Abstract
Snakes possess a unique sensory system for detecting infrared radiation, enabling them to generate a ‘thermal image’ of predators or prey. Infrared signals are initially received by the pit organ, a highly specialized facial structure that is innervated by nerve fibers of the somatosensory system. How this organ detects and transduces infrared signals into nerve impulses is not known. Here we use an unbiased transcriptional profiling approach to identify TRPA1 channels as infrared receptors on sensory nerve fibers that innervate the pit organ. TRPA1 orthologues from pit bearing snakes (vipers, pythons, and boas) are the most heat sensitive vertebrate ion channels thus far identified, consistent with their role as primary transducers of infrared stimuli. Thus, snakes detect infrared signals through a mechanism involving radiant heating of the pit organ, rather than photochemical transduction. These findings illustrate the broad evolutionary tuning of TRP channels as thermosensors in the vertebrate nervous system.
Venomous pit vipers detect warm-blooded prey through their ability to sense infrared (750 nm – 1 mm wavelength) radiation. Superimposition of thermal and visual images within the snake’s brain enables it to track animals with great precision and speed. Biophysical studies suggest that this system is exquisitely sensitive, such that vipers can detect prey at distances up to 1 meter. Infrared sensation may also be important for predator avoidance and thermoregulatory behavior 1–3.
The Western Diamondback rattlesnake (Crotalus atrox) is a highly evolved viper whose ability to detect infrared radiation (IR) is unmatched by other snakes. IR detection is mediated by specialized loreal pit organs located between the eye and nostril on either side of the viper’s face (Fig. 1a) 4. Suspended within each of these hollow chambers is a thin membrane that serves as an infrared antenna (Fig. 1b). The membrane is rich in mitochondria, highly vascularized, and densely innervated by primary afferent nerve fibers from the trigeminal branch of the somatosensory system (SFig. 1a) 5–8. These fibers convey infrared signals from the pit organ to the optic tectum of the brain, where they converge with input from other sensory modalities 9–11. Some members of the non-venomous Pythonidae and Boidae families (pythons and boas, respectively) also detect IR, albeit with 5–10 fold lower sensitivity compared to Crotalinae vipers 3,12,13. Pythons and boas possess labial pit organs, which are distributed over the snout and lack the complex architecture seen in loreal pits of vipers (SFig. 1b). Nonetheless, they are similarly vascularized and innervated by trigeminal fibers, but at lower density 14–16, 5. Thus, relative sensitivities of these snake species to IR likely reflect molecular and anatomical differences of this specialized sensory system. While the role of the pit organ as an infrared sensor is well established, fundamental questions remain regarding its mechanism of stimulus detection. For example, does the membrane, itself, contain the infrared sensor, or is the sensor expressed by the closely apposed nerve fibers? What is the molecular identity of the infrared sensor, and can its intrinsic biophysical characteristics account for the physiological properties of the pit organ? Do Crotalinae, Pythonidae, and Boidae snakes use similar molecular strategies for sensing IR?
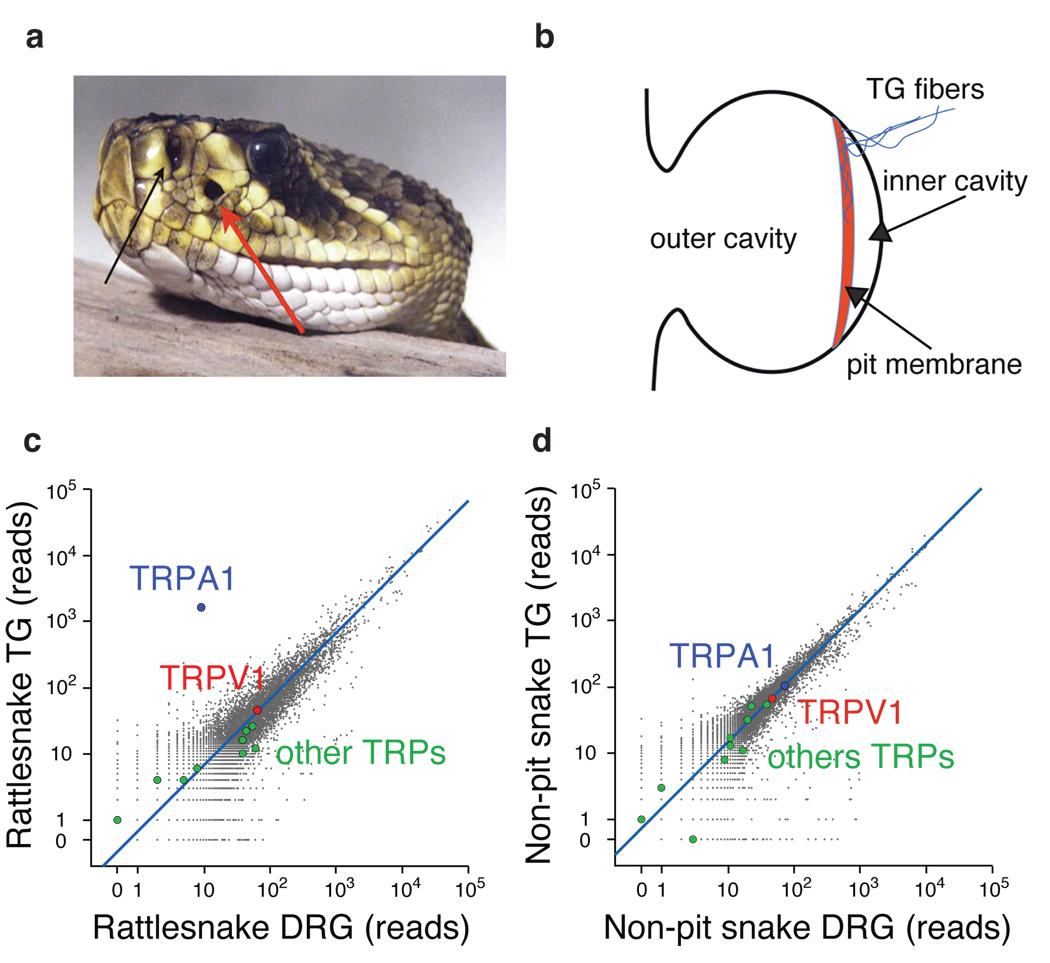
Figure 1. Anatomy of the pit organ and comparison of gene expression in snake sensory ganglia.
a Rattlesnake head showing location of nostril and loreal pit organ (black and red arrows, respectively)(from Wikimedia Commons). b Schematic of pit organ structure showing innervation of pit membrane suspended within hollow cavity. (c – d) Number of mRNA-Seq reads from snake ganglia that align to the chicken proteome. TRPA1 and TRPV1 are highlighted, as are other TRP channels. Blue line indicates expected number of sequencing reads for genes with similar expression levels in the two samples based on total number of aligned reads from each. Signals < 20 reads are within statistical noise and therefore scored as non-expressed sequences. Rattlesnake refers to C. atrox and non-pit refers to a combination of Texas Rat (Elaphe obsolete lindheimerii) and Western Coachwhip (Masticophis flagellum testaceus) snakes.
