Summary
The mammalian Ajuba LIM proteins (Ajuba, LIMD1, WTIP) are cytosolic adapter proteins that exhibit the potential to communicate cell adhesive events with nuclear responses to remodel epithelia [1] [2]. Determining their role(s) in vivo, however, has been challenging due to overlapping tissue expression and functional redundancy. Thus, we turned to Drosophila where a single gene, CG11063 or djub, exists. Drosophila containing djub mutant loss-of-function alleles or depleted of dJub by RNAi identify djub as an essential gene for development and novel regulator of epithelial organ size as a component of the conserved Hippo pathway, which has been implicated in both tissue size control and cancer development [3-5] [6-9]. djub-deficient tissues were small, had decreased cell numbers as a result of increased apoptosis and decreased proliferation, due to downregulation of DIAP1 and cyclin E. This phenocopies tissues deficient for Yorkie (Yki), the downstream target of the Hippo pathway. djub genetically interacts with the Hippo pathway, and epistasis suggests that djub lies downstream of hpo. In mammalian and Drosophila cells, Ajuba LIM proteins/dJub interact with LATS/Wts and WW45/Sav to inhibit phosphorylation of YAP/Yki. This work describes a novel role for the Ajuba LIM proteins as negative regulators of the Hippo signaling pathway.
Keywords: LIM proteins, Hippo pathway, Drosophila
Results and Discussion
The Drosophila orthologue of mammalian Ajuba LIM proteins, dJub, regulates organ size
In Drosophila there is a single orthologue of the mammalian Ajuba subfamily of LIM proteins encoded by the CG11063 locus in the X chromosome [10]. CG11063 exhibits greater sequence similarity to the three mammalian Ajuba subfamily proteins, than to dZyx, the Drosophila orthologue of the Ajuba-related Zyxin subfamily of LIM proteins (Fig. S1gA). We designate CG11063 as dJub (Drosophila Ajuba LIM proteins).
To determine the in vivo function(s) of djub in Drosophila we generated two dJub RNAi lines: djub-RNAi 22.5 and djub-RNAi 18.1 (Fig. S1A). Ubiquitous expression of either, using GAL4/UAS and actin-GAL4, resulted in pharate lethality, suggesting that djub is an essential gene. Both RNAi constructs yielded similar phenotypes in all subsequent assays. Since djub-RNAi 22.5 consistently induced stronger phenotypes we use RNAi 22.5 when referring to dJub RNAi.
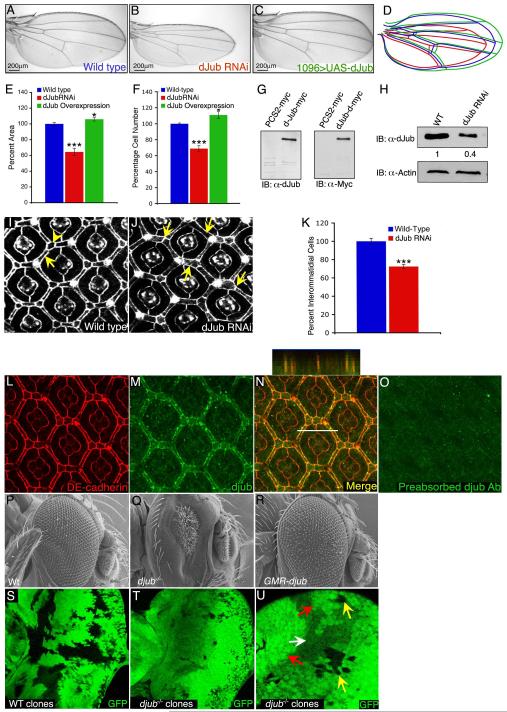
Since Ajuba LIM proteins are abundant in mammalian epithelia [11] and implicated in epithelia functions [1] [2], we selectively depleted dJub in larval wing and eye imaginal disc epithelium. dJub RNAi expression in the wing, decreased wing size to 65% of wild type (Fig. 1B and E). Western blot analysis revealed a 60% reduction of dJub protein level by the RNAi (Fig. 1H). The small wing phenotype was due to decreased cell number, not cell size, and wing patterning appeared unaffected (Fig. 1B,F). Similarly, expression of dJub RNAi in the pupal eye epithelium resulted in a 25% reduction in interommatidial cells, without disruption to ommatidial patterning (Fig. 1J,K). These RNAi phenotypes were specific for dJub depletion, as overexpression of a wt djub transgene in dJub RNAi-expressing cells partially