Abstract
Purpose
S100A8 is a member of the S100 protein family containing 2EF-hand calcium-binding motifs. S100 proteins are involved in the regulation of a number of cellular processes such as cell cycle progression and differentiation. Altered expression of this protein is associated with various diseases and cancers. The present study aimed to evaluate whether S100A8 has prognostic value for non-muscle-invasive bladder cancer (NMIBC).
Materials and Methods
A total of 103 primary NMIBC samples obtained by transurethral resection were evaluated. mRNA levels were examined by real-time reverse transcriptase polymerase chain reaction (RT-PCR) analysis. The results were compared with clinico-pathological parameters. The Kaplan-Meier method was applied to plot the curves for progression-free survival. The multivariate Cox regression model was used to identify the independent prognostic factors for progression.
Results
mRNA expression levels of S100A8 were significantly related to the progression of NMIBC. Kaplan-Meier estimates demonstrated significant differences in tumor progression according to the level of S100A8 expression (log-rank test, p<0.001). The multivariate Cox regression model revealed that the S100A8 mRNA expression level (hazard ratio: 12.538; 95% confidence interval: 2.245-70.023, p=0.004) was an independent predictor for disease progression of NMIBC.
Conclusions
Expression levels of S100A8 might be a useful prognostic marker for disease progression of NMIBC.
Keywords: Urinary bladder neoplasms, S100A8 protein, Polymerase chain reaction, Biological tumor markers
INTRODUCTION
Bladder cancer (BC), the incidence and mortality of which increases directly with age, is the second most common urological malignancy in Korea and is about 5 times as common in men as in women [1]. Over 70% of human BCs are non-muscle-invasive bladder cancer (NMIBC) that can be treated by transurethral resection (TUR). However, after TUR about three-quarters of patients confront tumor recurrence within 2 years, and 20-30% of patients experience progression to muscle-invasive bladder cancer (MIBC) even with complete TUR and intravesical therapy with bacillus Calmette-Guérin (BCG) [2]. Useful prognostic variables such as grade, stage, tumor diameter, and presence of carcinoma in situ (CIS) and various biological makers have been proposed to assess the prognosis of BC [3-6]. But the efficacy of these variables is inadequate to accurately predict the heterogeneous behavior of BC, and new reliable molecular indicators are required.
The S100 proteins belong to the calcium-binding EF-hand motif superfamily and play essential roles in epithelial tissues, where they are involved in a wide range of cellular processes including transcription, proliferation, and differentiation [7,8]. At least 16 genes of the S100 family, including the gene coding for S100A8, are clustered on human chromosome 1q21, a frequent target region for chromosomal rearrangements that occur during tumor development [9,10]. S100A8, one of the S100 calcium binding proteins, was described to be up-regulated in many cancers including BC and has been implicated in regulating cell proliferation and metastatic processes [11-14].
It is very important to select the aggressive features of patients with NMIBC for adequate management, for example, early radical cystectomy has a superior 5-year survival rate in comparison with bladder-sparing surgery [15]. To our knowledge, few studies have evaluated the prognostic value of S100A8 as a marker of disease progression in NMIBC. So, we performed real-time reverse transcriptase polymerase chain reaction (RT-PCR) to quantify mRNA expression levels of S100A8 in NMIBC tissues and assessed its prognostic value for tumor recurrence and progression in NMIBC.
MATERIALS AND METHODS
1. Patients and tissue samples
We used bladder cancer specimens harvested between 1995 and 2007 from 103 patients at our institution with primary BC in whom transitional cell carcinoma had been diagnosed histologically. Tumors were staged and graded according to standard criteria; tumor grade was determined by the 1973 WHO classification [4,16]. To reduce confounding factors that might affect the analyses, and to make the study population more homogeneous, we excluded any patients diagnosed with a concomitant carcinoma in situ. Collection and analysis of all samples were approved by the Institutional Review Board (IRB approved protocol number 2006-01-001), and informed consent was obtained from each subject.