In principle, an infrared receptor could detect photons directly, similar to photochemical activation of opsins in the eye, or indirectly through heating of tissue within the pit, leading to activation of a thermoreceptor 1. Because the pit receives direct input from the somatosensory, rather than the visual system 9, it seems likely that infrared signals are detected through a thermotransduction, rather than phototransduction mechanism. Consistent with this, heat-activated membrane currents from rattlesnake trigeminal neurons have been described, although their functional properties have not been extensively characterized 17, 6.
Snakes - particularly pit vipers - are inconvenient subjects for physiological and behavioral studies. They are also genetically intractable organisms for which annotated genomic information is scarce, limiting molecular studies of IR detection. We therefore used transcriptome profiling to identify pit-enriched sensory transducers, yielding the snake orthologue of the ‘wasabi receptor’, TRPA1, as a candidate IR detector. This channel is highly enriched in trigeminal neurons that innervate the pit and, when heterologously expressed, exhibits robust heat sensitivity. Thus, TRPA1 has been evolutionarily selected to function as a specialized and highly sensitive heat receptor in the pit, whereas in mammals it functions primarily as a detector of chemical irritants and inflammatory agents 18. Our results demonstrate that the pit membrane serves as a passive antenna for radiant heat, transducing thermal energy to heat-sensitive channels on embedded nerve fibers.
Exploiting specialization of pit vipers
In most sensory systems, specialized receptor cells detect relevant stimuli and transmit signals to adjacent nerve fibers. In the somatosensory system, however, bare nerve endings are themselves detectors of thermal, mechanical, or chemical stimuli 19. Indeed, trigeminal ganglia (TG) of pit bearing snakes are unusually large compared to those of mammals and send a thick bundle of afferents directly to the pit on the ipsilateral side of the face (SFig. 1a) 20,21. We therefore reasoned that snake TG should express proteins dedicated to pit function, and that such proteins should be less abundant in dorsal root ganglia (DRG), which provide somatosensory input to the trunk. Because mammalian TG and DRG gene expression profiles are more-or-less equivalent 22,23, marked differences in snakes should reflect functional specialization associated with IR detection. Remarkably, a pair-wise comparison of transcriptomes from rattlesnake TG versus DRG revealed a single gene encoding an orthologue of the TRPA1 ion channel (Fig.1c). Whereas other members of the TRP channel family (e.g. the capsaicin- and heat-activated receptor, TRPV1) showed equivalent expression in these ganglia, TRPA1 was enriched 400-fold in TG.
If TRPA1 is of unique functional importance to infrared sensing, then snakes lacking pit organs (non-pit species) should not show a disparity in TRPA1 expression between TG and DRG. Indeed, transcriptomes from two non-pit species, Texas Rat and Western Coachwhip snakes, showed no obvious outliers for either ganglion (Fig. 1d). Consistent with this, transcriptome comparison from TG of rattlesnake versus non-pit snakes again identified TRPA1 as the only differentially expressed gene (SFig. 1c). In striking contrast to non-pit snakes and other vertebrates, TRPA1 was absent from rattlesnake DRG (SFig. 1d), further supporting a specific role for this channel in TG/pit function. Lastly, we did not detect opsin-like sequences in TG of any snake species examined.
Unique expression of TRPA1 in viper TG
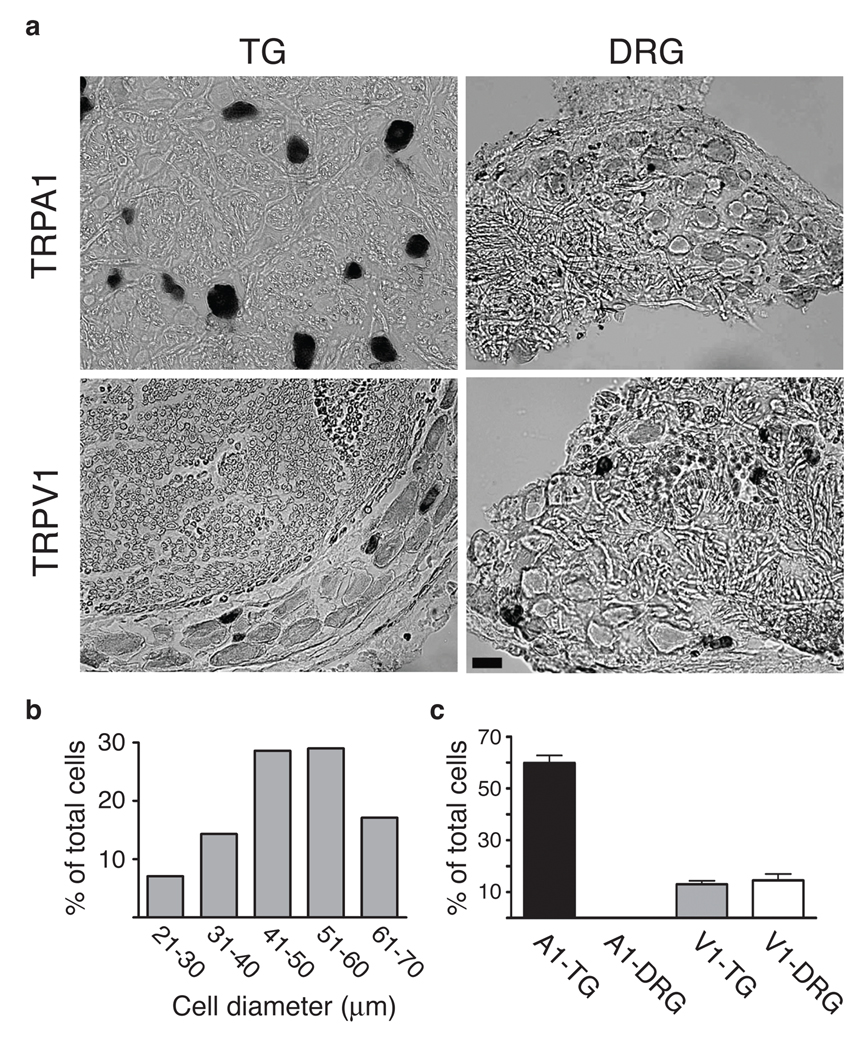
Vertebrate somatosensory ganglia contain anatomically and functionally diverse neuronal subpopulations 24. In general, neurons having the largest soma diameters are involved in detection of innocuous sensations, such as light touch, whereas small to medium diameter neurons constitute the majority of nociceptors that detect noxious stimuli. In mammals, TRPA1 is expressed by ~25% of all somatosensory neurons, preferentially nociceptors that also express TRPV1 25,26. We observed a very different anatomical profile in rattlesnakes, where most TG neurons were medium-to-large diameter, the majority (59.9±9.7%) of which expressed TRPA1 (Fig. 2a,b) (also see below). Consistent with our transcriptome analysis, no TRPA1 signal was observed in rattlesnake DRG (Fig. 2a,c). We also examined the distribution of TRPV1, which in rodent TG or DRG is expressed by 40–60% of neurons, predominantly nociceptors 26,27. In rattlesnake TG or DRG, TRPV1 was expressed by only 13±4.1% and 14.5±5.7% of neurons, respectively, most with small diameters (Fig. 2a,c). Thus, pit viper TG is unique among vertebrates, reflecting adaptation for IR detection.
