Abstract
Microbial fuel cell (MFC) systems employ the catalytic activity of microbes to produce electricity from the oxidation of organic, and in some cases inorganic, substrates. MFC systems have been primarily explored for their use in bioremediation and bioenergy applications; however, these systems also offer a unique strategy for the cultivation of synergistic microbial communities. It has been hypothesized that the mechanism(s) of microbial electron transfer that enable electricity production in MFCs may be a cooperative strategy within mixed microbial consortia that is associated with, or is an alternative to, interspecies hydrogen (H2) transfer. Microbial fermentation processes and methanogenesis in ruminant animals are highly dependent on the consumption and production of H2in the rumen. Given the crucial role that H2 plays in ruminant digestion, it is desirable to understand the microbial relationships that control H2 partial pressures within the rumen; MFCs may serve as unique tools for studying this complex ecological system. Further, MFC systems offer a novel approach to studying biofilms that form under different redox conditions and may be applied to achieve a greater understanding of how microbial biofilms impact animal health. Here, we present a brief summary of the efforts made towards understanding rumen microbial ecology, microbial biofilms related to animal health, and how MFCs may be further applied in ruminant research.
Introduction
Contemporary livestock production is distinctly linked to a variety of microbial processes that directly impact: (1) the efficiency by which ruminants convert available feedstuffs into energy for maintenance and growth; (2) the health of ruminants within production systems; and (3) the environmental consequences of ruminant production. Microbial communities in the gastrointestinal tract influence the efficiency of nutrient utilization through microbial digestion of fiber, starch, and protein in the rumen and the persistence and shedding of pathogenic bacteria and food safety pathogens such as Salmonella enterica and Escherichia coli O157:H7 in the hindgut. Ecology of viral and bacterial pathogens in the respiratory tract influences the incidence of respiratory disease, the most economically significant disease of ruminants [1]. Finally, the impacts of ruminant production on the environment are largely dictated by complex microbial communities related to enteric methane (CH4) production, emissions of hydrogen sulfide (H2S), and ammonia (NH3) from intensive production environments and dispersion of relevant human and ruminant pathogens in the production ecosystem.
A variety of therapeutic and preventive measures are available to address specific bacteria and viruses that pose a threat to the well-being of the animal or the safety of food derived for human consumption. However, considering the role of these pathogens independent of their environment, and/or their synergistic interactions within a respective microbial community, may limit the success of any applied interventions. Knowledge regarding the specific ecological niches of constituents within these complex microbial communities and microbial interactions associated with shifts in microbial diversity or the onset of infectious and metabolic disease is largely limited to those organisms that can be isolated in monoculture using available microbiological methods. Estimates suggest that less than 1% of all microbial species have been identified in culture and less than 10% of all rumen microbes have been identified using routine methods. Alternative technological platforms must be employed to illuminate more subtle shifts in microbial populations relevant to ruminant production and interactions therein.
Microbial fuel cells (MFCs) are an additional tool that can facilitate study of the physiological roles of microbes in complex ecosystems [2]. To date, there has been limited application of MFCs in ruminant health and production research [3]. In this review, we present an overview of MFCs and identify several potential applications of this technology to advancing knowledge of ruminant microbial ecology particularly as it relates to animal health, production, and environmental impact.
Microbial Fuel Cells
Microbial fuel cells are a developing technology that has been utilized for studying microbial physiology in terms of electron (and proton) transfer [4–6]. They provide a medium for the study of complex microbial systems like those encountered in ruminant production, as well as the opportunity for developing novel approaches to altering the dynamics within those systems. When combined with molecular approaches including genomics and metagenomics, MFCs have the potential to profoundly expand the existing body of knowledge regarding phylogenetic and functional diversity in complex microbial communities. Microbial fuel cell systems have been explored for identifying organisms that have unique metabolic functions within mixed consortia sampled from different environments [7–10] and to divert energy away from methanogenesis in favor of other forms of anaerobic respiration [11–13].
Microbial fuel cells also offer another cultivation strategy that enables the simultaneous exploration of biofilm and planktonic populations [2, 14, 15]. Biofilms are formed by individual bacterial species and/or consortia of species that lead to densely packed microbial populations densely contained within a protein and polysaccharide matrix secreted by its bacterial constituents [16–19]. As such, biofilms represent one of the most complex and dynamic microbial architectures. The ability to study biofilms in concert with planktonic communities is a particularly intriguing advantage when studying rumen microbial ecology, given that mixed populations of microorganisms may have varied community structures throughout the rumen and at different times during digestion [19–22].
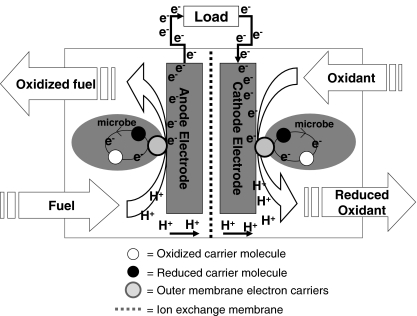
Biofilms form naturally, or artificially, on MFC components [23]. Microbial fuel cells exploit the energy metabolism of microbes that electrically interact with the conductive surfaces in the system called electrodes (Fig. 1). The electrical interaction is facilitated by direct contact with the electrode surface in the form of an electrochemically active biofilm or may also occur by way of extracellular chemical electron shuttles (or mediators) that are reduced by cells in the medium and re-oxidized at the electrode surface. Microbes within a biofilm or planktonic culture will enzymatically extract electrons from organic components, or in some cases H2 [24] and/or H2S [25], in the surrounding media and transfer them to the electrode, which serves as an electron acceptor for biological respiration and/or maintenance. Completion of the MFC reaction takes place in a physically separate but electronically and ionically linked compartment where different bacterial biofilms use the cathode electrode as a source for energy during the reduction of oxidants such as nitrate, sulfate, fumarate, or oxygen [26–30].
Figure 1.
Microbial fuel cell schematic for wastewater management operating with microbes as catalysts for fuel oxidation at the anode electrode and oxidant reduction at the cathode electrode. If sludge is used as the fuel and oxygen as the oxidant, then the net reaction, without nitrification, is: C18H19O9N + H+ → 8H2O + 18CO2 + NH+4 [132]
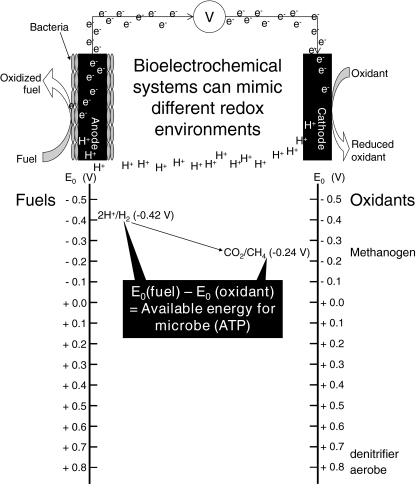
Microbial fuel cells facilitate a redox environment that can be precisely controlled by electron flow and may serve as ideal tools for cultivating microorganisms as biofilms and/or active planktonic cultures. The redox potentials recorded within an environment correspond to the thermodynamic parameters that govern specific chemical and biological reactions [31]. A schematic representation of redox potentials and microbial processes is shown in Fig. 2. Microbial energy metabolism is impacted by redox potential because growth is limited by the amount of energy that can be gained through the coupling of oxidation and reduction reactions [32]. The greater difference between the oxidation and reduction potentials, the more energy an organism can gain to facilitate growth and/or maintenance. However, microorganisms have adapted to nearly every redox environment corresponding to both high and low energy yields. Therefore, to gain insights relative to microbial energy metabolism in different environments, it is critical to understand the mechanisms of energy flow and transformation in reactive biofilms and suspended cultures. Current flow in a MFC can affect the redox energy available for microbial growth and begin to affect the metabolic activity of the microbial community [6]. In the case of rumen microbial ecology, it may be desirable to employ MFC systems that can facilitate carbon dioxide (CO2) reduction and H2 oxidation, conditions that are ideal for the competitive consumption of H2 in the rumen (Fig. 2). In this way, MFC systems may be utilized to select for existing consortia of microorganisms that can outcompete methanogens during H2 consumption [13]. Manipulating the redox environment of rumen microorganisms may be achieved by operating the MFC with external loads, i.e., increasing or decreasing the electron flow rates (current); or by applying a constant potential to the system such that the electron acceptor, or donor, available to the reactive community does not fluctuate. In the case of applied potentiostatic conditions, the system is no longer being operated as a dynamic MFC; however, precision control of the redox environment can be achieved. The engineered devices that can facilitate potentiostatic operations are more generally referred to as bioelectrochemical systems (BES) [5, 33–35]. Bioelectrochemical systems provide highly controlled environments that enable experimental analysis of microbial energy metabolism [36–40]. Utilizing BES, researchers can explore enrichment techniques to cultivate desirable microbes for improving rumen efficiency and also investigate potential electrochemical interventions to treat pathogenic biofilms.