rescued both wing and eye phenotypes (Fig. S1D,H). Furthermore, overexpression of human LIMD1 (most closely related to dJub) rescued the dJub RNAi wing phenotype (Fig. S1G,H), suggesting that this function of Ajuba LIM proteins is conserved between Drosophila and mammals. dJub and hLIMD1 overexpression in wings and eyes resulted in a modest increase in size, due to increased cell number (Fig. 1C,E,F,R). In pupal eye epithelium dJub localized to adherens junctions (AJs), predominantly interommatidial cells, co-localizing with DE-cadherin in a punctate pattern (Fig. 1L-O). The HA-LIMD1 transgene also localized to AJs in wing larval disc epithelia (Fig. S1I). This cellular localization of dJub is similar to that of mammalian Ajuba LIM proteins in mammalian epithelia [1].
Figure 1. dJub regulates tissue size by controlling cell number.
(A-C) Wings from wt female flies (A), female flies expressing dJub RNAi (B), or female flies expressing dJub-mCherry transgene (C). 1096-Gal4 was used to drive RNAi or transgene expression. (D) Outlines of the wings in panels A-C. (E, F) Quantification of relative wing areas (E) and cell numbers (F) of genotypes in A-C. Area and cell number measurements were taken from the wing region located between veins L4 and L5, and wt defined as 100% (N=20 for each). (G) Extracts of mammalian HEK293 cells transfected with myc-dJub immunoblotted with dJub antiserum (left) or Myc antiserum (right). (H) Immunoblot analysis of dJub protein levels in wt or dJub RNAi-expressing larval eye imaginal discs. Actin serves as loading control. Relative amount of dJub protein is indicated below each lane. Mid-pupal wt eyes (I) or GMR-Gal4 driven dJub RNAi expressing eyes (J) stained for DE-cadherin. Secondary (arrows) and tertiary (arrowheads) interommatidial cells are highlighted. Loss of interommatidial cells in dJub RNAi expressing pupal eyes denoted by arrows (J). (K) Quantification of relative numbers of interommatidial cells in wt versus dJub RNAi pupal eye. Interommatidial cells were counted in 20 fields, each containing a cluster of at least 7 ommatidia. (L-O) Mid-pupal wt eyes stained for DE-cadherin (L), dJub (M), and merged image (N). Z-stack analysis of line in N is shown above panel N (N). (O) Immunostaining with dJub antiserum preabsorbed with immunizing peptide. (P-R) Scanning electron micrographs (SEMs) of female adult eyes. WT (Q), djubI generated via the EGUF-Hid method, which results in eyes composed almost entirely of mutant tissue (Q), and GMR-gal4 driven overexpression of UAS-dJub-mCherry transgene (R). (S-U) Female third-instar larval eye imaginal discs containing wt (S) or djubI mutant (T, U) clones marked by the absence of GFP expression (black). (U) Enlarged view of djubI and wt twin spot clones. Yellow arrow identifies djubI clones, red arrow identifies wt twin spot clone containing two copies of Ubi-GFP, and white arrow identifies tissue carrying one copy of Ubi-GFP. In all experiments wings and eyes were dissected from female flies. In graphs data are shown as mean percentages +/− standard deviation, with N= 20 for each genotype. (***) Represents p-value ≤ 0.001 and (*) represents p-value ≤ 0.05. Anterior is to the left for all larval imaginal discs.
