the two isolates in liquid LB and examined them by phase-contrast microscopy; both exhibited characteristic swimming motility (data not shown), although only one of the two isolates also generated a swimming zone on 0.3 % agar LB (Fig. 4). The first isolate (Fig. 4a) failed to swarm on 0.5 % agar LB but was able to swarm on 0.5 % agar M8 similar to PAO1 (data not shown). Swarming of this isolate on LB required a lower agar concentration than is routinely needed for this assay. In contrast, the second isolate (Fig. 4b) was capable of swarming on both 0.5 % agar LB (Fig. 4d) and 0.5 % agar M8 (data not shown).
Fig. 4.
Swarming motility on LB plates. Two clinical isolates displayed colony spread consistent with swarming when assayed on 0.3 % agar LB plates. The first isolate (a) displayed no swimming, while the second (b) displayed both a swimming zone and a swarming morphology on the agar surface. Bars, 1 cm. (c) PAO1 does not swarm on 0.5 % agar LB (48 h post-inoculation), while the clinical isolate pictured in (b) shows extensive swarming on 0.5 % agar LB at this time (d).
Swarming isolates show diminished biofilm formation
Biofilm formation provides bacteria with a means of persistently colonizing either living or inert surfaces within a human host. Laboratory strains of P. aeruginosa that fail to twitch or swim form abnormal biofilms (Klausen et al., 2003; O'Toole & Kolter, 1998). Consistent with this observation, swim-positive/twitch-positive isolates scored higher in the in vitro static biofilm assay (median 1.05) than all other isolates (Table 2; P <0.001, Kruskal–Wallis). However, the measured extent of surface attachment varied among isolates with similar motility phenotypes, suggesting that additional factors influence biofilm formation (Table 2).
The relationship between swarming and biofilm formation is debated in the P. aeruginosa literature (Merritt et al., 2007; Shrout et al., 2006). Both behaviours are nutritionally regulated surface activities that are altered in the absence of motility organelles. Since we observed that the absence of either twitching or swimming potentially altered both biofilm and swarming behaviours, we restricted our subsequent analysis to isolates that were positive for both swimming and twitching. Based upon published observations of laboratory strains (Merritt et al., 2007; Yeung et al., 2009), we hypothesized that strains that did not swarm would score higher than swarming isolates in our biofilm assay. We did indeed observe an inverse relationship between swarming motility and biofilm formation among the 154 swim-positive/twitch-positive isolates that grew on M8 swarming plates, as swarm-negative isolates scored significantly higher than swarm-positive isolates in the biofilm assay (Table 3; P=0.009, Kruskal–Wallis).
Table 3.
Phenotype associated with swarming motility among twitch-positive/swim-positive isolates
| Quartile 1 (n=48) | Quartile 2 (n=35) | Quartile 3 (n=30) | Quartile 4* (n=41) | |
|---|---|---|---|---|
| Swarming diameter† | 0–0.09 | 0.1–0.3 | 0.31–0.69 | >0.7 |
| Biofilm formation‡ | 1.41 | 1.02 | 0.98 | 0.88 |
| PopD secretion, median§ | 16 | 19 | 12 | 51 |
| T3SS-positive|| | 25 (52 %) | 18 (53 %) | 15 (50 %) | 35 (85 %) |
*T3SS activity was increased in quartile 4 (P=0.001, Kruskal–Wallis).
†Swarming diameter is normalized relative to PAO1.
‡Biofilm formation is normalized relative to PAO1. Biofilm formation differs across quartiles (P=0.009, Kruskal–Wallis).
§Median expressed as percentage of PA103 control.
||Positive defined as ≥15 % of PA103 control.