Figure 2.
Redox energy and MFC schematic [31]
Microbial Fuel Cells and Potential Applications for Ruminant Microbial Ecology
Nutrition
The rumen is a complex ecosystem comprised of bacteria, archaea, protozoa, and fungi that are specifically adapted to allow ruminants to digest a variety of feedstuffs including forages and grains. The microbial enzymes break down starch and fiber constituents through anaerobic fermentation resulting in the production of volatile fatty acids (VFA) that are, in turn, used by the ruminant as an energy source. The adult dairy cow rumen is thought to contain approximately 1016 bacteria, 1014 Archaea, 1011 protozoa, and 108 fungi [41]. These broad groups of microbes have specific roles in the rumen with bacteria and protozoa contributing to the majority of starch and fiber digestion. Archaea, consisting primarily of methanogens, remove H2 to complete an anaerobic fermentation process that results in CH4 production. The fungi play variable roles depending on ration composition [42–44]. Within the rumen, microbial communities exist as biofilms living on tissue surfaces, biofilms associated with particulate matter, and planktonic cells free floating in the rumen fluids. Biofilm formation and microbiome establishment is an integral step in the digestion of recalcitrant carbohydrates [45]. Many fiber-associated biofilms form and are strongly adhered within minutes of forage entering the rumen [46]. Ruminal biofilms often contain primary and secondary carbohydrate digesters as well as bacteria that utilize intermediary metabolites [47]. This suggests communalism is one of the important life strategies of these species consortia. Species diversity is greater in adhered biofilm microbiomes than in either the fiber associated or planktonic communities. Biofilm consortia appear essential to efficient utilization of the dietary matrix in ruminants. As such, if not all potential sites for microbial activity are regularly sampled, and thoroughly explored, there exists another opportunity for under estimation of rumen microbial diversity.
The specific roles and interactions of microbial species in the rumen relative to feed digestion and production efficiency have not been comprehensively studied at the community level. This is due in large part to the fact that routine/traditional culture-based microbiological methods that have dominated the field of rumen microbiology have not allowed for the culturing of the bulk of the species present in the rumen which need very stringent growth conditions [48, 49]. Therefore, much of the work to date on ruminant microbial ecology has been restricted to a few species, most of which are bacteria, and which, based on our limited current understanding of the extent of microbial diversity in the rumen, may not represent the majority of the rumen microbial biomass. This biased representation of community diversity and abundance of uncultured microbes contributes to significant gaps in our understanding of the microbial species and their roles in ruminant digestion. For example, the most commonly described bacteria are those that favor growth in laboratory conditions [50]. These species may only be preferentially selected during perturbation of the rumen ecosystem as may be encountered during rapid diet changes including adaptation to high grain rations and may not reflect microbial distributions in steady states. Essentially, the properties that support their growth during transitional states in vivo would be expected to increase the probability of detectable growth in vitro, thus differentially selecting for these organisms under laboratory conditions.
A more comprehensive appreciation of the diversity of rumen microbial communities will have profound impact on our understanding of nutrient digestion, production efficiency, ruminant metabolic diseases, and environmental impacts of cattle production. Given the inherent advantage of precision redox control, MFCs may play a new, and significant, role in cultivating rumen microorganisms.
Evaluation to date of rumen microbial communities using molecular methods has begun to identify some important features of this ecosystem. Archaea and Eukarya comprise approximately 0.5% to 3% and 1% to 16% of the rumen metagenome, respectively [42, 51]. The Eukarya, and specifically protozoa, play an important role in digestion despite contributing a relatively small proportion of the metagenome size. In addition, because of the size of protozoa relative to other microbes, they represent approximately 50% of total microbial nitrogen, an important component of total protein nutrition [42, 52]. Archaea in the rumen are generally thought to be methanogens [42, 44, 53], although evidence of Archaea species within the rumen that do not cluster with known methanogens has been reported [54].
Several studies have examined differences in diversity of rumen microbial communities associated with differences in ration composition. Several such studies focused on protozoa and Archaea populations. Generally, substantial differences can be observed in the rumen communities of cattle fed the same diet [51, 55, 56]. In addition, distinct microbial communities appear to develop within fractions of the rumen content with differences in microbial communities observed between liquid and fiber-adherent fractions [47, 51, 57, 58]. As expected, rumen microbial communities differ between cattle fed forage diets and those fed diets containing increasing proportions of grain [47, 50, 56, 59]. However, studies have been inconsistent in measuring the relative extent of diversity with some results supporting greater diversity in corn-based diets [56] whereas others have found greater diversity in forage diets [47]. These differences may be attributed to differences in ration composition, specific gene targets, or sampling methods.
The associated restrictions to sampling rumen populations from live animals may add to the underestimation of rumen microbial diversity, given that rumen microbial communities exist as biofilms living on tissue surfaces, biofilms associated with particulate matter, and planktonic cells free floating in the rumen fluids. If not all potential sites for microbial growth are regularly sampled, there exists another opportunity for underestimating microbial diversity. Importantly, shifts in diversity of microbial populations may be observed overtime including changes within several hours following feeding [44, 60]. Inconsistent changes have been observed in microbial communities following addition of monensin sodium (i.e., Rumensin®, Elanco Animal Health, Greenfield, IN), an ionophore, to the ration [61]. Bacterial genetic profiles have also been observed to cluster with respect to feed efficiency traits correlated with shifts in VFA composition [62]. To better understand the complex relationship between rumen microbial ecology and imposed dietary and/or environmental changes, it is necessary, although challenging, to temporally investigate all rumen components that feature high levels of microbial activity.
Commonly, ruminant nutrition research has emphasized evaluating performance outcomes, selected metabolic processes, and products of ruminal digestion independent of the microbial constituents of the rumen. Therefore, the application of genomic and metagenomic sequencing to rumen microbial populations presents a tremendous opportunity for more comprehensive understanding of ruminant nutrition, metabolism, and nutrient utilization. A better, in situ, characterization of how microbial activity relates to nutrient utilization provides the opportunity to cultivate, isolate, and challenge microbial populations that will benefit ruminant nutrition. This knowledge base can be expanded by integrating MFCs and metatranscriptomic approaches to further elucidate microbial diversity under different specified redox conditions and characterize the genetic components within a given community that may offer metabolic advantages. One potential outcome of this technology integration is the identification of new ways to modify the rumen microbial population to enhance adaptation to varying diets. Modification of the rumen microbial population may significantly decrease the incidence of metabolic disorders that commonly occur during adaptation to new diets such as indigestion, bloat, and acidosis.
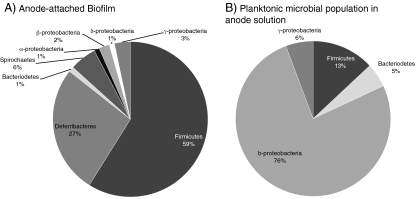
Microbial fuel cell research has shown that the system microbial composition will change relative to substrate composition and electrochemical operating conditions [9, 10, 13, 14, 30, 33, 37, 39, 40, 63–74]; however, more research is needed to explore these affects on rumen microbial consortia. Rumen fluids have been utilized as inocula for MFCs for the purpose of identifying biocatalysts that could be used to produce electricity from cellulose [3]. Rismani-Yazdi and coauthors conducted a study using one MFC system operated under different conditions to identify electrogenic communities that could utilize cellulose as the primary electron donor. DNA extraction from the anode-associated biofilms and suspended microbial consortia were independently analyzed using the 16S rRNA gene sequences from these two samples, and population differences were observed between the biofilm and suspended cultures. Firmicutes was found to be the dominant phylum in the anode-associated biofilm, while β-proteobacteria were the dominant phylum in the suspended culture (Fig. 3a, b). Interestingly, both of these phyla are known to play a role in digestion and it has recently been demonstrated that some Firmicutes strains play a significant, and previously unconsidered, role during anodic electron transfer in thermophilic MFC systems [75]. It is unknown how current flow in an MFC system directly affects the enrichment of microbial communities harvested from rumen environments; however, results from MFC systems inoculated with other environmental consortia strongly suggest that there is a correlation between electron flow and microbial diversity [2, 5, 6, 12, 39, 40, 70, 73, 76, 77].
Figure 3.