We generated djub mutant alleles using FLP-FRT based methods [12]. Two distinct, yet overlapping, deficiencies of the djub locus were made (Fig. S2A). Both deficiencies yielded identical results for all phenotypic studies. Flies hemizygous for each deficiency died at the late embryonic to first instar larval stage. Female flies (heterozygous for djubI or djubII) expressed 50% level of dJub protein (Fig. S2C). Ubiquitous expression of a wt djub transgene rescued lethality of both alleles, confirming that the loss of djub, and not flanking genetic material, was responsible and that djub is an essential gene.
When dJub was deleted in the eye, using eyeless-FLP (EGUF/hid) to produce eyes composed of over 90% djubI mutant cells [13] adult eyes were severely reduced in size (Fig. 1Q). Mosaic analysis of djubI and wt twin-spot clones in eye and wing imaginal discs resulted in djubI mutant clones (Fig 1T,U yellow arrows) that were significantly smaller than wt twin-spot clones (Fig 1S,U red arrow). To verify that growth defects were specific to loss of djub function, we induced djubI clones throughout the wing imaginal disc while simultaneously expressing wt mCherry-tagged djub transgene only in the posterior half of the wing disc using engrailed-gal4. In the anterior compartment djubI clones were small and few in number (Fig. S2E). In contrast, the posterior compartment contained more and larger clones, similar to wt clones induced in a wt background (Fig. S2D-F).
djub mutant clones exhibit reduced proliferation and increased apoptosis
The growth phenotype of djubI clones could result from decreased cell proliferation and/or increased apoptosis. In wt larval eye discs undifferentiated cells lie anterior to the morphogenetic furrow and undergo asynchronous cell divisions (Fig. S1J, white arrow). Posterior to the furrow cells differentiate or undergo one more cell division – the second mitotic wave (Fig. S1J, yellow arrow), after which they differentiate or die [14, 15]. Bromodeoxyuridine (BrdU) labeling of wt and djubI eye discs, generated via the EGUF-Hid method, revealed that djubI eye discs displayed a strong reduction in cells undergoing asynchronous division anterior to the furrow (Fig. S1J’, white arrow) and a near complete loss of the second mitotic wave (Fig. S1J’, yellow arrow).
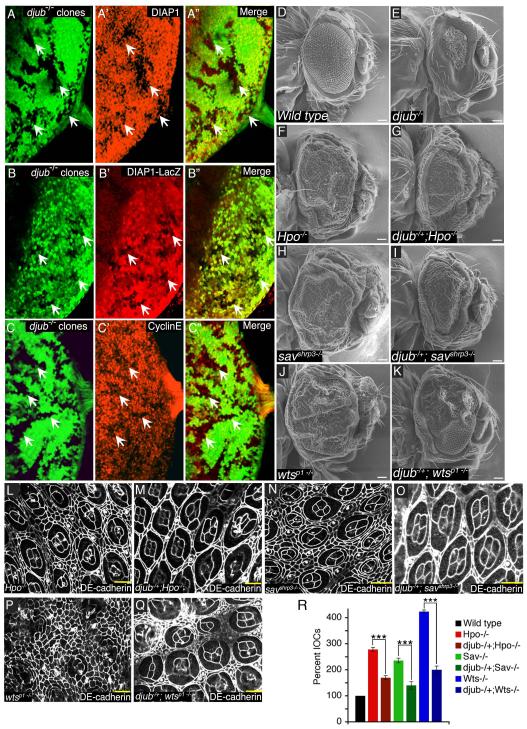
During eye development apoptosis determines the final number of cells in the eye [16]. Staining eye discs for activated caspase-3 revealed that djubI eye discs contained increased number of caspase-3-positive cells (Fig. S1K’). When the caspase inhibitor P35 was coexpressed with djubI the small eye phenotype was partially rescued (Fig. S3B). Relative to wt, djubI clones exhibited decreased levels of Drosophila inhibitor of apoptosis-1 (DIAP1) (Fig. 2A) and Cyclin E (Fig. 2C). dJub controlled transcription of DIAP1 as djubI clones expressed less lacZ, under control of the diap1 gene promoter (Fig. 2B). These data indicate that dJub regulates organ size by inhibiting apoptosis and promoting cell proliferation by influencing DIAP1 and Cyclin E expression, respectively. djub deletion did not affect adherens junction organization, as determined by DE-cadherin staining of djubI clones (Fig. S3C).