Swarm-positive isolates secrete increased levels of proteases
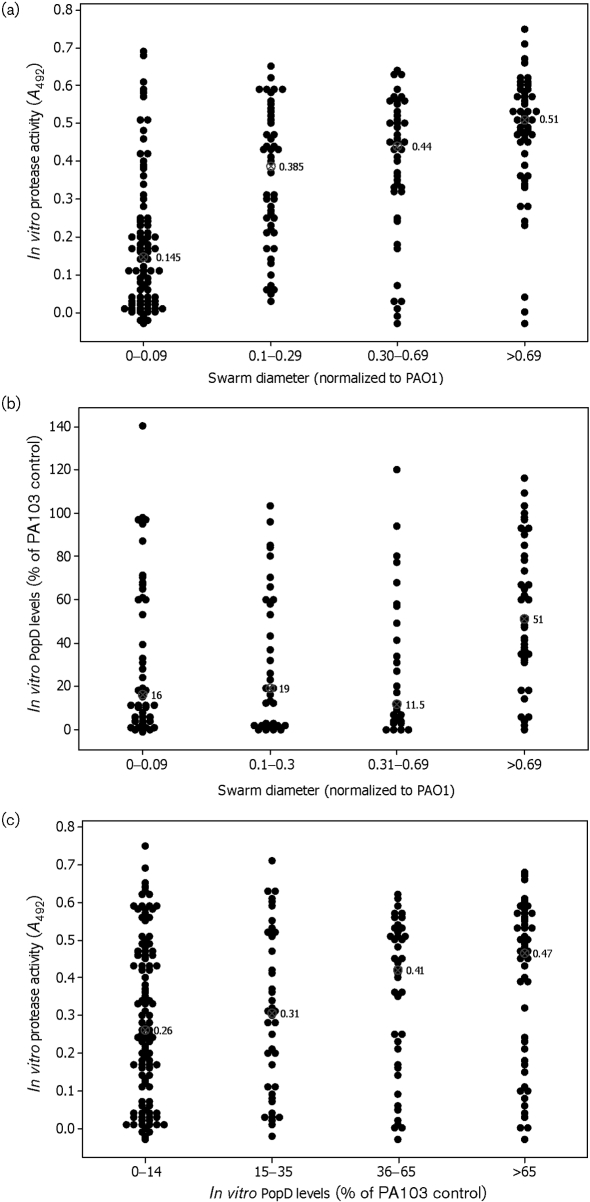
The transcription of genes encoding the extracellular proteases LasB, alkaline protease and protease IV is upregulated in swarming P. aeruginosa (Overhage et al., 2008). We measured casein hydrolytic activity present in bacterial culture supernatants after 24 h of planktonic growth, and asked whether secreted protease activity was correlated with bacterial swarming. Isolates that displayed large swarm diameters exhibited increased secreted caseinolytic activity relative to isolates that displayed reduced or absent swarming (Fig. 5a; P <0.001, Kruskal–Wallis). Because protease secretion was assayed under conditions that do not promote swarming, this result indicates that protease secretion is inherently greater in strains capable of swarming and not simply upregulated during swarming. To determine whether protease activity is required for swarming, we tested a number of transposon insertions in protease genes [lasA (PA1871), lasB (PA3724), aprA (PA1249) and protease IV (PA4175)] from the PAO1 transposon library for their ability to swarm. No single protease mutant was defective for swarming (data not shown).
Fig. 5.
Relationship between swarming, protease activity and type 3 secretion. (a) Swarming is positively correlated with in vitro protease activity (P <0.001, Kruskal–Wallis). (b) Isolates with the largest swarming zones secrete more PopD than isolates with decreased or absent swarming in an in vitro assay for T3SS (P <0.001, Kruskal–Wallis). (c) Isolates that secrete the highest amount of PopD display more active protease activity (P=0.06, Kruskal–Wallis). The median is shown for each group of isolates; values are expressed as percentages of the PA103 control.