Genus-level diversity based on clone libraries obtained from MFCs inoculated with rumen fluids a anode-associated biofilm; b suspended population in anode compartment. Charts are adapted from tables reported in Rizmani-Yazdi et al. [3]
Environmental Impacts
Microbial CH4 production in the rumen (and to a lesser degree the large intestine) is intimately associated with production efficiency. Carbon lost to CH4 is ultimately an energy loss to the animal [78]. These energy loses are estimated to be between 2% and 12% of the gross energy intake depending on diet [79]. Methane is also a greenhouse gas that contributes to global warming, and CH4 production resulting from the global livestock sector accounts for 37% of anthropogenic CH4 emissions [80]. Archaea predominantly utilize H2 and CO2 to produce CH4. The removal of H2 is a critical step to maintain fermentative activity by the other groups [81], and more importantly, there is a stoichiometric balance between VFA and CH4 production [82]. Therefore, alterations in CH4 production have the potential to increase energy in the form of VFAs in the rumen [83]. Most estimates of enteric CH4 emissions have been derived from cattle fed high-forage diets or grazing pastures [84–87] or high-concentrate rations containing barley as the predominant grain source [84, 88]. However, there has been limited exploration of how microbial ecology is linked to enteric CH4, and particularly little has been reported about using advanced molecular platforms to investigate this relationship [78]. Significant associations have been identified between CH4 production and a variety of feed additives [87, 88], managed grazing systems [86], and residual feed intake [83, 89, 90]. Importantly, the effects are often highly correlated with feed intake, which may be substantially altered by feed additives such as ionophores. Microbial fuel cells may offer another avenue for affecting enteric CH4 and VFA production by way of impacting H2 utilization. Hydrogen may be used as an electron donor for electrogenic microbial communities and, therefore, facilitate conditions that are more favorable for propionate and acetate production.
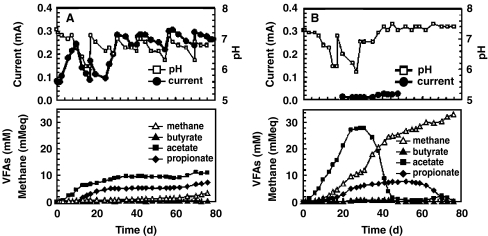
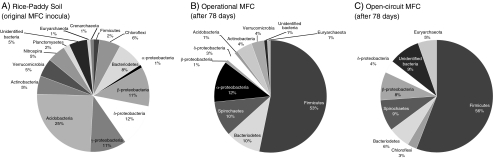
Microbial fuel cells have been used to study microbial diversity associated with methanogenesis in environmental systems. For example, Ishii and coauthors inoculated two MFCs with soil samples from CH4 producing rice-paddy fields [12]. One MFC was operated at open circuit (no current flow), and the other was operated with a 510-Ω resistor connecting the anode and cathode electrodes (high current flow). Ishii et al. found that current producing conditions in the MFC corresponded to a significant decrease in methanogenesis and temporally increasing concentrations of acetate and propionate (Fig. 4a). The MFC that was kept at open circuit (no current flow) showed increasing CH4 production and variable trends of VFA concentrations (Fig. 4b). Microbial diversity was examined in the original inocula, and from both MFC systems, using 16S rRNA gene sequencing. DNA was extracted directly from the rice-paddy soil and, after 78 days, the anode-attached biofilms and suspended microbes (mixed together) from both the open circuit and operational MFC systems. The sequencing results showed that the microbial diversity of the rice-paddy soil was significantly decreased during the MFC residence time. Acidobacteria represented the dominant genus in the soil samples. However, different phylotypes of Firmicutes became the dominant genus in the MFC systems (Fig. 5a–c).
Figure 4.
Current vs. time (top) and corresponding temporal measurements of volatile fatty acids (VFA) and methane production (bottom) for a MFC operated at high-current flow conditions and b MFC held at open-circuit conditions. Figure adapted from Ishii et al. [12]
Figure 5.
Genus-level diversity based on clone libraries obtained from a rice-paddy soil; b operational MFC; and c open-circuit MFC. Charts are adapted from tables reported in Ishii et al. [12]
The results from this study strongly suggest that MFC systems are useful tools for studying mixed CH4-producing consortia and selecting for organisms within these communities that out-compete methanogens. Similar methods could be employed using rumen-derived inocula to examine the microbiological basis for variable enteric CH4 production in ruminants associated with base diet and feed additives. Despite the fact that enteric CH4 is not currently regulated as part of control programs for greenhouse gas emissions [91], it is important to recognize the potential impact of ruminant digestion on the production of CH4 and other targeted gases, particularly since CH4 production represents a substantial inefficiency in conversion of dietary substrate to energy to support maintenance and growth. It may be possible to one day utilize operational MFCs, maintained across a constant load, in vivo to significantly decrease methanogenesis and simultaneously produce more VFAs that are in turn available for animal utilization. Much more research is needed to explore this conjecture and verify that in vivo MFCs would not adversely impact animal health; however, with the rapid advancement of cost–effective biocompatible materials and improved MFC system design, it is not unimaginable to envision a MFC architecture that could be directly used in the rumen to enhance VFA production and decrease methanogenesis. Alternatively, utilizing MFC’s to study rumen microbial ecology coupled with metagenomics may reveal novel probiotic candidates that may have similar impact on methanogenesis. Exploiting these two benefits would significantly impact the efficiency of cattle production and simultaneously decrease the emissions of harmful greenhouse gasses.
Infectious Disease
Infectious diseases in livestock remain a substantial threat to animal well-being despite vast improvements in therapeutic and preventive approaches. In dairy and beef cattle production, the most common and economically important infectious diseases include bacterial mastitis, viral and bacterial respiratory infections, and viral and bacterial enteric disease [92, 93]. Much of the research devoted to these diseases has focused on the major etiologic agents with limited consideration for how potential interactions with other pathogenic and nonpathogenic microbes may influence persistence and pathogenicity.
Many of the economically important infectious diseases of ruminants are not caused by a single etiologic agent. For example, bovine respiratory disease complex is associated with viral pathogens including bovine viral diarrhea virus, bovine herpesvirus type-1, parainfluenza virus type-3, bovine respiratory syncytial virus, and bacterial pathogens including Mannheimia haemolytica, Pasteurella multocida, Histophilus somni, and Mycoplasma bovis. Co-infection with viral and bacterial pathogens is common, and viral components often increase susceptibility to bacterial pathogens (e.g., immunosuppressive nature of bovine viral diarrhea infection) [94]. Similarly complex etiologies are observed with neonatal scours in cattle where viruses, bacteria, and parasites may cause disease in calves of varying ages as microbial populations in the juvenile gastrointestinal system evolve [95].
Of particular relevance to application of MFCs in the study of ruminant disease is the role of biofilms in establishing infection. The biofilm contributes to the persistence of pathogenic bacteria by aiding evasion of host defense mechanisms and may facilitate transfer of genetic material conferring specific drug resistance or virulence properties among biofilm consortia [19]. The contributions of biofilms to pathogen persistence and infection have been examined for S. enterica serovar Typhimurium [96], Pseudomonas aeruginosa and Staphylococcus spp. isolated from clinical cases of mastitis [97–102], H. somni in cardiopulmonary tissue and respiratory disease [100, 103], and Mycobacterium avium subsp. paratuberculosis, the causative agent of Johne's disease in ruminants that has also been linked to Crohn's disease in humans [104–108]. In addition, biofilm formation has been reported as a prominent factor in the pathogenesis of bloat, a metabolic disease of ruminants associated with impaired eructation of gas from the rumen and has been described with food safety pathogens including E. coli O157:H7 [109] and Enterococcus faecalis [110]. Biofilms have also been implicated as contributing to co-infection with multiple pathogenic bacteria as may be observed in bovine respiratory disease complex [103]. MFCs offer another resource for studying biofilm dynamics related to ruminant diseases. Because biofilms can be cultivated in MFC systems under controlled conditions, they may be explored relative to different redox parameters and further analyzed using techniques such as stable isotope probing ([111, 112]; McLean et al. 2009, in preparation), metagenomics [113], and metatranscriptomics [114]. A combined physiological and genomic approach to studying pathogenic biofilms, enabled by MFC systems may contribute to a better understanding of the populations that cause certain diseases and uncover more information about their metabolism, gene regulation, synergistic relationships, and susceptibility to various treatments.
Waste Management
Waste in the form of feces, urine, and associated overland transport in runoff or slurry pumping are significant challenges to continued development of intensive livestock production throughout the world. The quantities of waste may be substantial; an average dairy cow produces in excess of 35 kg of fecal and urine waste daily, and contemporary dairy production facilities may have thousands of cows in production [115]. A variety of methods have been developed for managing waste streams from livestock production systems. Horizontal-flow biofilm reactors have been implemented for remediation of wastewater prior to spreading on agricultural lands as a means of carbon and nitrogen removal [116, 117]. Anaerobic digesters have also been employed to reduce waste volumes and produce CH4. Methane produced from these systems is typically burned to produce heat, which is used to maintain the necessary temperature profile in the digester, and electricity through the process of cogeneration. Biogas production through anaerobic digestion has been a cost–effective solution for waste management; however, these systems produce a significant amount of secondary biomass, which can be challenging to dispose, and represent a loss of usable energy.