Figure 2. Expression of TRPA1 and TRPV1 in rattlesnake sensory ganglia.
a In situ hybridization showing expression of TRPA1 or TRPV1 in tissue sections from rattlesnake TG or DRG, as indicated. Scale bar = 20 µm. b Quantification of neuronal cell size (diameter) determined from histological sections of rattlesnake TG (n = 70 cells from 5 independent sections). c Quantitative analysis of cells within TG or DRG that express TRPA1 or TRPV1 transcripts (mean ± s.d.; n = 448 neurons from 11 independent sections for TRPA1 and 151 neurons from 5 independent sections for TRPV1).
Snake TRPA1 is a heat-activated channel
Mammalian TRPA1 is activated by allyl isothiocyanate (AITC), the pungent agent from wasabi and other mustard plants 25,28. AITC and other electrophilic irritants gate the channel through an unusual mechanism involving covalent modification of cysteine residues within the cytoplasmic N-terminus 29,30. Rattlesnake and rat snake TRPA1 exhibit 81% identity with one another and 63% identity with human TRPA1 and contain three conserved N-terminal cysteines required for activation by electrophiles (SFig. 2). Indeed, when expressed in HEK293 cells, TRPA1 from either snake species responded to AITC, demonstrating functionality of the cloned channels (Fig. 3a).
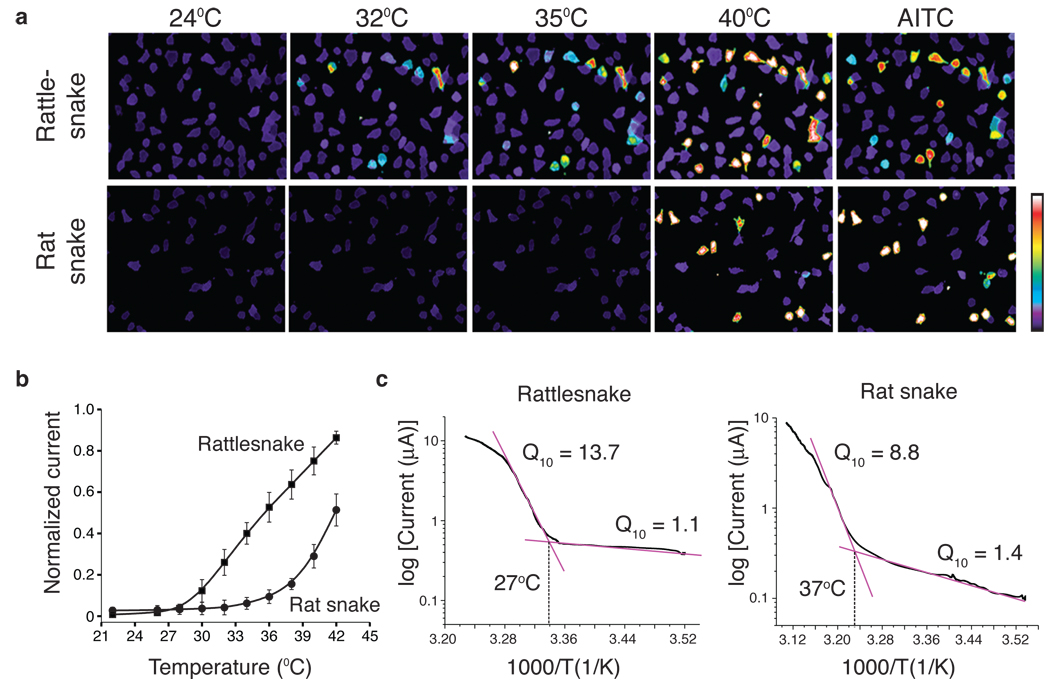
Figure 3. Functional analysis of snake TRPA1 channels.
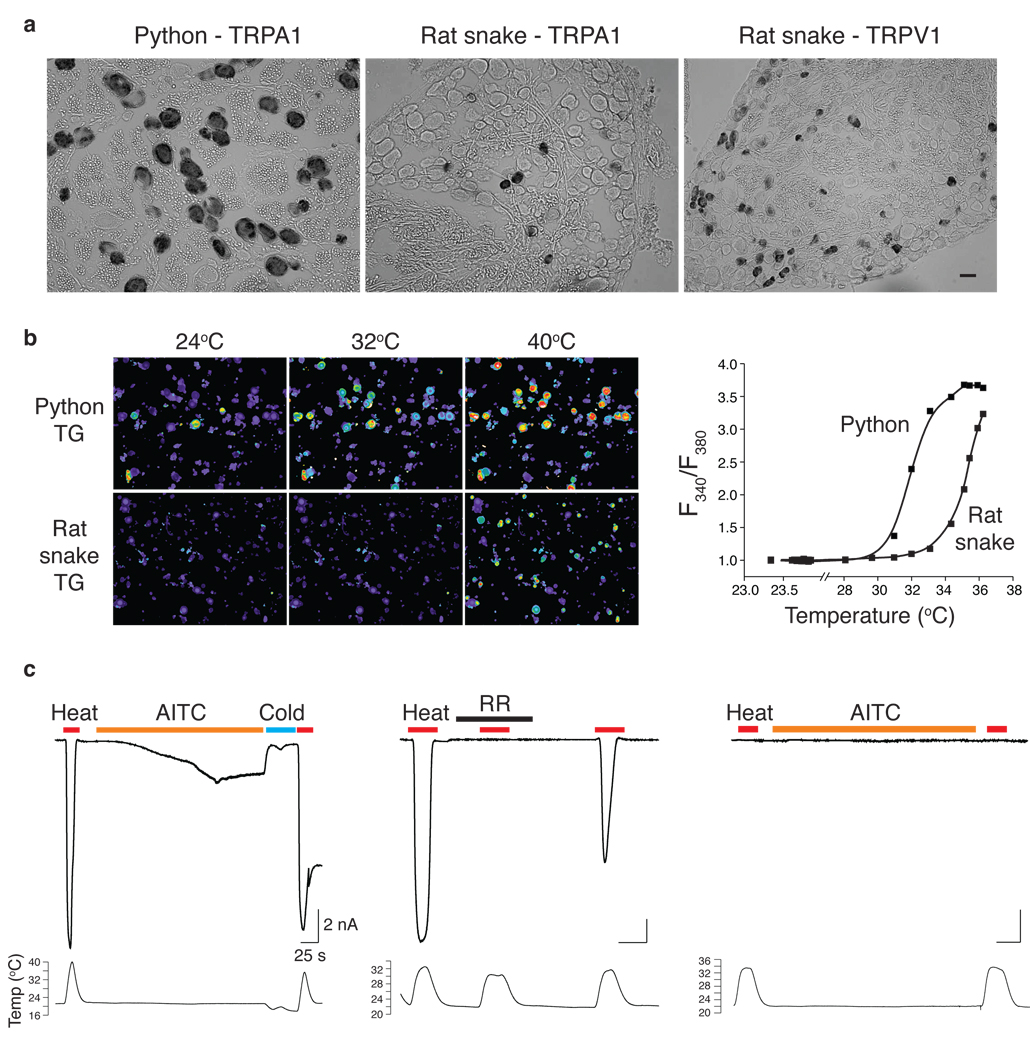
a HEK293 cells expressing cloned rattlesnake or rat snake TRPA1 channels were analyzed for heat- or mustard oil (AITC; 200 µM; 24°C)-evoked responses using calcium imaging; color bar indicates relative change in fluorescence ratio, with purple and white denoting lowest and highest cytoplasmic calcium, respectively (n ≥ 105 neurons per species). b Relative heat response profiles of rattlesnake and rat snake channels expressed in oocytes (response at each temperature was normalized to maximal response at 45°C (Vh = +80 mV; n ≥ 6). c Arrhenius plots show thermal thresholds and Q10 values for baseline and evoked responses of rattlesnake and rat snake TRPA1 channels, as indicated (temperature ramp of 1°C/sec).