Swarming isolates are more likely to be T3SS-positive
T3SS gene expression was induced in actively swarming bacteria (Overhage et al., 2008). We therefore examined whether swim-positive/twitch-positive isolates were more likely to be T3SS-positive in vitro if they were swarm-positive. Eighty-five per cent (35/41) of swim-positive/twitch-positive bacteria that showed the largest swarming colony diameters (>0.7 normalized to PAO1) were T3SS-positive compared with bacteria that showed either no (25/48 T3SS-positive, 52 %) or reduced (18/35 T3SS-positive, 53 %) swarming diameters (Table 3; P=0.001, Kruskal–Wallis). The median amount of PopD secreted by bacteria with the greatest swarming colony diameters (51 % of PA103 control) was also significantly greater than that secreted by bacteria that demonstrated no (16 % of PA103 control) or little (19 % of PA103 control) swarming (Fig. 5b; P <0.001, Kruskal–Wallis). Since bacteria that scored the highest in swarming motility were more likely to secrete both proteases and T3SS effectors, we also examined whether there was a direct correlation between protease secretion and T3SS activity in vitro. Indeed, isolates with higher in vitro T3SS activity showed greater secreted caseinolytic activity than T3SS-negative bacteria (Fig. 5c; P=0.06, Kruskal–Wallis).
Since swarming was inversely correlated with biofilm formation and positively associated with T3SS, we hypothesized there would be an inverse relationship between T3SS and biofilm formation, as reported for laboratory strains harbouring mutations in the sensor kinases GacS, RetS or LadS (Goodman et al., 2004; Kuchma et al., 2005; Laskowski & Kazmierczak, 2006; Ventre et al., 2006). However, most clinical isolates were capable of both biofilm formation and T3SS activity when assayed in vitro (Table 4), and we observed no inverse relationship between biofilm formation and the frequency of T3SS-positive isolates (P=0.3, chi-square). Most clinical isolates, therefore, retain the ability to respond to the environmental cues that elicit one or the other of these behaviours in vitro.
Table 4.
Biofilm formation does not predict T3SS activity among swim-positive/twitch-positive isolates
| Quartile 1 | Quartile 2 | Quartile 3 | Quartile 4 | |
|---|---|---|---|---|
| Biofilm score (n)* | <0.75 (40) | 0.75–1.04 (40) | 1.05–1.59 (40) | ≥1.6 (39) |
| PopD secretion, median† | 32 | 14.5 | 40 | 20 |
| T3SS-positive (n=94)‡ | 26/40 (65 %) | 20/40 (50 %) | 26/40 (65 %) | 23/39 (59 %) |
*Biofilm formation is normalized to PAO1. The number of isolates in each quartile is indicated.
†Median is expressed as a percentage of the PA103 control.
‡Positive defined as ≥15 % of PA103 control; percentage of T3SS-positive strains across quartiles not significantly different (P=0.3, chi-square).
Several authors have described phenotypes that suggest co-ordinated regulation between the T3SS and tfp (Beatson et al., 2002; Whitchurch et al., 2005) or between T3SS and flagella (Soscia et al., 2007). We examined whether T3SS was associated with tfp-dependent twitching and flagella-dependent swimming motility in our clinical isolates. There was no apparent relationship between T3SS and flagellar motility in our isolates, as 48 % (16/33) of swim-negative isolates were T3SS-positive compared with 55 % (113/204) of swim-positive isolates (P=0.46, chi-square test). However, twitch-positive isolates were significantly more likely to be T3SS-positive, 60 % (108/180), compared with twitch-negative isolates, 38 % (21/56) (P=0.003, chi-square test).
Relationship of swarming motility to virulence factors of P. aeruginosa
We have collected and examined P. aeruginosa isolates from 237 unique individuals hospitalized at a tertiary care hospital. The isolates were obtained from patients whose clinical presentations ranged from no signs of acute infection to life-threatening acute infection and sepsis. The isolates displayed diverse motility, biofilm and protein secretion phenotypes. A primary goal of our work was to analyse swarming motility in a large sample of clinical isolates, as the role of swarming in pathogenesis is poorly understood. Additionally, we examined whether relationships between swarming motility and other phenotypes associated with virulence initially proposed from studies of laboratory strains could also be observed in a large set of clinical isolates. The results of this analysis support several important conclusions.