Microbial fuel cells can also play a more “traditional” role in the cattle production environment relative to waste management. Much of the MFC research to date has been dedicated to development of these systems for bioenergy, bioremediation, and wastewater treatment [118]. Researchers have begun exploring MFC systems as tools for treating, and recovering energy from, swine wastewater [119–121]. Kim and coauthors found that MFC systems, inoculated with swine manure wastewater as the sole source of microbes and substrate, could remove over 99% of ten chemicals associated with odor and 84% of the organic matter in less than 11 days of operation [119].
Energy recovery from waste streams has also been investigated using microbial electrolysis cells (MECs), a technology originating from MFCs that utilizes biocatalysts in combination with an external power source to drive electrolysis reactions [122]. These systems have been utilized to produce H2 gas from acetate and swine waste, and preliminary data have shown the effective production of clean burning alternative fuel and the simultaneous decrease in overall waste footprint [123, 124]. Methane is also evolved from MEC systems, which decreases the overall production of H2; however, emerging research efforts have suggested that MECs may be used as a “polishing step” for the treatment of effluent from anaerobic digesters [125]. Using MEC systems, CH4 and H2 can be produced under low organic loading conditions and at room temperature, a significant advantage over traditional anaerobic digesters.
Microbial activity plays an important role in all of the described approaches for waste management in livestock production including methanogenesis from anaerobic digestion and aerobic or anaerobic fermentation of waste. As expected, shifts in bacterial diversity are observed during transition through potential mitigation strategies [126].
In a H2 production-based system, mitigating growth of methanogenic microbes should increase net H2 yields. Much research is presently ongoing to understanding how these relationships occur and can be systemically controlled [127]. A greater understanding of CH4 mitigation at this level may also benefit our development of cultivation strategies to affect rumen microbial diversity. Development of novel waste mitigation strategies that employ MFCs as experimental platforms to study microbial population dynamics or primary treatment devices may help to address challenges in waste management in livestock production. In addition, MFCs could be applied directly to recover energy in the form of electricity that could be utilized to offset consumption of energy derived from external sources.
Conclusions
Ruminant production systems are faced with a variety of challenges. Chief among them are the need to: (1) improve the efficiency by which grains and forages are converted to food and fiber to meet increasing global demand; (2) complete the conversion of natural resources into consumable products in a sustainable manner with limited environmental impact; and (3) achieve these goals while supporting and improving animal health and well-being. Microbial communities play an integral role in all of these challenges such that the health, efficiency, and environmental impact of a ruminant cannot be distinguished from that of its inherent microbial community. For these reasons, it is essential to develop methods for characterizing complex microbial communities and their associated dynamics. Research to date has largely focused on a narrow proportion of ruminant microbes, in part due to limitations of culture-based methods in microbiology. These limitations have further contributed to an inability to fully characterize microbial diversity and how it may change in time or in response to nutrient composition and concentration. The capacity to examine linkages between ruminants and microbial communities in ecological contexts will be necessary to derive the next evolution of interventions that may enhance production and mitigate environmental impacts.
Developing tools for improving ruminant production systems will have a positive impact on global society. The direct relationship between increased nutrient utilization efficiency and decreased enteric CH4 production creates the opportunity to define solutions for this dual purpose, which would have significant benefit to mitigating global warming and increasing global food supplies. For example, the US Environmental Protection Agency estimated that the enteric CH4 resulting from livestock production accounted for 32% of the global, non-CO2, agricultural emissions in the year 2000; and this level is expected to increase by more than 30% by the year 2020. Further, CH4 is approximately 21 times more powerful at warming the atmosphere than CO2 over a 100-year period. Fortunately, CH4 is also known to only have a 12-year chemical lifetime in the atmosphere (relative to CO2, which has a 100-year lifetime). The immediate decrease of anthropogenic emissions of CH4 has therefore become a feasible near-term target for the mitigation of global warming [128].
Decreasing enteric CH4 emissions during ruminant production would have the added benefit of increasing feed efficiency, therefore, requiring fewer animals to produce the same protein resource. The Food and Agriculture Organization of the United Nations (FOA) has reported that diets in developing countries are changing as incomes rise, contributing to an increased consumption of meat and dairy products. Between 1964–1966 and 1997–1999, per capita meat consumption in developing countries rose by 150%, and by 2030, per capita consumption of livestock products could rise by another 44% [129]. The FOA further speculates that demand for livestock products will grow faster than production in developing countries, creating a growing trade deficit. Meat products are expected to rise steeply, from 1.2 million tons a year (1997–1999) to 5.9 million tons in 2030, and it is reported that the increasing share of livestock production will likely come from industrial enterprises. In recent years, production from this sector has grown twice as fast as that from more traditional mixed farming systems and more than six times faster than from grazing systems [129].
Given the predicted increase in global industrial livestock production, it will be essential to develop interventions for preventing disease among the increasing ruminant population. As has been discuss previously, microbial biofilm-related infections such as BRD and bloat have an enormous impact on ruminant health and production economics. The development of new interventions to prevent and treat such diseases is yet another benefit that can be realized from understanding microbial ecology as it relates to ruminants.
Recent advances in molecular and microbiological platforms are beginning to facilitate alternative research approaches that contribute to more comprehensive understanding of the phylogenetic and functional diversity in microbial communities and the consequences of perturbations within these systems. As has been discussed, MFCs and related technologies are an example of an emerging platform that contributes to a greater understanding of microbial ecology and complex microbial systems. Several examples of how these systems may apply to ruminant research have been proposed within this review. MFCs have been widely utilized to isolate microbes that have unique energy metabolisms and thrive in extreme environments; therefore, it is easy to consider that these systems may be additionally exploited to study the complex microbial relationships that exist in the rumen. For example, Archaeal populations have been among the microbial constituents targeted in MFC research and are more thoroughly addressed here with respect to environmental impacts and nutrient inefficiency in production attributed to carbon loss through enteric CH4 production. MFC or BES systems could be utilized to explore the Archaeal populations that exist in the rumen, and perhaps even be designed as in situ devises to specifically enrich for microbial consortia that will increase feed efficiency and decrease methanogenesis. However, one of the more immediate benefits that MFCs could provide is the ability to cultivate and study biofilms under defined redox environments that may be set to mimic the rumen or conditions that may arise during an infection.
The applications of MFCs and the ability to cultivate targeted microbial populations that are described here are by no means comprehensive and cannot be considered feasible without incorporating advanced tools for characterizing the molecular diversity of mixed microbial communities. The coupling of metagenomic and metatranscriptomic techniques with new cultivation strategies and research methodologies has been described elsewhere, and a full discussion is outside the scope of this manuscript [52, 130, 131]. However, it is exciting to consider how MFCs may complement the growing arsenal of advanced tools that contribute to a greater understanding of the microbial world. The research opportunities provided by MFC technology extend beyond the generation of electricity and represent a unique opportunity to study and control the impact of microbial ecology and physiology on complex systems such as those found in ruminant biology.
Acknowledgements
The authors would like to thank the Texas Cattle Feeders Association for hosting the planning session that led to the development of this manuscript and Drs. Garry Adams and Eric Eisenstadt for their insightful comments and suggestions. Funding for this effort was provided by the J. Craig Venter Institute.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Orianna Bretschger, Phone: +1-858-2001824, Email: obretschger@jcvi.org.