All tumors were macro-dissected, typically within 15 minutes of surgical resection. Each bladder cancer specimen was confirmed as being representative by analysis of adjacent tissue in fresh frozen sections from TUR specimens and was then frozen in liquid nitrogen and stored at -80℃ until RNA was extracted.
A second TUR was performed 2-4 weeks after the initial resection when it was incomplete or when a high-grade or T1 tumor was detected [4]. Patients who had multiple tumors or large tumors (≥3 cm of diameter) or high-grade NMIBC received one cycle of intravesical BCG treatment [4,17]. Response to treatment was assessed by cystoscopy and urinary cytology. Patients who were free of disease in 3 months after treatment were assessed every 3 months for the first 2 years and every 6 months thereafter [4,17]. We defined recurrence as the recurrence of primary NMIBC with a lower or the same pathologic stage, and progression as disease with a higher TNM stage.
2. RNA extraction and construction of cDNA
One milliliter TRIzol (Invitrogen, Carlsbad, USA) was added to BC tissue and homogenized in a 5 ml glass tube. The homogenate was transferred to a 1.5 ml tube and was mixed with 200µl chloroform. After incubation for 5 min at 4℃, the homogenate was centrifuged for 13 min at 13,000 g at 4℃. The upper aqueous phase was transferred to a clean tube and 500µl isopropanol was added, followed by incubation for 60 min at 4℃C. The tube was then centrifuged for 8 min at 13,000 g and 4℃. Then, the upper aqueous phase was discarded, mixed with 500µl of 75% ethanol, and centrifuged for 5 min at 13,000 g and 4℃. After the upper aqueous layer was discarded, the pellet was dried at room temperature, dissolved with diethylpyrocarbonate (DEPC)-treated water, and stored at -80℃.
The quality and integrity of RNA were confirmed by agarose gel electrophoresis and ethidium bromide staining, followed by visual examination under ultraviolet light. cDNA was then prepared from 1µg of random priming by using a First-Strand cDNA Synthesis Kit (Amersham Biosciences Europe GmbH, Freiburg, Germany) according to the manufacturer's protocol.
3. Real-time PCR
To quantify the expression levels of S100A8, real-time PCR amplification was performed with a Rotor Gene 6000 instrument (Corbett Research, Mortlake, Australia). Real-time PCR assays using SYBR Premix EX Taq (TAKARA BIO INC., Otsu, Japan) were carried out in micro-reaction tubes (Corbett Research, Mortlake, Australia).
The primers used for amplifying S100A8 (185 bp) were 5'-CAT CGA CGT CTA CCA CAA GT-3' and 5'-GAA TGA GGA ACT CCT GGA AG-3'. The PCR reaction was performed in a final volume of 10µl, consisting of 5µl of 2x SYBR premix EX Taq buffer, 0.5µl of each 5'- and 3'- primer (10 pmol/µl), and 1µl of the sample cDNA. The product was purified with a QIAquick Extraction kit (QIAGEN, Hilden, Germany), quantified with a spectrometer (Perkin Elmer MBA2000, Fremont, USA), and sequenced with an automated laser fluorescence sequencer (ABI PRISM 3100 Genetic Analyzer, Foster City, USA). The known concentration of the product was 10-fold serially diluted from 100 pg/µl to 0.1 pg/µl. The dilution series of PCR products were used for establishing the standard curve of real-time PCR.
The real-time PCR conditions were 1 cycle at 96℃ for 20 seconds, followed by 40 cycles of 2 seconds at 96℃ for denaturation, 15 seconds at 60℃ for annealing, and 15 seconds at 72℃ for extension. The melting program was performed at 72-95℃ with a heating rate of 1℃ per 45 seconds. Spectral data were captured and analyzed by using Rotor-Gene Real-Time Analysis Software 6.0 Build 14 (Corbett Research, Mortlake, Australia). All samples were run in triplicate. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was applied as an endogenous RNA reference gene. Gene expression was normalized to the expression of GAPDH.