Figure 2. djub affects expression of DIAP1 and Cyclin E and genetically interacts with the Hippo pathway.
(A-C) Female third-instar larval eye imaginal discs containing djubI clones (GFP –ve, yellow arrows) generated by Eyeless-flp and stained for DIAP1 (A’), DIAP1-lacZ (B’), or Cyclin E (C’) expression. Anterior is to the left for all larval eye imaginal discs. (D-K) SEMs of adult female Drosophila eyes of wt (E) and Hippo pathway mutants (F,H,J) and djubI (E) or Hippo pathway mutants containing a deletion of a single copy of djub (G,I,K), as indicated. Mid-pupal eye dissection of Hippo pathway mutants (L,N,P) or Hippo pathway mutants containing a deletion of a single copy of djub (M,O,Q), as indicated, and stained for DE-cadherin to identify interommatidial cells. Scale bars in (D-K) equal 100μm and (L-Q) equal 10μm. (R) Quantification of interommatidial cell numbers in 10 random fields containing 10 ommatidia each of genotypes (D,F-K). Data are shown as mean percentages +/− standard deviation and (***) represents p-value ≤ 0.001.
DJub genetically interacts with the Hippo pathway
The djub loss of function phenotype resembles yorkie (yki), which encodes a transcriptional coactivator, the activity of which is antagonized by the Hippo signaling pathway. Active Yki promotes proliferation and inhibits apoptosis by facilitating transcription of Cyclin E and DIAP1 [17, 18]. Activated Hpo kinase, in association with Salvador (Sav), a WW-repeat scaffolding protein, phosphorylates and activates Wts kinase, which in association with Mob-as-tumor-suppressor protein (Mats) phosphorylates Yki. Phosphorylation of Yki directs Yki to the cytoplasm inhibiting its transcriptional regulation [8]. Given the similarity between djub and yki loss of function phenotypes we hypothesized that dJub governed organ size by affecting Yki activity either directly, or indirectly by inhibiting Hippo pathway function.
Hippo pathway mutants (hpo, sav, wts) produce overgrown adult eyes (Fig. 2F,H,J) and pupal eyes with increased interommatidial cells (Fig. 2L,N,P,R and Fig. S3D,E,F) [19, 20]. Removing a copy of djub reduced the magnitude of hpo and sav phenotypes (Fig. 2G,I and M,O,R), and modestly affected the wts phenotype (Fig. 2K and Fig. 2Q,R). In a reciprocal manner, a 50% reduction in Wts suppressed the dJub RNAi small wing phenotype (Fig. S4A-E), while a 50% reduction of Yki enhanced this phenotype (Fig. S4F-J). Taken together these suggest the possibility that djub and the Hippo pathway genetically interact. If so, then djub specifically interacted with the Hippo pathway as no genetic interactions were observed between djub and myc or components of the Insulin receptor signaling pathway, known to regulate organ size by affecting cell size (Fig. S4K). DJub localization to AJs was unaltered in wts, hpo, and sav mutant pupal eyes (Fig. S3D-F).