The role of swarming motility in human colonization and infection has not been previously addressed. In our study, P. aeruginosa swarming motility was associated with neither increased nor decreased risk of infection (M. Ledizet and others, unpublished). However, a majority of clinical isolates were capable of swarming motility in vitro. The isolates displayed varied patterns of tendril formation and a range of colony sizes as illustrated in Fig. 1. Swarming motility is promoted by the production of bacterial wetting agents, such as HAAs and mono- and di-rhamnolipids (Caiazza et al., 2005; Deziel et al., 2003; Tremblay et al., 2007). In our series, the majority of swim-positive isolates that failed to swarm did not produce detectable wetting agents, which may have contributed to this motility defect.
Biofilm formation is a behaviour of sessile bacteria; nonetheless, it shares many features with swarming motility. Both are community behaviours that bacteria exhibit on surfaces. Common sets of bacterial surface structures (flagella, tfp, EPS) and secreted products (rhamnolipids) influence both biofilm and swarming behaviours, and common nutritional cues affect the extent and structure of both swarming colonies and biofilms (Shrout et al., 2006). However, it is not clear whether swarming and biofilm formation represent bacterial commitment to more or less exclusive modes of behaviour, as is suggested by some experimental observations. Phenotypes such as increased EPS formation suppress swarming and increase biofilm formation among laboratory isolates (Caiazza et al., 2007). Furthermore, swarming-deficient mutants identified in the PA14 transposon insertion mutant library illustrate an inverse relationship between swarming motility and biofilm formation even in the absence of obvious swimming or twitching motility defects (Yeung et al., 2009).
Although we observed a range of behaviours in our clinical isolates, in general non-swarming isolates scored higher in the biofilm assay than isolates with the largest swarming diameters (Table 3). This held true after we controlled for both twitching and swimming phenotypes, which can affect biofilm formation and swarming (Klausen et al., 2003; O'Toole & Kolter, 1998). Many of the swarm-negative strains did not produce detectable wetting agents such as rhamnolipids in vitro. As rhamnolipids are associated with biofilm dispersion, their absence could contribute to the increased biofilm formation observed for these swarm-negative isolates (Boles et al., 2005; Irie et al., 2005). Only seven isolates (0.3 %) scored higher than the reference strain PAO1 in assays for both biofilm formation and swarming motility. These data are consistent with the hypothesis that swarming and biofilm formation represent distinct and largely non-overlapping mechanisms for surface colonization, rather than a continuum of behaviours, and suggest that clinical isolates exhibit a bias for one or the other behaviour that persists when they are analysed ex vivo.
Laboratory strains secrete type 3 exotoxins on swarming plates, suggesting that the environmental cues that trigger swarming behaviour may also activate the T3SS (Overhage et al., 2008). While our data do not explicitly test this hypothesis, isolates with the largest swarming zones were significantly more likely to be T3SS-positive than all other isolates. Likewise, swarm-positive isolates were also found to secrete significantly more proteases than swarm-negative isolates. These data are consistent with the hypothesis that a common set of environmental signals may trigger swarming, T3SS and protease secretion. Specific isolates may respond to these cues more robustly than other isolates.
We observed that the majority of clinical isolates could both form static biofilms and secrete T3SS effectors in vitro. These two behaviours are associated with distinct gene expression profiles (Goodman et al., 2004). Laboratory strains in which the sensor kinases LadS, GacS or RetS or the response regulator GacA have been mutated can be locked into one mode of behaviour or the other (Goodman et al., 2004; Kuchma et al., 2005; Laskowski & Kazmierczak, 2006; Ventre et al., 2006). However, it is not surprising that isolates in which these proteins are presumably functional demonstrate both biofilm formation and T3SS activity in response to the appropriate environmental cues. This is distinct from the situation described for chronically colonized patients with CF, where the ability to secrete T3SS effectors is likely selected against during bacterial adaptation to the CF airway (Smith et al., 2006). Most isolates from the current study retain the ability to carry out behaviours typically associated with acute infection and persistence, suggesting that the ability to respond to a range of environments – rather than specialization for a particular type of behaviour – is a general characteristic of clinical P. aeruginosa isolates from non-CF patients.