Jason B. Osterstock, Phone: +1-806-6775600, Email: jbosterstock@ag.tamu.edu
William E. Pinchak, Phone: +1-940-5529941, Email: bpinchak@ag.tamu.edu
Karen E. Nelson, Phone: +1-301-7957565, Email: kenelson@jcvi.org
References
- 1.Griffin D. Economic impact associated with respiratory disease in beef cattle. Vet Clin North Am Food Anim Pract. 1997;13:367–377. doi: 10.1016/s0749-0720(15)30302-9. [DOI] [PubMed] [Google Scholar]
- 2.Aelterman P, Rabaey K, De Schamphelaire L, Clauwaert P, Boon N, Verstraete W. Microbial fuel cells as an engineered ecosystem. In: Wall JD, Harwood CS, Demain AL, editors. Bioenergy. Washington, DC: ASM; 2008. [Google Scholar]
- 3.Rismani-Yazdi H, Christy AD, Dehority BA, Morrison M, Yu Z, Tuovinen OH. Electricity generation from cellulose by rumen microorganisms in microbial fuel cells. Biotechnol Bioeng. 2007;97:1398–1407. doi: 10.1002/bit.21366. [DOI] [PubMed] [Google Scholar]
- 4.Schröder U. Anodic electron transfer mechanisms in microbial fuel cells and their energy efficiency. Phys Chem Chem Phys. 2007;9:2619–2629. doi: 10.1039/b703627m. [DOI] [PubMed] [Google Scholar]
- 5.Aelterman P, Freguia S, Keller J, Verstraete W, Rabaey K. The anode potential regulates bacterial activity in microbial fuel cells. Appl Microbiol Biotechnol. 2008;78:409–418. doi: 10.1007/s00253-007-1327-8. [DOI] [PubMed] [Google Scholar]
- 6.Cheng KY, Ho G, Cord-Ruwisch R. Affinity of microbial fuel cell biofilm for the anodic potential. Environ Sci Technol. 2008;42:3828–3834. doi: 10.1021/es8003969. [DOI] [PubMed] [Google Scholar]
- 7.Choi Y, Jung E, Park H, Paik SR, Jung S, Kim S. Construction of microbial fuel cells using thermophilic microorganisms, Bacillus licheniformis and Bacillus thermoglucosidasius. Bull Korean Chem Soc. 2004;25:813–818. [Google Scholar]
- 8.Clauwaert P, van der Ha D, Verstraete W. Energy recovery from energy rich vegetable products with microbial fuel cells. Biotechnol Lett. 2008;30:1947–1951. doi: 10.1007/s10529-008-9778-2. [DOI] [PubMed] [Google Scholar]
- 9.Ishii Si, Shimoyama T, Hotta Y, Watanabe K. Characterization of a filamentous biofilm community established in a cellulose-fed microbial fuel cell. BMC Microbiology. 2008;8:6–18. doi: 10.1186/1471-2180-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jung S, Regan J. Comparison of anode bacterial communities and performance in microbial fuel cells with different electron donors. Appl Microbiol Biotechnol. 2007;77:393–402. doi: 10.1007/s00253-007-1162-y. [DOI] [PubMed] [Google Scholar]
- 11.Kaku N, Yonezawa N, Kodama Y, Watanabe K. Plant/microbe cooperation for electricity generation in a rice paddy field. Appl Microbiol Biotechnol. 2008;79:43–49. doi: 10.1007/s00253-008-1410-9. [DOI] [PubMed] [Google Scholar]
- 12.Ishii Si, Hotta Y, Watanabe K. Methanogenesis versus electrogenesis: morphological and phylogenetic comparisons of microbial communites. Biosci Biotechnol Biochem. 2008;72:286–294. doi: 10.1271/bbb.70179. [DOI] [PubMed] [Google Scholar]
- 13.Prathap P, César IT, Hyung-Sool L, Rosa K-B, Bruce ER. Syntrophic interactions among anode respiring bacteria (ARB) and Non-ARB in a biofilm anode: electron balances. Biotechnol Bioeng. 2009;103:513–523. doi: 10.1002/bit.22267. [DOI] [PubMed] [Google Scholar]
- 14.Lee J, Phung NT, Chang IS, Kim BH, Sung HC. Use of acetate for enrichment of electrochemically active microorganisms and their 16S rDNA analyses. FEMS Microbiol Lett. 2003;223:185. doi: 10.1016/S0378-1097(03)00356-2. [DOI] [PubMed] [Google Scholar]
- 15.Franks AE, Nevin KP, Jia H, Izallalen M, Woodard TL, Lovley DR. Novel strategy for three-dimensional real-time imaging of microbial fuel cell communities: monitoring the inhibitory effects of proton accumulation within the anode biofilm. Energy and Environmental Science. 2009;2:113–119. [Google Scholar]
- 16.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyd A, Chakrabarty AM. Pseudomonas aeruginosa biofilms: role of the alginate exopolysaccharide. J Ind Microbiol. 1995;15:162–168. doi: 10.1007/BF01569821. [DOI] [PubMed] [Google Scholar]
- 18.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial Biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 19.Nadell CD, Xavier JB, Foster KR. The sociobiology of biofilms. FEMS Microbiol Rev. 2009;33:206–224. doi: 10.1111/j.1574-6976.2008.00150.x. [DOI] [PubMed] [Google Scholar]
- 20.Santegoeds CM, Muyzer G, de Beer D. Biofilm dynamics studied with microsensors and molecular techniques. Water Sci Technol. 1998;37:125–129. [Google Scholar]
- 21.Ramos C, Licht TR, Sternberg C, Krogfelt KA, Molin S. Microbial growth in biofilms: part B, special environments and physiochemical aspects. In: Doyle RJ, editor. Methods in enzymology. San Diego: Academic; 2001. pp. 21–42. [DOI] [PubMed] [Google Scholar]
- 22.Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Feng Y, Ren N, Wang H, Lee H, Li N, Zhao Q. Accelerated start-up of two-chambered microbial fuel cells: effect of anodic positive poised potential. Electrochimica Acta. 2009;54:1109–1114. [Google Scholar]
- 24.Lovley DR, Phillips EJP, Lonergan DJ. Hydrogen and formate oxidation coupled to dissimilatory reduction of iron and manganese by Alteromonas putrefaciens. Appl Environ Microbiol. 1989;55:700–706. doi: 10.1128/aem.55.3.700-706.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rabaey K, VandeSompel K, Maignien L, Boon N, Aelterman P, Clauwaert P, DeSchamphelaire L, Pham HT, Vermeulen J, Verhaege M, Lens P, Verstraete W. Microbial fuel cells for sulfide removal. Environ Sci Technol. 2006;40:5218–5224. doi: 10.1021/es060382u. [DOI] [PubMed] [Google Scholar]
- 26.Gregory KB, Bond DR, Lovley DR. Graphite electrodes as electron donors for anaerobic respiration. Environ Microbiol. 2004;6:596–604. doi: 10.1111/j.1462-2920.2004.00593.x. [DOI] [PubMed] [Google Scholar]
- 27.Bergel A, Féron D, Mollica A. Catalysis of oxygen reduction in pem fuel cell by seawater biofilm. Electrochem Commun. 2005;7:900–904. [Google Scholar]
- 28.He Z, Angenent Largus T. Application of bacterial biocathodes in microbial fuel cells. Electroanalysis. 2006;18:2009–2015. [Google Scholar]
- 29.Clauwaert P, Rabaey K, Aelterman P, DeSchamphelaire L, Pham TH, Boeckx P, Boon N, Verstraete W. Biological denitrification in microbial fuel cells. Environ Sci Technol. 2007;41:3354–3360. doi: 10.1021/es062580r. [DOI] [PubMed] [Google Scholar]
- 30.Chen G-W, Choi S-J, Lee T-H, Lee G-Y, Cha J-H, Kim C-W. Application of biocathode in microbial fuel cells: cell performance and microbial community. Appl Microbiol Biotechnol. 2008;79(3):379–388. doi: 10.1007/s00253-008-1451-0. [DOI] [PubMed] [Google Scholar]
- 31.Brock TD, Madigan MT. Biology of microorganisms. Englewood Cliffs: Prentice-Hall; 1991. [Google Scholar]
- 32.Nealson KH, Belz A, McKee B. Breathing metals as a way of life: geobiology in action. Antonie Van Leeuwenhoek International Journal of General and Molecular Microbiology. 2002;81:215–222. doi: 10.1023/a:1020518818647. [DOI] [PubMed] [Google Scholar]
- 33.Rabaey K, Rodriguez J, Blackall LL, Keller J, Gross P, Batstone D, Verstraete W, Nealson KH. Microbial ecology meets electrochemistry: electricity-driven and driving communities. ISME J. 2007;1:9–18. doi: 10.1038/ismej.2007.4. [DOI] [PubMed] [Google Scholar]
- 34.Rozendal RA, Leone E, Keller J, Rabaey K. Efficient hydrogen peroxide generation from organic matter in a bioelectrochemical system. Electrochem Commun. 2009;11:1752–1755. [Google Scholar]
- 35.Rabaey K, Angenent L, Schroder U, Keller J. Bioelectrochemical systems from extracellular electron transfer to biotechnological application. London: IWA; 2009. [Google Scholar]