Epistatic analysis suggests that djub acts upstream of wts and yki but downstream of hpo
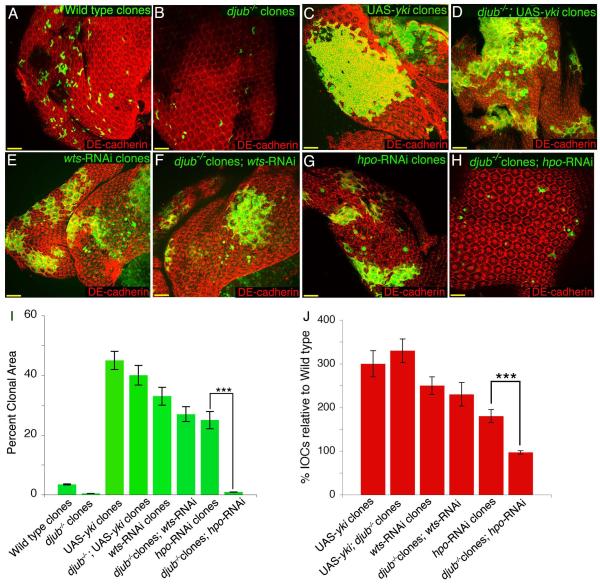
To confirm that djub indeed genetically interacts with the Hippo pathway we performed genetic epistasis experiments between djub and yki, wts and hpo. MARCM pupal eye clones of djubI alone result in small clones (Fig. 3B, quantified in I, Fig. S2H), whereas MARCM clones overexpressing Yki or depleted of Wts or Hpo exhibit increased clonal area (Fig. 3C,E,G, quantified in I) as well as overproliferation of interommatidial cells [17, 21-23] (Fig. S2I,K,M, quantified in Fig. 3J). djubI mutant MARCM clones overexpressing Yki displayed a phenotype identical to overexpression of Yki alone (Fig. 3D, quantified in I and Fig. S2J, quantified in Fig. 2J). djubI mutant MARCM clones depleted of Wts, resembled wts RNAi clones (Fig. 3F, quantified in I and Fig. S2L, quantified in Fig. 2J), however, removing djub in hpo RNAi MARCM clones resulted in a djubI-like phenotype (Fig. 3H, quantified in I and Fig. S2N, quantified in Fig. 2J). This epistatic analysis suggested that djub acts downstream to hpo but upstream of wts and yki, but since the core Hippo pathway proteins (Hpo, Sav, Wts, and Mats) are thought to function as a complex, a precise epistatic relationship is difficult to conclude.
Figure 3. djub is epistatic to hpo based on clonal area and interommatidial cell numbers.
(A-H) Female mid-pupal eyes stained for DE-cadherin (red). Wt, GFP positive MARCM clones (A); djubI MARCM clones (GFP +ve) (B); MARCM clones overexpressing Yki (GFP +ve) (C); MARCM clones mutant for djubI and overexpressing Yki (GFP +ve) (D); MARCM clones expressing wts RNAi (GFP +ve) (E); MARCM clones mutant for djubI and expressing wts RNAi (GFP +ve) (F); MARCM clones expressing hpo RNAi (GFP +ve) (G); MARCM clones mutant for djubI and expressing hpo RNAi (GFP +ve) (H). (I) Quantification of the clonal area (GFP +ve) for each genotype as a percentage of the entire pupal eye area. (J) Quantification of the percent increase of interommatidial cells within the clonal area (GFP +ve) as compared to wild type (set at 100% IOCs) for each genotype. In graphs data are shown as mean percentages +/− standard deviation, with N= 10 for each genotype. (***) Represents p-value ≤ 0.001. Scale bars in (A-H) equal 20μm.