Acknowledgments
We thank Maria Lebron and Xiao Bai for technical assistance, and Roberta Wilkerson for assistance with patient enrolment. This publication was made possible by CTSA grant KL2 RR024138 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH roadmap for research and Yale Child Health Research Center grant K12 HD001401 (T. S. M.), the Patrick and Catherine Weldon Donaghue Medical Research Foundation (B. I. K.) and NIH grant R42 AI058659 (B. I. K.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases, the NCRR or NIH.
Abbreviations
CF, cystic fibrosis
EPS, exopolysaccharide
3-HAA, 3-(3-hydroxyalkanoyloxy)alkanoic acid
T3SS, type 3 secretion system
tfp, type IV pili
References
- Balloy, V., Verma, A., Kuravi, S., Si-Tahar, M., Chignard, M. & Ramphal, R. (2007). The role of flagellin versus motility in acute lung disease caused by Pseudomonas aeruginosa. J Infect Dis 196, 289–296. [DOI] [PubMed] [Google Scholar]
- Beatson, S. A., Whitchurch, C. B., Sargent, J. L., Levesque, R. C. & Mattick, J. S. (2002). Differential regulation of twitching motility and elastase production by Vfr in Pseudomonas aeruginosa. J Bacteriol 184, 3605–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles, B. R., Thoendel, M. & Singh, P. K. (2005). Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol Microbiol 57, 1210–1223. [DOI] [PubMed] [Google Scholar]
- Caiazza, N. C., Shanks, R. M. & O'Toole, G. A. (2005). Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J Bacteriol 187, 7351–7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazza, N. C., Merritt, J. H., Brothers, K. M. & O'Toole, G. A. (2007). Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189, 3603–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deziel, E., Lepine, F., Milot, S. & Villemur, R. (2003). rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149, 2005–2013. [DOI] [PubMed] [Google Scholar]
- Feldman, M., Bryan, R., Rajan, S., Scheffler, L., Brunnert, S., Tang, H. & Prince, A. (1998). Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect Immun 66, 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltman, H., Schulert, G. S., Khan, S., Jain, M., Peterson, L. & Hauser, A. (2001). Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147, 2659–2669. [DOI] [PubMed] [Google Scholar]
- Fonseca, A. P., Correia, P., Sousa, J. C. & Tenreiro, R. (2007). Association patterns of Pseudomonas aeruginosa clinical isolates as revealed by virulence traits, antibiotic resistance, serotype and genotype. FEMS Immunol Med Microbiol 51, 505–516. [DOI] [PubMed] [Google Scholar]
- Goodman, A. L., Kulasekara, B., Rietsch, A., Boyd, D., Smith, R. S. & Lory, S. (2004). A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell 7, 745–754. [DOI] [PubMed] [Google Scholar]
- Head, N. E. & Yu, H. (2004). Cross-sectional analysis of clinical and environmental isolates of Pseudomonas aeruginosa: biofilm formation, virulence, and genome diversity. Infect Immun 72, 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie, Y., O'Toole, G. A. & Yuk, M. H. (2005). Pseudomonas aeruginosa rhamnolipids disperse Bordetella bronchiseptica biofilms. FEMS Microbiol Lett 250, 237–243. [DOI] [PubMed] [Google Scholar]
- Kang, P. J., Hauser, A. R., Apodaca, G., Fleiszig, S. M., Wiener-Kronish, J., Mostov, K. & Engel, J. N. (1997). Identification of Pseudomonas aeruginosa genes required for epithelial cell injury. Mol Microbiol 24, 1249–1262. [DOI] [PubMed] [Google Scholar]
- Klausen, M., Heydorn, A., Ragas, P., Lambertsen, L., Aaes-Jorgensen, A., Molin, S. & Tolker-Nielsen, T. (2003). Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol 48, 1511–1524. [DOI] [PubMed] [Google Scholar]