- 36.Reimers C, Girguis P, Westall J, Newman D, Stecher H, Howell K, Alleau Y. Using electrochemical methods to study redox processes and harvest energy from marine sediments. Geochim Cosmochim Acta. 2005;69:A575–A575. [Google Scholar]
- 37.Kim BH, Park HS, Kim HJ, Kim GT, Chang IS, Lee J, Phung NT. Enrichment of microbial community generating electricity using a fuel-cell-type electrochemical cell. Appl Microbiol Biotechnol. 2004;63:672–681. doi: 10.1007/s00253-003-1412-6. [DOI] [PubMed] [Google Scholar]
- 38.Kim JR, Min B, Logan BE. Evaluation of procedures to acclimate a microbial fuel cell for electricity production. Appl Microbiol Biotechnol. 2005;68:23–30. doi: 10.1007/s00253-004-1845-6. [DOI] [PubMed] [Google Scholar]
- 39.White HK, Reimers CE, Cordes EE, Dilly GF, Girguis PR. Quantitative population dynamics of microbial communities in plankton-fed microbial fuel cells. ISME J. 2009;3:635–646. doi: 10.1038/ismej.2009.12. [DOI] [PubMed] [Google Scholar]
- 40.Reimers CE, Stecher HA, III, Westall JC, Alleau Y, Howell KA, Soule L, White HK, Girguis PR. Substrate degradation kinetics, microbial diversity, and current efficiency of microbial fuel cells supplied with marine plankton. Appl Environ Microbiol. 2007;73:7029–7040. doi: 10.1128/AEM.01209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Attwood GT, Kelly WJ, Altermann EH, Moon CD, Leahy S, Cookson AL. Application of rumen microbial genome information to livestock systems in the postgenomic era. Aust J Exp Agric. 2008;48:695–700. [Google Scholar]
- 42.Lin C, Raskin L, Stahl DA. Microbial community structure in gastrointestinal tracts of domestic animals: comarative analyses using rRNA-targeted oligonucleotide probes. FEMS Microbiol Ecol. 1997;22:281–294. [Google Scholar]
- 43.Kiyoshi T, Takafumi N, Hiroki M, Mutsumi N, Rustam IA. Phylogenetic analysis of archaeal 16S rRNA libraries from the rumen suggests the existence of a novel group of archaea not associated with known methanogens. FEMS Microbiol Lett. 2001;200:67–72. doi: 10.1111/j.1574-6968.2001.tb10694.x. [DOI] [PubMed] [Google Scholar]
- 44.Ziemer CJ, Sharp R, Stern MD, Cotta MA, Whitehead TR, Stahl DA. Comparison of microbial populations in model and natural rumens using 16S ribosomal RNA-targeted probes. Environ Microbiol. 2000;2:632–643. doi: 10.1046/j.1462-2920.2000.00146.x. [DOI] [PubMed] [Google Scholar]
- 45.Weimer PJ, Price NPJ, Kroukamp O, Joubert L-M, Wolfaardt GM, Van Zyl WH. Studies of the extracellular glycocalyx of the anaerobic cellulolytic bacterium Ruminococcus albus 7. Appl Environ Microbiol. 2006;72:7559–7566. doi: 10.1128/AEM.01632-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edwards JE, Huws SA, Kim EJ, Lee MRF, Kingston-Smith AH, Scollan ND. Advances in microbial ecosystem concepts and their consequences for ruminant agriculture. Animal. 2008;2:653–660. doi: 10.1017/S1751731108002164. [DOI] [PubMed] [Google Scholar]
- 47.Larue R, Yu ZT, Parisi VA, Egan AR, Morrison M. Novel microbial diversity adherent to plant biomass in the herbivore gastrointestinal tract, as revealed by ribosomal intergenic spacer analysis and rrs gene sequencing. Environ Microbiol. 2005;7:530–543. doi: 10.1111/j.1462-2920.2005.00721.x. [DOI] [PubMed] [Google Scholar]
- 48.Edwards JE, McEwan NR, Travis AJ, John Wallace R. 16S rDNA library-based analysis of ruminal bacterial diversity. Antonie Van Leeuwenhoek. 2004;86:263–281. doi: 10.1023/B:ANTO.0000047942.69033.24. [DOI] [PubMed] [Google Scholar]
- 49.Whitford MF, Forster RJ, Beard CE, Gong J, Teather RM. Phylogenetic analysis of rumen bacteria by comparative sequence analysis of cloned 16S rRNA genesß. Anaerobe. 1998;4:153–163. doi: 10.1006/anae.1998.0155. [DOI] [PubMed] [Google Scholar]
- 50.Tajima K, Arai S, Ogata K, Nagamine T, Matsui H, Nakamura M, Aminov RI, Benno Y. Rumen bacterial community transition during adaptation to high-grain diet. Anaerobe. 2000;6:273–284. [Google Scholar]
- 51.Brulc JM, Antonopoulos DA, Berg Miller ME, Wilson MK, Yannarell AC, Dinsdale EA, Edwards RE, Frank ED, Emerson JB, Wacklin P, Coutinho PM, Henrissat B, Nelson KE, White BA. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci U S A. 2009;106:1948–1953. doi: 10.1073/pnas.0806191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh B, Gautam SK, Verma V, Kumar M, Singh B. Metagenomics in animal gastrointestinal ecosystem: potential biotechnological prospects. Anaerobe. 2008;14:138–144. doi: 10.1016/j.anaerobe.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 53.Nicholson MJ, Evans PN, Joblin KN. Analysis of methanogen diversity in the rumen using temporal temperature gradient gel electrophoresis: identification of uncultured methanogens. Microb Ecol. 2007;54:141–150. doi: 10.1007/s00248-006-9182-1. [DOI] [PubMed] [Google Scholar]
- 54.Tajima K, Nagamine T, Matsui H, Nakamura M, Aminov RI. Phylogenetic analysis of archaeal 16S rRNA libraries from the rumen suggests the existence of a novel group of archaea not associated with known methanogens. FEMS Microbiol Lett. 2001;200:67–72. doi: 10.1111/j.1574-6968.2001.tb10694.x. [DOI] [PubMed] [Google Scholar]
- 55.Galbraith EA, Antonopoulos DA, White BA. Suppressive subtractive hybridization as a tool for identifying genetic diversity in an environmental metagenome: the rumen as a model. Environ Microbiol. 2004;6:928–937. doi: 10.1111/j.1462-2920.2004.00575.x. [DOI] [PubMed] [Google Scholar]
- 56.Kocherginskaya SA, Aminov RI, White BA. Analysis of the rumen bacterial diversity under two different diet conditions using denaturing gradient gel electrophoresis, random sequencing, and statistical ecology approaches. Anaerobe. 2001;7:119–134. [Google Scholar]
- 57.Briesacher SL, May T, Grigsby KN, Kerley MS, Anthony RV, Paterson JA. Use of DNA probes to monitor nutritional effects on ruminal prokaryotes and Fibrobacter succinogenes S85. J Anim Sci. 1992;70:289–295. doi: 10.2527/1992.701289x. [DOI] [PubMed] [Google Scholar]
- 58.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 59.Tajima K, Aminov RI, Nagamine T, Matsui H, Nakamura M, Benno Y. Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR. Appl Environ Microbiol. 2001;67:2766–2774. doi: 10.1128/AEM.67.6.2766-2774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krause DO, Dalrymple BP, Smith WJ, Mackie RI, McSweeney CS. 16S rDNA sequencing of Ruminococcus albus and Ruminococcus flavefaciens: design of a signature probe and its application in adult sheep. Microbiology. 1999;145:1797–1807. doi: 10.1099/13500872-145-7-1797. [DOI] [PubMed] [Google Scholar]
- 61.Stahl DA, Flesher B, Mansfield HR, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guan IL, Nkrumah JD, Basarab JA, Moore SS. Linkage of microbial ecology to phenotype: correlation of rumen microbial ecology to cattle's feed efficiency. FEMS Microbiol Lett. 2008;288:85–91. doi: 10.1111/j.1574-6968.2008.01343.x. [DOI] [PubMed] [Google Scholar]
- 63.Rabaey K, Boon N, Siciliano SD, Verhaege M, Verstraete W. Biofuel cells select for microbial consortia that self-mediate electron transfer. Appl Environ Microbiol. 2004;70:5473–5482. doi: 10.1128/AEM.70.9.5373-5382.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Phung NT, Lee J, Kang KH, Chang IS, Gadd GM, Kim BH. Analysis of microbial diversity in oligotrophic microbial fuel cells using 16s rdna sequences. FEMS Microbiol Lett. 2004;233:77. doi: 10.1016/j.femsle.2004.01.041. [DOI] [PubMed] [Google Scholar]
- 65.Chung K, Okabe S. Continuous power generation and microbial community structure of the anode biofilms in a three-stage microbial fuel cell system. Appl Microbiol Biotechnol. 2009;83:965–977. doi: 10.1007/s00253-009-1990-z. [DOI] [PubMed] [Google Scholar]