If TRPA1 is important for infrared sensing, then it should respond to thermal stimuli in a temperature range consistent with sensitivity of the pit, which detects changes in ambient temperature above ~30°C (ref 17). Indeed, rattlesnake TRPA1 was inactive at room temperature, but robustly activated above 28.0 ± 2.5°C (Fig. 3a and SFig. 3a). Interestingly, rat snake TRPA1 was also heat sensitive, albeit with a substantially higher threshold of 36.3 ± 0.6°C. To assess thermal response profiles in greater detail, we measured heat-evoked membrane currents in voltage-clamped Xenopus oocytes expressing snake channels. Consistent with calcium imaging data, rattlesnake TRPA1 showed extremely robust and steep responses to heat with a threshold of 27.6 ± 0.9°C (Q10 = 13.7), whereas the rat snake channel responded with a higher threshold of 37.2 ± 0.7°C (Q10 = 8.8) (Fig. 3b,c and SFig. 3b). Thus, although the rat snake channel is heat sensitive, its thermal response properties make it less well suited to act as an infrared sensor compared to the pit viper channel. Instead, TRPA1, in conjunction with TRPV1, may contribute to cutaneous and somatic thermosensation in non-pit snakes, consistent with the higher activation thresholds of rat snake versus rattlesnake TRPA1. The rattlesnake channel did not respond to cold (12°C) (not shown).
TRPA1 channels have been characterized from a number of vertebrate species, including fish 31, all of which are activated by AITC, but not heat (SFig. 4). TRPA1-like channels are also found in invertebrate organisms, including Drosophila melanogaster, whose genome contains three TRPA1 orthologues. One of these (dTRPA1) is heat sensitive 32,33, and in our experimental conditions shows a thermal threshold of 33.7 ± 1.0°C (SFig. 5). Relative to rat TRPA1, which responds to AITC with an EC50 of 11 µM, the rattlesnake and rat snake orthologues are less sensitive, showing robust responses at concentrations ≥ 500 µM and with significantly slower activation. The relative sensitivities of these channels to heat versus AITC are clearly shown by comparing current-voltage profiles (SFig. 3b). This inverse relationship between heat and AITC sensitivity likely underscores the relative contribution of TRPA1 to thermo- versus chemo-sensation in different organisms. Taken together, our bioinformatics, anatomical, and functional results strongly suggest that TRPA1 serves as an IR detector in the pit viper.
Ancient snakes use TRPA1 to sense IR
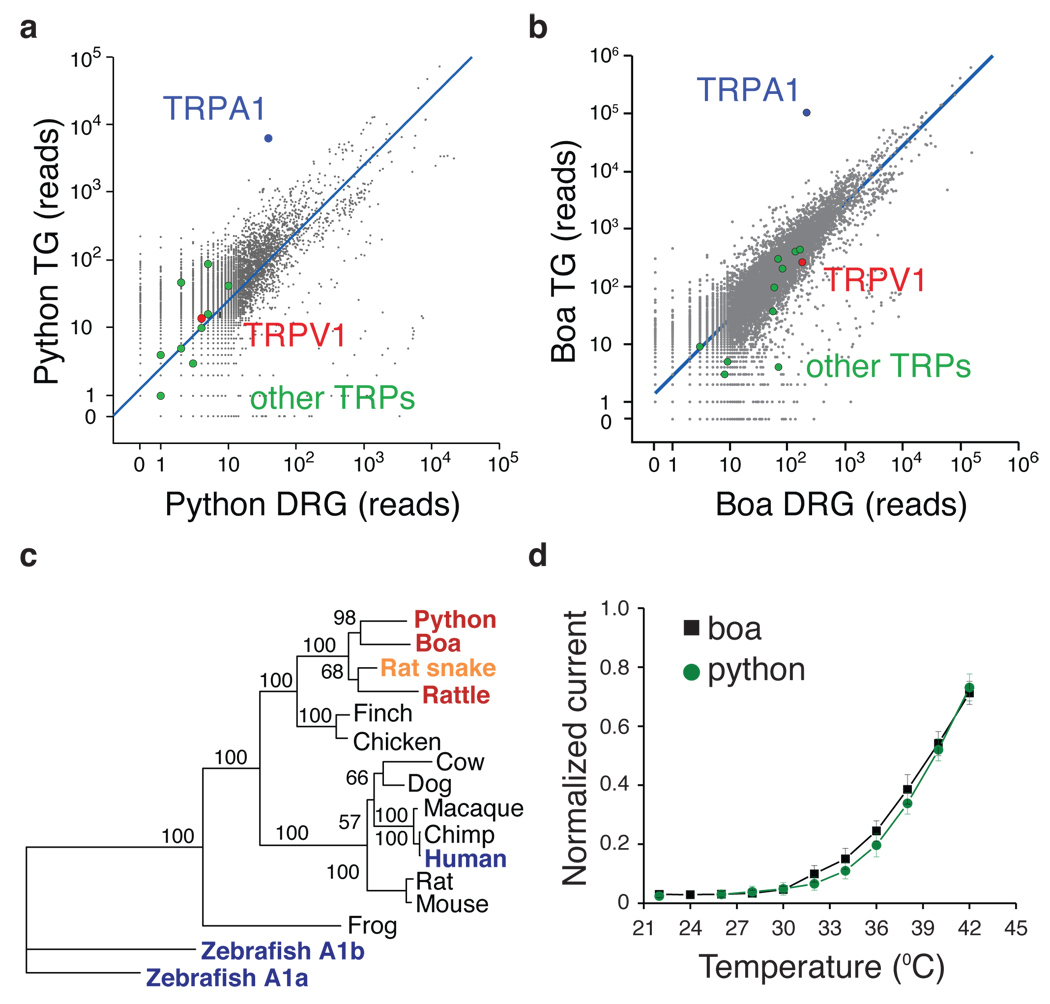
Ancient (pythons and boas) and modern (pit vipers) snakes are separated by a long evolutionary distance (>30 million years) and show substantial differences in pit architecture and sensitivity 34, 35. We therefore asked whether they use the same molecule to detect heat. In sensory ganglia of Royal python (Python regius) and Amazon tree boa (Corallus hortulanus) TRPA1, again, stood out as the major differentially expressed transcript, being 65- and 170-fold more abundant in TG versus DRG for pythons and boas, respectively (Fig. 4a,b). Moreover, comparison of transcript ratios from rattlesnake and python showed that TRPA1 stands alone as a highly TG-specific molecule (SFig. 6a). In contrast to pit vipers, TRPA1 was expressed in DRG of python and boa, but only at relatively modest levels, comparable to that of other TRP channels. Surprisingly, TRPV1 transcripts were not observed above background levels in pythons (Fig. 4a), suggesting that TRPA1 or another heat-sensitive channel underlies somatic thermosensation in this species.