Ajuba LIM proteins/dJub associate with LATS/Wts and WW45/Sav in mammalian and Drosophila cells, and influence YAP activity in mammalian cells
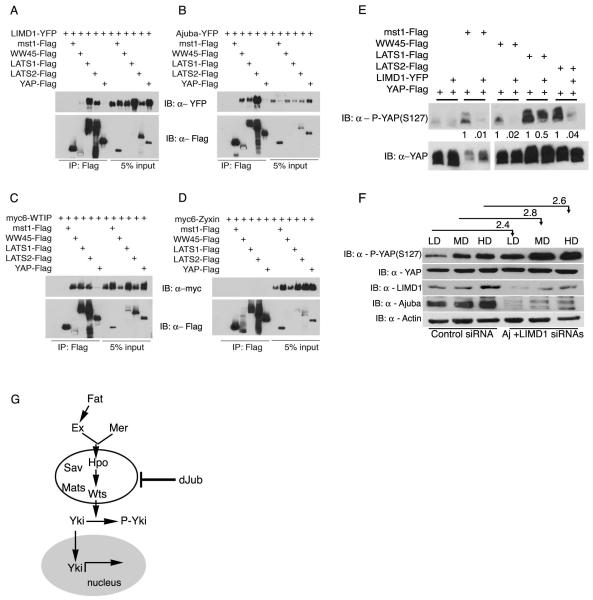
The Hippo pathway is highly conserved between Drosophila and vertebrates [5, 8, 23, 24], and human LIMD1 rescues the cell growth defects of dJub depleted Drosophila wings (Fig. S1G, H). To determine whether Ajuba LIM proteins physically and functionally interact with Hippo pathway components in cells, we tested whether the mammalian homologs of dJub (Ajuba, LIMD1 and WTIP) associated with (i.e., co-immunoprecipitated) mammalian orthologues of Hippo pathway members in human epithelial cells. All three Ajuba subfamily members associated strongly with LATS1/2, and Ajuba and WTIP associated with WW45, but none associated with MST1/2 or YAP (Fig. 4A-C). The interaction between LATS and WW45 and Ajuba family proteins was specific as Zyxin, a closely related LIM protein, failed to associate with either LATS or WW45 (Fig. 4D). In Drosophila S2 cells dJub associated with Wts and Sav but not Hpo (Fig. S4L). A weak association between dJub and Yki was noted but this was >10 fold less than that observed for Wts and Sav, and may be nonspecific as transfected Yki was massively overexpressed in S2 cells (Fig. S4L).
Figure 4. Ajuba LIM proteins associate with components of the Hippo pathway in mammalian cells and influence YAP phosphorylation.
(A-D) HEK293 cells were co-transfected with LIMD1-YFP (A), Ajuba-YFP (B), Myc-WTIP (C), or Myc-Zyxin (D) and Flag-tagged Mst1, Lats1/2, WW45 or YAP, as indicated. Cell lysates were immunoprecipitated for each Hippo pathway member (anti-Flag), and bound products Immunoblotted (IB) for the presence of each LIM protein (anti-YFP or anti-Myc). Immunoblots of input controls (5%) are shown on the right side of each panel. (E) HEK293 cells were transfected with the indicated member of the Hippo pathway in the absence or presence of LIMD1-YFP. Levels of phospho-S127-YAP (upper panel) or total YAP (lower panels) were then determined by immunoblot (IB) analysis. Relative amounts of phospho-S127-YAP with respect to total YAP protein is indicated below each lane. (F) MDCK cells were transfected with control Luc siRNA (lanes 1-3) or Ajuba and LIMD1 siRNAs (lanes 4-6) and then plated at low (LD), medium (MD) and high density (HD). Amount of S127-YAP phosphorylation relative to total YAP for each density within control and Ajuba/LIMD1 depleted cells was determined by immunoblotting. The relative level of YAP phosphorylation for each density between control and Ajuba/LIMD1 depleted cells was determined and is indicated above the lanes. (G) Working model, based upon results herein, for how Ajuba LIM proteins could influence Hippo pathway signaling.