- 66.Mohan SV, Mohanakrishna G, Sarma PN. Effect of anodic metabolic function on bioelectricity generation and substrate degradation in single chambered microbial fuel cell. Environ Sci Technol. 2008;42:8088–8094. doi: 10.1021/es8012529. [DOI] [PubMed] [Google Scholar]
- 67.Chae K-J, Choi M-J, Lee J-W, Kim K-Y, Kim IS. Effect of different substrates on the performance, bacterial diversity, and bacterial viability in microbial fuel cells. Bioresour Technol. 2009;100:3518–3525. doi: 10.1016/j.biortech.2009.02.065. [DOI] [PubMed] [Google Scholar]
- 68.Liu Z, Li H, Liu J, Su Z. Effects of inoculation strategy and cultivation approach on the performance of microbial fuel cell using marine sediment as bio-matrix. J Appl Microbiol. 2008;104:1163–1170. doi: 10.1111/j.1365-2672.2007.03643.x. [DOI] [PubMed] [Google Scholar]
- 69.Freguia S, Rabaey K, Yuan Z, Keller J. Electron and carbon balances in microbial fuel cells reveal temporary bacterial storage behavior during electricity generation. Environ Sci Technol. 2007;41:2915–2921. doi: 10.1021/es062611i. [DOI] [PubMed] [Google Scholar]
- 70.Virdis B, Rabaey K, Yuan ZG, Rozendal RA, Keller J. Electron fluxes in a microbial fuel cell performing carbon and nitrogen removal. Environ Sci Technol. 2009;43:5144–5149. doi: 10.1021/es8036302. [DOI] [PubMed] [Google Scholar]
- 71.Back JH, Kim MS, Cho H, Chang IS, Lee J, Kim KS, Kim BH, Park YI, Han YS. Construction of bacterial artificial chromosome library from electrochemical microorganisms. FEMS Microbiol Lett. 2004;238:65. doi: 10.1016/j.femsle.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 72.Jia Y-H, Tran H-T, Kim D-H, Oh S-J, Park D-H, Zhang R-H, Ahn D-H. Simultaneous organics removal and bio-electrochemical denitrification in microbial fuel cells. Bioprocess Biosyst Eng. 2008;31:315–321. doi: 10.1007/s00449-007-0164-6. [DOI] [PubMed] [Google Scholar]
- 73.Aelterman P, Versichele M, Marzorati M, Boon N, Verstraete W. Loading rate and external resistance control the electricity generation of microbial fuel cells with different three-dimensional anodes. Bioresour Technol. 2008;99:8895–8902. doi: 10.1016/j.biortech.2008.04.061. [DOI] [PubMed] [Google Scholar]
- 74.Venkata Mohan S, Veer Raghavulu S, Sarma PN. Influence of anodic biofilm growth on bioelectricity production in single chambered mediatorless microbial fuel cell using mixed anaerobic consortia. Biosens Bioelectron. 2008;24:41–47. doi: 10.1016/j.bios.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 75.Wrighton KC, Agbo P, Warnecke F, Weber KA, Brodie EL, DeSantis TZ, Hugenholtz P, Andersen GL, Coates JD. A novel ecological role of the Firmicutes identified in thermophilic microbial fuel cells. ISME J. 2008;2:1146–1156. doi: 10.1038/ismej.2008.48. [DOI] [PubMed] [Google Scholar]
- 76.Holmes DE, Bond DR, O’Neil RA, Reimers CE, Tender LR, Lovley DR. Microbial communities associated with electrodes harvesting electricity from a variety of aquatic sediments. Microb Ecol. 2004;48:178–190. doi: 10.1007/s00248-003-0004-4. [DOI] [PubMed] [Google Scholar]
- 77.Erable B, Roncato MA, Achouak W, Bergel A. Sampling natural biofilms: a new route to build efficient microbial anodes. Environ Sci Technol. 2009;43:3194–3199. doi: 10.1021/es803549v. [DOI] [PubMed] [Google Scholar]
- 78.Ellis JL, Dijkstra J, Kebreab E, Bannink A, Odongo NE, McBride BW, France J. Aspects of rumen microbiology central to mechanistic modelling of methane production in cattle. Journal of Agricultural Science. 2008;146:213–233. [Google Scholar]
- 79.Johnson KA, Johnson DE. Methane emissions from cattle. J Anim Sci. 1995;73:2483–2492. doi: 10.2527/1995.7382483x. [DOI] [PubMed] [Google Scholar]
- 80.Gill M, Smith P, Wilkinson JM (2009) Mitigating climate change: the role of domestic livestock. Animal. Published online by Cambridge University Press. doi:10.1017/S1751731109004662 [DOI] [PubMed]
- 81.Janssen PH, Kirs M. Structure of the archaeal community of the rumen. Appl Environ Microbiol. 2008;74:3619–3625. doi: 10.1128/AEM.02812-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Van Soest PJ. Nutritional ecology of the ruminant. Corvallis: O & B; 1982. [Google Scholar]
- 83.Zhou M, Hernandez-Sanabria E, Guan LL. Assessment of the microbial ecology of ruminal methanogens in cattle with different feed efficiencies. Appl Environ Microbiol. 2009;75:6524–6533. doi: 10.1128/AEM.02815-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Beauchemin KA, McGinn SM. Methane emissions from feedlot cattle fed barley or corn diets. J Anim Sci. 2005;83:653–661. doi: 10.2527/2005.833653x. [DOI] [PubMed] [Google Scholar]
- 85.Beauchemin KA, McGinn SM. Methane emissions from beef cattle: effects of fumaric acid, essential oil, and canola oil. J Anim Sci. 2006;84:1489–1496. doi: 10.2527/2006.8461489x. [DOI] [PubMed] [Google Scholar]
- 86.DeRamus HA, Clement TC, Giampola DD, Dickison PC. Methane emissions of beef cattle on forages: efficiency of grazing management systems. J Environ Qual. 2003;32:269–277. doi: 10.2134/jeq2003.2690. [DOI] [PubMed] [Google Scholar]
- 87.McGinn SM, Beauchemin KA, Coates T, Colombatto D. Methane emissions from beef cattle: effects of monensin, sunflower oil, enzymes, yeast, and fumaric acid. J Anim Sci. 2004;82:3346–3356. doi: 10.2527/2004.82113346x. [DOI] [PubMed] [Google Scholar]
- 88.Jordan E, Kenny D, Hawkins M, Malone R, Lovett DK, O'Mara FP. Effect of refined soy oil or whole soybeans on intake, methane output, and performance of young bulls. J Anim Sci. 2006;84:2418–2425. doi: 10.2527/jas.2005-354. [DOI] [PubMed] [Google Scholar]
- 89.Hegarty RS, Goopy JP, Herd RM, McCorkell B. Cattle selected for lower residual feed intake have reduced daily methane production. J Anim Sci. 2007;85:1479–1486. doi: 10.2527/jas.2006-236. [DOI] [PubMed] [Google Scholar]
- 90.Nkrumah JD, Okine EK, Mathison GW, Schmid K, Li C, Basarab JA, Price MA, Wang Z, Moore SS. Relationships of feedlot feed efficiency, performance, and feeding behavior with metabolic rate, methane production, and energy partitioning in beef cattle. J Anim Sci. 2006;84:145–153. doi: 10.2527/2006.841145x. [DOI] [PubMed] [Google Scholar]
- 91.EPA . Mandatory reporting of greenhouse gasses. Washington: Federal Register; 2009. [Google Scholar]
- 92.Dargatz D. Feedlot 99, Part III: health management and biosecurity in U.S. Feedlots. Washington, DC: United States Department of Agriculture, National Animal Health Monitoring System; 1999. [Google Scholar]
- 93.USDA . Dairy 2007. Fort Collins: USDA, Animal and Plant Health Inspection Service, Veterinary Services, National Animal Health Monitoring System; 2007. [Google Scholar]
- 94.Peterhans E, Jungi TW, Schweizer M. BVDV and innate immunity. Biologicals. 2003;31:107–112. doi: 10.1016/s1045-1056(03)00024-1. [DOI] [PubMed] [Google Scholar]
- 95.Navarre CB, Belknap EB, Rowe SE. Differentiation of gastrointestinal diseases of calves. Vet Clin North Am Food Anim Pract. 2000;16:37–57. doi: 10.1016/S0749-0720(15)30136-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ngwai YB, Adachi Y, Ogawa Y, Hara H. Characterization of biofilm-forming abilities of antibiotic-resistant Salmonella typhimurium DT104 on hydrophobic abiotic surfaces. J Environ Qual. 2006;39:278–291. [PubMed] [Google Scholar]