Figure 4. Analysis of TRPA1 from python and boa.
a – b Transcriptome profiling of ancient snakes. Number of mRNA-Seq reads from python and boa ganglia that align to chicken proteome, as described in Fig. 1. c Phylogenetic tree of TRPA1 channel protein sequences with bootstrap values from 100 trials. Red denotes heat-sensitive channels with lower thermal threshold compared to rat snake (orange). Blue indicates non-heat sensitive channels according to this study. d Relative heat response profiles for python and boa TRPA1 as measured in oocytes (recorded and normalized as described in Fig 3b).
Dendrogram analysis of snake TRPA1 channels shows that they constitute a closely related subfamily of heat-sensitive orthologues (Fig. 4c). Moreover, the position of boa and python sequences supports the hypothesis that these species represent an evolutionarily ancient branch of snakes that is independent of modern snakes such as pit vipers or rat snakes.
Expression of cloned python and boa TRPA1 in oocytes showed that both are heat-activated channels with modest sensitivity to AITC (Fig. 4d, SFig. 6b,c). Interestingly, we found that python and boa channels exhibited a slightly higher thermal threshold compared to rattlesnake TRPA1 (32.7 ± 1.3°C and 29.6 ± 0.7°C, respectively, versus 27.6 ± 0.9°C for rattlesnake), consistent with differential sensitivity of these snakes to IR. As in the case of the rattlesnake channel, python and boa TRPA1 were substantially more sensitive to heat than chemical agonists, as evidenced by relatively small response to AITC (SFig. 6b,c).
Endogenous TRPA1 subserves IR detection
To assess the contribution of TRPA1 to neuronal heat sensitivity, we chose pythons as a convenient (i.e. non-venomous) pit bearing species for functional studies. Anatomically, python TG resemble those of rattlesnakes, consisting primarily of large and medium diameter neurons, most of which (73.1 ± 7.8%) express TRPA1 (Fig. 5a and SFig. 7a,b). Consistent with this, a majority (78.2 ± 14.0%) of neurons from python TG were heat sensitive and exhibited a threshold of 28.0 ± 2.2°C (Fig. 5b). Moreover, all heat-sensitive neurons responded to AITC (500 µM)(not shown), confirming expression of functional TRPA1 channels in these cells. No capsaicin sensitive neurons were observed in python TG cultures, consistent with our bioinformatics profile showing lack of TRPV1 in these ganglia.
Figure 5. Functional analysis of snake sensory neurons.
a Expression of TRPA1 or TRPV1 transcripts in python and rat snake TG (scale bar = 40 µm). b Thermal sensitivity of python and rat snake TG neurons as measured by calcium imaging. Temperature ramps (24 to 46°C) were applied by continuous perfusion to assess thresholds (color scale as in Fig. 3a). Corresponding temperature-response profiles are shown at right (n = 5 and 26 neurons, respectively). Thresholds (28.0 ± 0.7 and 36.2 ± 0.6; P < 0.0001) were determined from average of 43 and 89 neurons from python and rat snake, respectively (10 independent fields each). c Patch-clamp recordings from python neurons showing robust heat- and AITC-evoked currents that were suppressed by cold (left) and blocked by ruthenium red (RR, 10 µM) (center)(n > 45). A minority of neurons was insensitive to heat and AITC (right)(n > 8).
TG from control rat snake more closely resembled those of mammals in the relative proportion of small, medium, and large diameter neurons. Compared to pit bearing species, TRPA1 was expressed by a restricted cohort (13.3 ± 5.7%) of rat snake TG neurons that included mostly small and medium diameter cells (Fig. 5a and SFig. 7a). Unlike pythons, rat snake TG contained a significant proportion (27.3 ± 4.4%) of TRPV1-positive neurons (SFig. 7a), suggesting that both TRPA1 and TRPV1 contribute to heat sensation in this species. Neurons from rat snake TG also showed a lower prevalence (~20%) of heat and AITC sensitivity compared to pythons, and responders were confined to the medium/small diameter subpopulation (SFig. 7b). Importantly, rat snake neurons responded at higher temperatures, binning into two distinct populations with thresholds of 36.2 ± 1.8°C and 38.7 ± 1.4°C (P < 0.025), the former being AITC-sensitive and the latter capsaicin-sensitive (Fig. 5b and SFig. 7c). Taken together, our results suggest that TRPA1 underlies infrared and somatic heat sensation in pit bearing snakes, whereas TRPA1 and TRPV1 contribute to somatic thermosensation in rat snakes. Furthermore, the functional properties and tissue distribution of rattlesnake TRPV1 (SFig. 8 and Fig. 2a,c) make it a likely candidate for mediating somatic thermosensation in this species.
Finally, patch clamp recording verified the presence of heat-sensitive membrane currents in snake neurons. The majority of python TG neurons showed enormous heat-evoked currents bearing the hallmarks of TRPA1 channels, including blockade by ruthenium red, inward rectification, and desensitization (Fig. 5c and SFig. 7d). Like heterologously expressed python TRPA1, these responses were attenuated (~50%) by the mammalian TRPA1 antagonist HC-030031 (not shown). Consistent with our calcium imaging results, heat-sensitive python TG neurons also responded to AITC and showed a thermal threshold of 29.5 ± 1.7°C. A more restricted population of medium-diameter neurons were insensitive to heat or AITC (Fig. 5c), although they showed robust action potential firing upon depolarization (not shown). In contrast with pythons, the activation threshold for heat-sensitive rat snake neurons was substantially higher (35.6 ± 1.2°C) (SFig. 7d).
Discussion
Four vertebrate families possess specialized sensory organs devoted to IR detection: pit viper, python, and boa families of snakes, as well as vampire bats 2,36. Here we delineate the mechanism whereby three of these families sense IR, starting with the pit viper as the paragon of this unique sensory modality. We accomplished this by taking an unbiased transcriptome profiling approach in which minimal assumptions were made in regard to the molecular specialization of the pit and associated neural structures. This represents a powerful, sensitive, and quantitative version of the classic plus-minus screen for identifying organ-specific genes. We exploited this technology to address a problem vexed by a paucity of tissue and lack of genomic information.
In vertebrates, temperature sensation is mediated through activation of TRP channels that detect heat or cold 37. In invertebrate organisms, such as flies (Drosophila), activation of TRP channels also contributes to temperature detection 32,33, whereas in worms (C. elegans), thermosensation is suggested to involve a phototransduction-like pathway involving activation of cyclic nucleotide-gated channels 38,39. Our analysis suggests that the pit organ detects IR through a TRP channel-based process, rather than an opsin-like pathway, consistent with thermal, rather than photochemical signal transduction.