To determine if these protein interactions were functionally relevant we asked whether Ajuba LIM proteins affected YAP phosphorylation [8, 18, 25]. In cells transfected with MST1, WW45, LATS1/2 alone there was variable increase in phospho-YAP levels, however, when co-transfected with LIMD1 phospho-YAP levels were decreased in all instances (Fig. 4E). Overexpression of dJub in drosophila imaginal discs did not change the level or subcellular localization pattern of Yki or other Hippo pathway targets namely, Ex and Diap1. This may be because only a small (10%) increase in wing size occurs in wings overexpressing dJub (Fig. 1C, E, F). In another approach, Ajuba and LIMD1 were RNAi-depleted in MDCK cells, and phospho-YAP levels determined in cells at differing density [7]. Analysis of MDCK cells depleted of all three Ajuba LIM proteins was not possible as cells died, like drosophila cells lacking dJub. Compared to control MDCK cells, in cells depleted of Ajuba and LIMD1 basal phospho-YAP levels were increased 2.5 fold at all densities (Fig. 4F). These results demonstrated that mammalian Ajuba LIM proteins/dJub specifically associate with LATS/Wts and WW45/Sav in cells, and in mammalian cells these associations antagonize the phosphorylation of YAP.
Our work indicates that the Ajuba LIM proteins influence organ growth through negative regulation of the Hippo signaling pathway in flies, and likely mammals. Genetic and biochemical data suggest that Ajuba LIM proteins/dJub likely interface with the Hippo pathway at the level of LATS/Wts. Prior work has described an interaction between Ajuba and LATS at centrosomes that influences mitotic centrosome/spindle organization [26]. Whether this contributes to the djubI phenotype we cannot exclude, as mitotic damage can lead to apoptotic cell death.
Precisely how Ajuba LIM proteins (dJub) influence LATS/Wts mediated inactivation of YAP/Yki remains to be determined, but possibilities include: inhibition of LATS/Wts activation by upstream kinases (MST/Hpo), inhibition of the ability of LATS/Wts to phosphorylate YAP/Yki, or affecting the subcellular localization of LATS/Wts or WW45/Sav and thus their access to the Hippo pathway. The regulatory relationship between Ajuba LIM proteins/dJub and LATS/Wts may not be simply unidirectional as LATS has been shown to phosphorylate Ajuba [26].
Ajuba LIM proteins are components of AJs. Upstream members of the Hippo pathway, including atypical cadherins (Fat, dachsous), Expanded and Merlin, also localize to AJs, leading to the hypothesis that AJs are nodal points for initiation/regulation of Hippo signaling [25, 27-32]. How these upstream components activate MST/Hpo kinase is unknown. The Hippo pathway is thought to regulate cell contact growth inhibition [7]. In sub-confluent non-contacted cells, Ajuba LIM proteins are cytosolic while YAP is nuclear and cells proliferate [7]. When cells achieve confluence Ajuba proteins are recruited to AJs while YAP is phosphorylated and re-localized to the cytosol and cell proliferation ceases. Whether these events are related is not known, but given that Ajuba proteins associate with and inhibit LATS/Wts-mediated phosphorylation of YAP raises the possibility that the recruitment of Ajuba LIM proteins/dJub to AJ in confluent cell cultures may “release” LATS/Wts allowing for Hippo pathway mediated YAP/Yki phosphorylation, inactivation, and growth arrest.
Experimental Procedures
For experimental procedures see supplemental data, online.