- 97.Fox LK, Zadoks RN, Gaskins CT. Biofilm production by Staphylococcus aureus associated with intramammary infection. Vet Microbiol. 2005;107:295–299. doi: 10.1016/j.vetmic.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 98.Melchior MB, Fink-Gremmels J, Gaastra W. Extended antimicrobial susceptibility assay for Staphylococcus aureus isolates from bovine mastitis growing in biofilms. Vet Microbiol. 2007;125:141–149. doi: 10.1016/j.vetmic.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 99.Melchior MB, Vaarkamp H, Fink-Gremmels J. biofilms: a role in recurrent mastitis infections? Vet J. 2006;171:398–407. doi: 10.1016/j.tvjl.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 100.Oliveira M, Nunes SF, Carneiro C, Bexiga R, Bernardo F, Vilela CL. Time course of biofilm formation by Staphylococcus aureus and Staphylococcus epidermidis mastitis isolates. Vet Microbiol. 2007;124:187–191. doi: 10.1016/j.vetmic.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 101.Sela S, Hammer-Muntz O, Krifucks O, Pinto R, Weisblit L, Leitner G. Phenotypic and genotypic characterization of Pseudomonas aeruginosa strains isolated from mastitis outbreaks in dairy herds. J Dairy Res. 2007;74:955–962. doi: 10.1017/S0022029907002610. [DOI] [PubMed] [Google Scholar]
- 102.Vautor E, Abadie G, Pont A, Thiery R. Evaluation of the presence of the bap gene in Staphylococcus aureus isolates recovered from human and animals species. Vet Microbiol. 2008;127:407–411. doi: 10.1016/j.vetmic.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 103.Sandal I, Shao JQ, Annadata S, Apicella MA, Boye M, Jensen TK, Saunders GK, Inzana TJ. Histophilus somni biofilm formation in cardiopulmonary tissue of the bovine host following respiratory challenge. Microbes Infect. 2009;11:254–263. doi: 10.1016/j.micinf.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 104.Behr MA, Schurr E. Mycobacteria in Crohn's disease: a persistent hypothesis. Inflammatory Bowel Diseases. 2006;12:1000–1004. doi: 10.1097/01.mib.0000228183.70197.dd. [DOI] [PubMed] [Google Scholar]
- 105.Behr MA, Kapur V. The evidence for Mycobacterium paratuberculosis in Crohn's disease. Curr Opin Gastroenterol. 2008;24:17–21. doi: 10.1097/MOG.0b013e3282f1dcc4. [DOI] [PubMed] [Google Scholar]
- 106.El-Zaatari FA, Osato MS, Graham DY. Etiology of Crohn's disease: the role of Mycobacterium avium paratuberculosis. Trends Mol Med. 2001;7:247–252. doi: 10.1016/s1471-4914(01)01983-9. [DOI] [PubMed] [Google Scholar]
- 107.C-w Wu, Schmoller SK, Bannantine JP, Eckstein TM, Inamine JM, Livesey M, Albrecht R, Talaat AM. A novel cell wall lipopeptide is important for biofilm formation and pathogenicity of Mycobacterium avium subspecies paratuberculosis. Microb Pathog. 2009;46:222–230. doi: 10.1016/j.micpath.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huggett J, Dehda K, Zumla A, Rook G. Crohn's disease and MAP. Lancet. 2004;364:2178–2179. doi: 10.1016/S0140-6736(04)17585-5. [DOI] [PubMed] [Google Scholar]
- 109.Skandamis PN, Stopforth JD, Ashton LV, Geornaras I, Kendall PA, Sofos JN. Escherichia coli O157:H7 survival, biofilm formation and acid tolerance under simulated slaughter plant moist and dry conditions. Food Microbiol. 2009;26:112–119. doi: 10.1016/j.fm.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 110.Macovei L, Ghosh A, Thomas VC, Hancock LE, Mahmood S, Zurek L. Enterococcus faecalis with the gelatinase phenotype regulated by the fsr operon and with biofilm-forming capacity are common in the agricultural environment. Environ Microbiol. 2009;11:1540–1547. doi: 10.1111/j.1462-2920.2009.01881.x. [DOI] [PubMed] [Google Scholar]
- 111.Dumont MG, Murrell JC. Stable isotope probing - linking microbial identity to function. Nat Rev Microbiol. 2005;3:499–504. doi: 10.1038/nrmicro1162. [DOI] [PubMed] [Google Scholar]
- 112.Madsen EL. The use of stable isotope probing techniques in bioreactor and field studies on bioremediation. Curr Opin Biotechnol. 2006;17:92–97. doi: 10.1016/j.copbio.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 113.Kalyuzhnaya MG, Lapidus A, Ivanova N, Copeland AC, McHardy AC, Szeto E, Salamov A, Grigoriev IV, Suciu D, Levine SR, Markowitz VM, Rigoutsos I, Tringe SG, Bruce DC, Richardson PM, Lidstrom ME, Chistoserdova L. High-resolution metagenomics targets specific functional types in complex microbial communities. Nat Biotechnol. 2008;26:1029–1034. doi: 10.1038/nbt.1488. [DOI] [PubMed] [Google Scholar]
- 114.Gilbert JA, Field D, Huang Y, Edwards R, Li W, Gilna P, Joing I. Detection of large numbers of novel sequences in the metatranscriptome of complex marine microbial communties. PLoS ONE. 2008;3:e3042. doi: 10.1371/journal.pone.0003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Miner JR, Humenik FJ, Overcash MR. Managing livestock wastes to preserve environmental quality. Ames: Iowa State University Press; 2000. [Google Scholar]
- 116.Clifford E, Rodgers M, de Paor D. Dairy washwater treatment using a horizontal flow biofilm system. Water Sci Technol. 2008;58:1879–1888. doi: 10.2166/wst.2008.526. [DOI] [PubMed] [Google Scholar]
- 117.Rodgers M, de Paor D, Clifford E. Dairy washwater treatment using a horizontal flow biofilm system. J Environ Manag. 2008;86:114–120. doi: 10.1016/j.jenvman.2006.11.027. [DOI] [PubMed] [Google Scholar]
- 118.Rittmann BE. Opportunities for renewable bioenergy using microorganisms. Biotechnol Bioeng. 2008;100:203–212. doi: 10.1002/bit.21875. [DOI] [PubMed] [Google Scholar]
- 119.Kim JR, Dec J, Bruns MA, Logan BE. Removal of odors from swine wastewater by using microbial fuel cells. Appl Environ Microbiol. 2008;74:2540–2543. doi: 10.1128/AEM.02268-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Min B, Kim J, Oh S, Regan JM, Logan BE. Electricity generation from swine wastewater using microbial fuel cells. Water Res. 2005;39:4961. doi: 10.1016/j.watres.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 121.Jung Rae K, Yi Z, John MR, Bruce EL. Analysis of ammonia loss mechanisms in microbial fuel cells treating animal wastewater. Biotechnol Bioeng. 2008;99:1120–1127. doi: 10.1002/bit.21687. [DOI] [PubMed] [Google Scholar]
- 122.Cheng S, Logan BE. Sustainable and efficient biohydrogen production via electrohydrogenesis. Proc Natl Acad Sci U S A. 2007;104:18871–18873. doi: 10.1073/pnas.0706379104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wagner RC, Regan JM, Oh SE, Zuo Y, Logan BE. Hydrogen and methane production from swine wastewater using microbial electrolysis cells. Water Res. 2009;43:1480–1488. doi: 10.1016/j.watres.2008.12.037. [DOI] [PubMed] [Google Scholar]
- 124.Liu H, Grot S, Logan BE. Electrochemically assisted microbial production of hydrogen from acetate. Environ Sci Technol. 2005;39:4317–4320. doi: 10.1021/es050244p. [DOI] [PubMed] [Google Scholar]
- 125.Clauwaert P, Verstraete W. Methanogenesis in membraneless microbial electrolysis cells. Appl Microbiol Biotechnol. 2009;82:829–836. doi: 10.1007/s00253-008-1796-4. [DOI] [PubMed] [Google Scholar]
- 126.McGarvey JA, Miller WG, Zhang R, Ma Y, Mitloehner F. Bacterial population dynamics in dairy waste during aerobic and anaerobic treatment and subsequent storage. Appl Environ Microbiol. 2007;73:193–202. doi: 10.1128/AEM.01422-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang AJ, Liu WZ, Cheng SA, Xing DF, Zhou JH, Logan BE. Source of methane and methods to control its formation in single chamber microbial electrolysis cells. Int J Hydrog Energy. 2009;34:3653–3658. [Google Scholar]
- 128.EPA . Global mitigation of non-CO2 greenhouse gases. Washington: National Service Center for Environmental Publications; 2006. [Google Scholar]
- 129.FAO . World agriculture: towards 2015/2030. Rome: United Nations; 2002. [Google Scholar]
- 130.Schmeisser C, Steele H, Streit WR. Metagenomics, biotechnology with non-culturable microbes. Appl Microbiol Biotechnol. 2007;75:955–962. doi: 10.1007/s00253-007-0945-5. [DOI] [PubMed] [Google Scholar]
- 131.Xu J. Microbial ecology in the age of genomics and metagenomics: concepts, tools, and recent advances. Mol Ecol. 2006;15:1713–1731. doi: 10.1111/j.1365-294X.2006.02882.x. [DOI] [PubMed] [Google Scholar]
- 132.Henze M, Harremoes P, JlC J, Arvin E. Wastewater treatment. Heidelberg: Springer; 2002. [Google Scholar]