Identification of snake TRPA1 as an infrared sensor is interesting from an evolutionary perspective because previously identified vertebrate TRPA1 orthologues function primarily as detectors of chemical irritants 18, and possibly cold 40,41. Thus, snake TRPA1 is functionally more like its invertebrate counterparts, despite their greater sequence diversity. Recent observations suggest that among TRP channels, TRPA1 orthologues show particularly rapid evolution in invertebrate species, where they display a range of heat sensitivities and contribute differentially to thermosensation 42,43. Our findings indicate that this functional diversification extends to vertebrate channels, as well. While the evolutionary relationship among snake species is a subject of ongoing study and debate 34,44, our phylogenetic analysis suggests that ancient and modern snakes have independently adapted TRPA1 as an infrared sensor through convergent evolution. The cloned rattlesnake channel is the most heat-sensitive (i.e. lowest thermal activation threshold and highest Q10), in keeping with the greater infrared acuity of pit vipers compared to pythons or boas 3. At the same time, differences in thermosensitivity among snake TRPA1 channels can differ by as little as 2°C (e.g. rattlesnake versus boa), suggesting that other cellular or anatomical factors contribute to physiologic and behavioral differences in stimulus detection. Finally, the relative contributions of TRPA1 and other heat sensitive channels (such as TRPV1) to somatic thermosensation likely differ among snake species, depending on thermal thresholds and expression patterns.
Sensory systems evolve rapidly to accommodate variations in environmental niche, such as those affecting climate and predator-prey relationships 45,46. TRPA1 channels have undergone particularly fascinating evolutionary perturbation and selection to function as thermo- or chemoreceptors in organisms of very different lineage, indicative of their unique physiological plasticity throughout the animal kingdom. Thus, TRPA1 and other TRP channels provide new genetic and physiologic markers with which to delineate evolutionary relationships among vertebrate and invertebrate species.
Methods Summary
cDNA libraries were sequenced on Illumina Genome Analyzer II and aligned to chicken RefSeq protein database. Unrooted phylogenetic tree was constructed from multiple sequence alignments using PhyML (version 3.0). Bootstrapping was performed with 100 trials. Adult snake tissue was fixed with paraformaldehyde for chromogenic in situ hybridization histochemistry. Rattlesnakes were provided by the Natural Toxins Research Center, Texas A&M University- Kingsville; boas, pythons, and rat snakes were obtained from Glades Herp Farm (Bushnell, Florida). Animal husbandry and euthanasia procedures were approved by the UCSF or University of Texas Institutional Animal Care and Use Committee. Cloned channels were transiently expressed in HEK293 cells and subjected to calcium imaging using Fura-2/AM ratiometric dye. Snake TG neurons were cultured as previously described 17. Oocytes from Xenopus laevis were cultured, injected with 5 ng of RNA, and analyzed 2–5 days postinjection by TEVC as described 47. Membrane currents were recorded under the whole-cell patch-clamp configuration and thermal stimulation applied with a custom-made Peltier device (Reid-Dan Electronics). Temperature thresholds represent the point of intersection between linear fits to baseline and the steepest component of Arrhenius profile, as described 48.
Methods
Deep Sequencing and Analysis
Sequencing libraries were prepared from poly A+ RNA using the Illumina mRNA-Seq Sample Prep Kit (RS-100-0801) according to the manufacturer's instructions. Libraries were then sequenced on the Illumina Genome Analyzer II using two 36-cycle sequencing kits (FC-104-3002) to read 80 nucleotides of sequence from a single end of each insert, by standard protocols. Between 2.4 million and 12.5 million inserts were sequenced for each sample.
Sequences were aligned to the chicken RefSeq protein database (NCBI version 2.1) using the blastx tool from NCBI Blast (version 2.2.21), which aligns a six-frame translation of each query against a protein database. The alignment was performed with a word size of 4 amino acids and a window size of 5; a maximum E value of 1e-5 was required. For each read that aligned to the chicken proteome, a set of optimal hits was collected based on alignments whose bit score was within 0.2 of the highest bit score reported for that sequencing read. Each RefSeq alignment for a given sequencing read was converted to an Entrez Gene identifier and redundant alignments for a single read (which correspond to alignments against different isoforms of the same protein) were collapsed. The number of optimally aligning reads was then counted for each gene; in some cases a single read counted towards multiple genes. Multiple sequence alignment was performed with MUSCLE v3.70 and the tree was built from the MSA using PhyML 3.0. The multiple sequence alignment of all TRPA1 amino acid sequences was constructed using MUSCLE (version 3.70) using the default parameters. The unrooted phylogenetic tree was constructed from this multiple sequence alignment using PhyML (version 3.0) with default parameters and maximum likelihood estimation of the gamma shape parameter and the fraction of invariant sites. Bootstrapping was performed with 100 trials.
In situ hybridization histochemistry
Adult snakes were euthanized with Beuthanasia-D (1 mL/4.5 kg body weight). TG and DRG tissue were dissected and fixed in 4% PFA in PBS for 5 days. Cryostat sections (12–15 µm thick) were processed and probed with a digoxigenin-labeled cRNA. Probes were generated by T7/T3 in vitro transcription reactions using a 2.9-kb fragment of TRPA1 cDNA (nucleotide 153–3024) and 1.9-kb fragment of TRPV1 cDNA (nucleotide 417–2387). Signal was developed with alkaline phosphatase-conjugated anti-digoxigenin Fab fragments according to the manufacturer’s instructions.
Channel cloning
Functional cDNAs were amplified from ssDNA, generated by reverse transcriptase reaction, using following primers: rattlesnake TRPA1 and non-pit TRPA1 and Royal python TRPA1 (5 ’-3’forward: GAATGACCAGGAGCTGTATC; 5’-3’reverse: AGCCAGCTTGACTGGAATTG); rattlesnake TRPV1 (5 ’-3’forward: CAGGTGAGGTGAGTCCTTCGTAAC; 5’–3’reverse: TGAATGACGCAGATGGGGGTC).
Calcium imaging
All tested channels were transiently expressed in HEK293 cells with the use of Lipofectamin 2000 (Invitrogen). Calcium imaging of HEK293 cells using Fura-2/AM was performed on coverslips coated with Matrigel (BD). Fluorecent images were acquired with Metaflour software (Molecular Device) and analyzed using Graph Pad Prizm 4.
Culture of sensory neurons
Snake were anaesthetized using isofluorane and then decapitated. TGs were isolated and cultured as previously described 17. Briefly, dissected ganglia were first placed in ice-cold DMEM/F12 solution. Cells were dissociated from trigeminal ganglia by treatment with collagenase (1mg/ml, 50 min, 28°C) and trypsin (10min, room temperature) followed by mechanical dissociation with plastic pipette. Dissociated cells were centrifuged at 500 rcf for 10 min and then diluted with DMEM/F12, 10%FBS, pen/strep and 2mM glutamine. Cells were plated onto the Matrigel precoated coverslips. Cells were maintained at 28°C in 7% CO2-93% air for 6–48hours.
Oocyte electrophysiology
Surgically extracted oocytes from Xenopus laevis (Nasco) were cultured and analyzed 2–5 days postinjection by TEVC as previously described 47. Oocytes were injected with 5 ng of RNA and whole cell currents measured after 24–72 h using a Geneclamp 500 amplifier (Axon Instruments, Inc). Microelectrodes were pulled from borosilicate glass capillary tubes to obtain resistances of 0.3–.07 MΩ. Bath solution contained 10 mM Hepes, 120 mM NaCl, 2 mM KCl, 0.2 mM EGTA, 1 mM CaCl2 and 2 mM MgCl2 buffered to a final pH of 7.4 with NaOH. Data were analyzed using pCLAMP10 software.