Supplementary Material
Acknowledgements
We thank T. Wolff, R. Fehon, G. Halder, H. McNeill, K. Irvine, DJ. Pan, H. Richardson, N. Dyson, B. Hay, N. Tapon, the Bloomington Drosophila Stock Center, the Vienna Drosophila RNAi Center and the Developmental Studies Hybridoma Bank for reagents. This work was supported by grants NIH CA85839 and GM080673 to GDL, and GM068048 to JBS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Marie H, Pratt SJ, Betson M, Epple H, Kittler JT, Meek L, Moss SJ, Troyanovsky S, Attwell D, Longmore GD, et al. The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with alpha-catenin. J Biol Chem. 2003;278:1220–1228. doi: 10.1074/jbc.M205391200. [DOI] [PubMed] [Google Scholar]
- 2.Langer EM, Feng Y, Zhaoyuan H, Rauscher FJ, 3rd, Kroll KL, Longmore GD. Ajuba LIM proteins are snail/slug corepressors required for neural crest development in Xenopus. Dev Cell. 2008;14:424–436. doi: 10.1016/j.devcel.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iida S, Hirota T, Morisaki T, Marumoto T, Hara T, Kuninaka S, Honda S, Kosai K, Kawasuji M, Pallas DC, et al. Tumor suppressor WARTS ensures genomic integrity by regulating both mitotic progression and G1 tetraploidy checkpoint function. Oncogene. 2004;23:5266–5274. doi: 10.1038/sj.onc.1207623. [DOI] [PubMed] [Google Scholar]
- 4.Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 5.Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 6.Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci U S A. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steinhardt AA, Gayyed MF, Klein AP, Dong J, Maitra A, Pan D, Montgomery EA, Anders RA. Expression of Yes-associated protein in common solid tumors. Hum Pathol. 2008;39:1582–1589. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Renfranz PJ, Siegrist SE, Stronach BE, Macalma T, Beckerle MC. Molecular and phylogenetic characterization of Zyx102, a Drosophila orthologue of the zyxin family that interacts with Drosophila Enabled. Gene. 2003;305:13–26. doi: 10.1016/s0378-1119(02)01173-3. [DOI] [PubMed] [Google Scholar]
- 11.Goyal RK, Lin P, Kanungo J, Payne AS, Muslin AJ, Longmore GD. Ajuba, a novel LIM protein, interacts with Grb2, augments mitogen-activated protein kinase activity in fibroblasts, and promotes meiotic maturation of Xenopus oocytes in a Grb2- and Ras-dependent manner. Mol Cell Biol. 1999;19:4379–4389. doi: 10.1128/mcb.19.6.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- 13.Stowers RS, Schwarz TL. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics. 1999;152:1631–1639. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- 15.Tomlinson A, Ready DF. Cell fate in the Drosophila ommatidium. Dev Biol. 1987;123:264–275. doi: 10.1016/0012-1606(87)90448-9. [DOI] [PubMed] [Google Scholar]
- 16.Baker NE. Cell proliferation, survival, and death in the Drosophila eye. Semin Cell Dev Biol. 2001;12:499–507. doi: 10.1006/scdb.2001.0274. [DOI] [PubMed] [Google Scholar]
- 17.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- 20.Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- 21.Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 22.Pantalacci S, Tapon N, Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- 23.Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 24.Tao W, Zhang S, Turenchalk GS, Stewart RA, St John MA, Chen W, Xu T. Human homologue of the Drosophila melanogaster lats tumour suppressor modulates CDC2 activity. Nat Genet. 1999;21:177–181. doi: 10.1038/5960. [DOI] [PubMed] [Google Scholar]
- 25.Reddy BV, Irvine KD. The Fat and Warts signaling pathways: new insights into their regulation, mechanism and conservation. Development. 2008;135:2827–2838. doi: 10.1242/dev.020974. [DOI] [PubMed] [Google Scholar]
- 26.Abe Y, Ohsugi M, Haraguchi K, Fujimoto J, Yamamoto T. LATS2-Ajuba complex regulates gamma-tubulin recruitment to centrosomes and spindle organization during mitosis. FEBS Lett. 2006;580:782–788. doi: 10.1016/j.febslet.2005.12.096. [DOI] [PubMed] [Google Scholar]
- 27.Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 28.Feng Y, Irvine KD. Fat and expanded act in parallel to regulate growth through warts. Proc Natl Acad Sci U S A. 2007;104:20362–20367. doi: 10.1073/pnas.0706722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 31.Tyler DM, Baker NE. Expanded and fat regulate growth and differentiation in the Drosophila eye through multiple signaling pathways. Dev Biol. 2007;305:187–201. doi: 10.1016/j.ydbio.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.