- Kohler, T., Curty, L. K., Barja, F., van Delden, C. & Pechere, J. C. (2000). Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol 182, 5990–5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchma, S. L., Connolly, J. P. & O'Toole, G. A. (2005). A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J Bacteriol 187, 1441–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski, M. A. & Kazmierczak, B. I. (2006). Mutational analysis of RetS, an unusual sensor kinase-response regulator hybrid required for Pseudomonas aeruginosa virulence. Infect Immun 74, 4462–4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Ledizet, M., Kar, K., Koski, R. A. & Kazmierczak, B. I. (2005). An indirect enzyme-linked immunosorbent assay for rapid and quantitative assessment of Type III virulence phenotypes of Pseudomonas aeruginosa isolates. Ann Clin Microbiol Antimicrob 4, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyczak, J. B., Cannon, C. L. & Pier, G. B. (2000). Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect 2, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Mattick, J. S. (2002). Type IV pili and twitching motility. Annu Rev Microbiol 56, 289–314. [DOI] [PubMed] [Google Scholar]
- Merritt, J. H., Brothers, K. M., Kuchma, S. L. & O'Toole, G. A. (2007). SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J Bacteriol 189, 8154–8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, T. S. & Kazmierczak, B. I. (2008). Pseudomonas aeruginosa exhibits sliding motility in the absence of type IV pili and flagella. J Bacteriol 190, 2700–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole, G. A. & Kolter, R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30, 295–304. [DOI] [PubMed] [Google Scholar]
- Overhage, J., Lewenza, S., Marr, A. K. & Hancock, R. E. (2007). Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 mini-Tn5-lux mutant library. J Bacteriol 189, 2164–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overhage, J., Bains, M., Brazas, M. D. & Hancock, R. E. (2008). Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J Bacteriol 190, 2671–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkins, M. D., Ceri, H. & Storey, D. G. (2001). Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol Microbiol 40, 1215–1226. [DOI] [PubMed] [Google Scholar]
- Potvin, E., Lehoux, D. E., Kukavica-Ibrulj, I., Richard, K. L., Sanschagrin, F., Lau, G. W. & Levesque, R. C. (2003). In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ Microbiol 5, 1294–1308. [DOI] [PubMed] [Google Scholar]
- Rashid, M. H. & Kornberg, A. (2000). Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 97, 4885–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrout, J. D., Chopp, D. L., Just, C. L., Hentzer, M., Givskov, M. & Parsek, M. R. (2006). The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol 62, 1264–1277. [DOI] [PubMed] [Google Scholar]
- Singh, P. K., Schaefer, A. L., Parsek, M. R., Moninger, T. O., Welsh, M. J. & Greenberg, E. P. (2000). Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407, 762–764. [DOI] [PubMed] [Google Scholar]
- Smith, E. E., Buckley, D. G., Wu, Z., Saenphimmachak, C., Hoffman, L. R., D'Argenio, D. A., Miller, S. I., Ramsey, B. W., Speert, D. P. & other authors (2006). Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 103, 8487–8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soscia, C., Hachani, A., Bernadac, A., Filloux, A. & Bleves, S. (2007). Cross talk between type III secretion and flagellar assembly systems in Pseudomonas aeruginosa. J Bacteriol 189, 3124–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay, J., Richardson, A. P., Lepine, F. & Deziel, E. (2007). Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ Microbiol 9, 2622–2630. [DOI] [PubMed] [Google Scholar]
- Ventre, I., Goodman, A. L., Vallet-Gely, I., Vasseur, P., Soscia, C., Molin, S., Bleves, S., Lazdunski, A., Lory, S. & other authors (2006). Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci U S A 103, 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch, C. B., Beatson, S. A., Comolli, J. C., Jakobsen, T., Sargent, J. L., Bertrand, J. J., West, J., Klausen, M., Waite, L. L. & other authors (2005). Pseudomonas aeruginosa fimL regulates multiple virulence functions by intersecting with Vfr-modulated pathways. Mol Microbiol 55, 1357–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung, A. T., Torfs, E. C., Jamshidi, F., Bains, M., Wiegand, I., Hancock, R. E. & Overhage, J. (2009). Swarming of Pseudomonas aeruginosa is controlled by a broad spectrum of transcriptional regulators, including MetR. J Bacteriol 191, 5592–5602. [DOI] [PMC free article] [PubMed] [Google Scholar]