Patch clamp recording
Membrane currents were recorded using gap free protocol at −60 mV under the whole-cell configuration of the patch-clamp technique using Axopatch 200B amplifier (Axon Instruments). Membrane currents were digitized online using a Digidata 1440A interface board and pCLAMP 10.2 software (Axon Instruments). Sampling frequency was set to 5 kHz, and the low-pass filter was set to 1 kHz. Patch electrodes were fabricated from borosilicate glass with a resistance of 2–4 MΩ. The bath solution contained (mM): 130 NaCl, 3 KCl, 1.2 MgSO4, 2 CaCl2, 10 HEPES, 10 Glucose adjusted to pH 7.4. The perfusion solution was the same as the bath solution but with 0.2 mM CaCl2 to reduce the desensitization process. The pipette solution contained (mM): 130 CsMeSO4, 20 CsCl, 9 NaCl, 0.2 EGTA, 10 HEPES, 1 MgATP, and adjusted to pH 7.2. Thermal stimulation was applied with a custom-made Peltier device (Reid-Dan Electronics) that heated or cooled the flowing perfusate stream. Temperature was measured using a thermistor placed adjacent to the cell.
Determination of thermal threshold
Temperature thresholds represent the point of intersection between linear fits to baseline and the steepest component of the Arrhenius profile. Values are derived from averages of individual curves; n ≥ 6. Arrhenius curve were obtained by plotting the current on a log scale against the reciprocal of the absolute temperature. Q10 was used to characterize the temperature dependence of the ionic current as calculated using the following equation:
where R2 is the current at the higher temperature T2 and R1 is the current at the lower temperature T1 (ref 48).
Supplementary Material
Acknowledgements
We thank A. Priel for advice and assistance with calcium imaging and electrophysiology, C. Chu for help with sequencing, J. Poblete for technical assistance, and the staff of the NTRC serpentarium for animal husbandry. We thank Paul Garrity for providing the dTRPA1 cDNA. This work was supported by a Ruth L. Kirschstein National Research Service Award (GM080853) (N.T.I.), a NIH Institutional Research Service Award in Molecular and Cellular Basis of Cardiovascular Disease (A.T.C.), the Howard Hughes Medical Institute (J.S.W.), and grants from the National Institutes of Health, including NCRR Viper grant P40 RR018300-06 (E.E.S. and J.C.P.) and NS047723 and NS055299 (D.J.).
Footnotes
Author contributions E.O.G., J.F.C-M. and N.T.I. designed and performed experiments and analyzed data. N.T.I. and J.S.W. developed analytical tools and analyzed data. Y.M.K., G.H. and A.T.C. performed experiments and/or provided reagents and analyzed data. E.E.S. and J.C.P. supervised snake husbandry and handling. E.O.G., Y.M.K., J.F.C-M. and D.J. wrote the manuscript with discussion and contributions from all authors. J.S.W. and D.J. provided advice and guidance throughout. D.J. initiated and supervised the project.
Deep sequencing data are archived under GEO accession number GSE19911. GenBank accession numbers are GU562965 (Python regius TRPA1), GU562966 (Elaphe obsoleta lindheimeri TRPA1), GU562967 (Crotalus atrox TRPA1), GU562968 (Crotalus atrox TRPV1), and GU562969 (Corallus hortulanus TRPA1).
Competing interests statement The authors declare no competing financial interests.
References
- 1.Bullock TH, Cowles RB. Physiology of an Infrared Receptor: The Facial Pit of Pit Vipers. Science. 1952;115:541–543. doi: 10.1126/science.115.2994.541-a. [DOI] [PubMed] [Google Scholar]
- 2.Campbell AL, Naik RR, Sowards L, Stone MO. Biological infrared imaging and sensing. Micron. 2002;33:211–225. doi: 10.1016/s0968-4328(01)00010-5. [DOI] [PubMed] [Google Scholar]
- 3.Ebert J. Dr. rer. nat. thesis. Rheinische Friedrich Wilhelms University of Bonn; 2007. Infrared sense in snakes - behavioural and anatomical examinations (Crotalus atrox, Python regius, Corallus hortulanus) [Google Scholar]
- 4.Barrett R, Maderson PFA, Meszler RM. In: Biology of Reptilia. Parsons TS, editor. Ch. 4. Academic Press; 1970. pp. 277–300. [Google Scholar]
- 5.Ebert J, Schmitz A. In: Herpetologia Bonnensis II. Vences M, Kohler J, Ziegler T, Bohme W, editors. 2006. pp. 215–217. [Google Scholar]
- 6.Terashima S, Liang YF. Temperature neurons in the crotaline trigeminal ganglia. J Neurophysiol. 1991;66:623–634. doi: 10.1152/jn.1991.66.2.623. [DOI] [PubMed] [Google Scholar]
- 7.Amemiya F, Ushiki T, Goris RC, Atobe Y, Kusunoki T. Ultrastructure of the crotaline snake infrared pit receptors: SEM confirmation of TEM findings. Anat Rec. 1996;246:135–146. doi: 10.1002/(SICI)1097-0185(199609)246:1<135::AID-AR15>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 8.Bleichmar H, De Robertis E. Submicroscopic morphology of the infrared receptor of pit vipers. Z Zellforsch Mikrosk Anat. 1962;56:748–761. doi: 10.1007/BF00336332. [DOI] [PubMed] [Google Scholar]
- 9.Hartline PH, Kass L, Loop MS. Merging of modalities in the optic tectum: infrared and visual integration in rattlesnakes. Science. 1978;199:1225–1229. doi: 10.1126/science.628839. [DOI] [PubMed] [Google Scholar]
- 10.Newman EA, Hartline PH. Integration of visual and infrared information in bimodal neurons in the rattlesnake optic tectum. Science. 1981;213:789–791. doi: 10.1126/science.7256281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molenaar GJ. The sensory trigeminal system of a snake in the possession of infrared receptors. II. The central projections of the trigeminal nerve. J Comp Neurol. 1978;179:137–151. doi: 10.1002/cne.901790109. [DOI] [PubMed] [Google Scholar]
- 12.de Cock Bunning T, Terashima S, Goris RC. Python pit organs analyzed as warm python pit organs analyzed as warm receptors. Cellular and Molecular Neurobiology. 1981;1:271–278. doi: 10.1007/BF00710682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warren JW, Proske U. Infrared receptors in the facial pits of the Australian python Morelia spilotes. Science. 1968;159:439–441. doi: 10.1126/science.159.3813.439. [DOI] [PubMed] [Google Scholar]
- 14.Kishida R, Amemiya F, Kusunoki T, Terashima S. A new tectal afferent nucleus of the infrared sensory system in the medulla oblongata of Crotaline snakes. Brain Res. 1980;195:271–279. doi: 10.1016/0006-8993(80)90064-5. [DOI] [PubMed] [Google Scholar]
- 15.Kishida R, de Cock Buning T, Dubbeldam JL. Primary vagal nerve projections to the lateral descending trigeminal nucleus in boidae (Python molurus and Boa constrictor) Brain Res. 1983;263:132–136. doi: 10.1016/0006-8993(83)91209-x. [DOI] [PubMed] [Google Scholar]
- 16.Noble GK, Schmidt A. The structure and function of facial and labial pits of snakes. Proc. Am Philos. Soc. 1937;77:263–288. [Google Scholar]
- 17.Pappas TC, Motamedi M, Christensen BN. Unique temperature-activated neurons from pit viper thermosensors. Am J Physiol Cell Physiol. 2004;287:C1219–C1228. doi: 10.1152/ajpcell.00040.2004. [DOI] [PubMed] [Google Scholar]
- 18.Bautista DM, et al. TRPA1 mediates the inflammatory actions of environmental irritants and proalgesic agents. Cell. 2006;124:1269–1282. doi: 10.1016/j.cell.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 19.Julius D, Basbaum AI. Molecular mechanisms of nociception. Nature. 2001;413:203–210. doi: 10.1038/35093019. [DOI] [PubMed] [Google Scholar]
- 20.Molenaar GJ. An additional trigeminal system in certain snakes possessing infrared receptors. Brain Res. 1974;78:340–344. doi: 10.1016/0006-8993(74)90560-5. [DOI] [PubMed] [Google Scholar]
- 21.Schroeder DM, Loop MS. Trigeminal projections in snakes possessing infrared sensitivity. J Comp Neurol. 1976;169:1–11. doi: 10.1002/cne.901690102. [DOI] [PubMed] [Google Scholar]
- 22.Eng SR, Dykes IM, Lanier J, Fedtsova N, Turner EE. POU-domain factor Brn3a regulates both distinct and common programs of gene expression in the spinal and trigeminal sensory ganglia. Neural Dev. 2007;2:3. doi: 10.1186/1749-8104-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woolf CJ, Ma Q. Nociceptors--noxious stimulus detectors. Neuron. 2007;55:353–364. doi: 10.1016/j.neuron.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 25.Jordt SE, et al. Mustard oils and cannabinoids excite sensory nerve fibres through the TRP channel ANKTM1. Nature. 2004;427:260–265. doi: 10.1038/nature02282. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi K, et al. Distinct expression of TRPM8, TRPA1, and TRPV1 mRNAs in rat primary afferent neurons with adelta/c-fibers and colocalization with trk receptors. J Comp Neurol. 2005;493:596–606. doi: 10.1002/cne.20794. [DOI] [PubMed] [Google Scholar]
- 27.Tominaga M, et al. The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron. 1998;21:531–543. doi: 10.1016/s0896-6273(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 28.Bandell M, et al. Noxious cold ion channel TRPA1 is activated by pungent compounds and bradykinin. Neuron. 2004;41:849–857. doi: 10.1016/s0896-6273(04)00150-3. [DOI] [PubMed] [Google Scholar]
- 29.Macpherson LJ, et al. Noxious compounds activate TRPA1 ion channels through covalent modification of cysteines. Nature. 2007;445:541–545. doi: 10.1038/nature05544. [DOI] [PubMed] [Google Scholar]
- 30.Hinman A, Chuang HH, Bautista DM, Julius D. TRP channel activation by reversible covalent modification. Proc Natl Acad Sci U S A. 2006;103:19564–19568. doi: 10.1073/pnas.0609598103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prober DA, et al. Zebrafish TRPA1 channels are required for chemosensation but not for thermosensation or mechanosensory hair cell function. J Neurosci. 2008;8:10102–10110. doi: 10.1523/JNEUROSCI.2740-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamada FN, et al. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454:217–220. doi: 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viswanath V, et al. Opposite thermosensor in fruitfly and mouse. Nature. 2003;423:822–823. doi: 10.1038/423822a. [DOI] [PubMed] [Google Scholar]
- 34.Krochmal AR, Bakken GS, LaDuc TJ. Heat in evolution's kitchen: evolutionary perspectives on the functions and origin of the facial pit of pitvipers (Viperidae: Crotalinae) J Exp Biol. 2004;207:4231–4238. doi: 10.1242/jeb.01278. [DOI] [PubMed] [Google Scholar]
- 35.Safer AB, Grace MS. Infrared imaging in vipers: differential responses of crotaline and viperine snakes to paired thermal targets. Behav Brain Res. 2004;154:55–61. doi: 10.1016/j.bbr.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 36.Kishida R, Goris RC, Terashima S, Dubbeldam JL. A suspected infrared-recipient nucleus in the brainstem of the vampire bat, Desmodus rotundus. Brain Res. 1984;322:351–355. doi: 10.1016/0006-8993(84)90132-x. [DOI] [PubMed] [Google Scholar]
- 37.Jordt SE, McKemy DD, Julius D. Lessons from peppers and peppermint: the molecular logic of thermosensation. Curr Opin Neurobiol. 2003;13:487–492. doi: 10.1016/s0959-4388(03)00101-6. [DOI] [PubMed] [Google Scholar]
- 38.Komatsu H, Mori I, Rhee JS, Akaike N, Ohshima Y. Mutations in a cyclic nucleotide-gated channel lead to abnormal thermosensation and chemosensation in C. elegans. Neuron. 1996;17:707–718. doi: 10.1016/s0896-6273(00)80202-0. [DOI] [PubMed] [Google Scholar]
- 39.Ramot D, MacInnis BL, Goodman MB. Bidirectional temperature-sensing by a single thermosensory neuron in C. elegans. Nat Neurosci. 2008;11:908–915. doi: 10.1038/nn.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Story GM, et al. ANKTM1, a TRP-like channel expressed in nociceptive neurons, is activated by cold temperatures. Cell. 2003;112:819–829. doi: 10.1016/s0092-8674(03)00158-2. [DOI] [PubMed] [Google Scholar]
- 41.Caspani O, Heppenstall PA. TRPA1 and cold transduction: an unresolved issue? J Gen Physiol. 2009;133:245–249. doi: 10.1085/jgp.200810136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang G, et al. Anopheles gambiae TRPA1 is a heat-activated channel expressed in thermosensitive sensilla of female antennae. Eur J Neurosci. 2009 doi: 10.1111/j.1460-9568.2009.06901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsuura H, Sokabe T, Kohno K, Tominaga M, Kadowaki T. Evolutionary conservation and changes in insect TRP channels. BMC Evol Biol. 2009;9:228. doi: 10.1186/1471-2148-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong S, Kumazawa Y. Complete mitochondrial DNA sequences of six snakes: phylogenetic relationships and molecular evolution of genomic features. J Mol Evol. 2005;61 doi: 10.1007/s00239-004-0190-9. [DOI] [PubMed] [Google Scholar]
- 45.Liman ER. Use it or lose it: molecular evolution of sensory signaling in primates. Pflugers Arch. 2006;453:125–131. doi: 10.1007/s00424-006-0120-3. [DOI] [PubMed] [Google Scholar]
- 46.Myers BR, Sigal YM, Julius D. Evolution of thermal response properties in a cold-activated TRP channel. PLoS One. 2009;4:e5741. doi: 10.1371/journal.pone.0005741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chuang HH, Neuhausser WM, Julius D. The super-cooling agent icilin reveals a mechanism of coincidence detection by a temperature-sensitive TRP channel. Neuron. 2004;43:859–869. doi: 10.1016/j.neuron.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 48.DeCoursey TE, Cherny VV. Temperature dependence of voltage-gated H+ currents in human neutrophils, rat alveolar epithelial cells, and mammalian phagocytes. J Gen Physiol. 1998;112:503–522. doi: 10.1085/jgp.112.4.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.