4. Statistical analysis
Because of its highly skewed distribution, the S100A8 mRNA data were examined as the natural log function and were subsequently back transformed for the interpretation of the model results [18]. Student's t-test was applied to assess the association of the mRNA expression level for the clinical variables. The optimal cutoff for the mRNA level was determined by using a receiver operating characteristics (ROC) curve. The Kaplan-Meier method was used to estimate the time to recurrence and progression, and differences were assessed by using log rank statistics. The prognostic value of the expression level of S100A8 in NMIBC was studied by use of multivariate Cox proportional hazards regression models. Statistical analysis was performed by using SPSS 12.0 software (SPSS Inc., Chicago, USA), and p-values of <0.05 were considered statistically significant.
RESULTS
1. Baseline characteristics
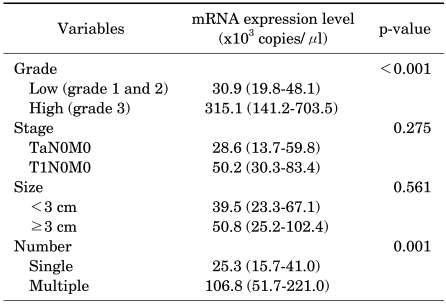
The mean follow-up period of the 103 primary NMIBC patients was 58.2 months (median, 52.2; range, 3.2-137.0). A total of 80 of 103 patients (77.7%) who were reported as having T1 or higher grade received repeat TUR for adequate tumor staging. During the follow-up period, 36 of 103 subjects (35.0%) experienced recurrence and 11 of 103 (10.7%) experienced progression. Two patients with Ta progressed into T1, and the other 9 patients into muscle-invasive or metastatic disease. The mean intervals of recurrence and progression were 44.0 months (median, 31.5; range, 3.0-137.0) and 55.8 months (median, 51.8; range, 3.2-137.0), respectively. Other clinical and pathological features of the enrolled patients are presented in Table 1.
TABLE 1.
Clinical and pathological features of the non-muscle-invasive bladder cancer
2. Quantification of the S100A8 mRNA expression level
Table 2 summarizes the expression levels of S100A8 according to the clinical factors associated with the prognosis of NMIBC. The mRNA expression levels of S100A8 in high-grade and multiple tumors were higher than in lower grade or single tumors, respectively (p<0.05). There was no significant difference in the expression level of S100A8 between patients without recurrence and those with recurrence (p=0.978). S100A8 mRNA levels were significantly higher in the patients with progression than in those without progression (p=0.047).
TABLE 2.
mRNA expression levels of S100A8 in non-muscle-invasive bladder cancer
3. Relations between mRNA expression levels of S100A8 mRNA and recurrence and progression
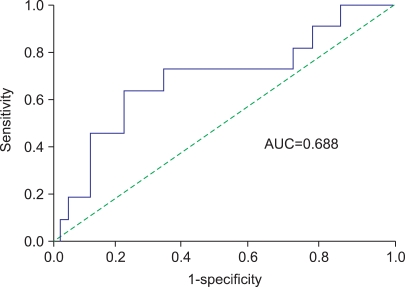
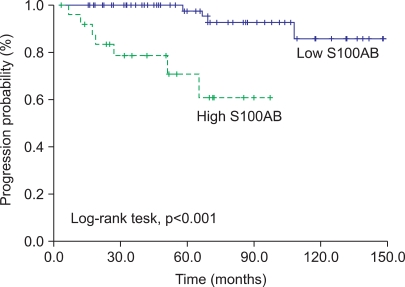
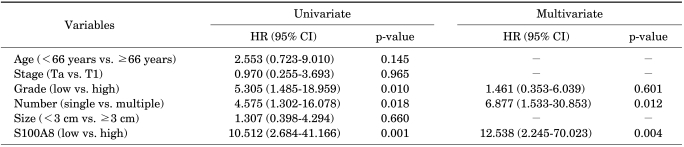
The associations between the S100A8 mRNA expression level and the time to recurrence and progression were analyzed. Recurrent-free survival was not related to the S100A8 mRNA expression level. The cutoff (171.2×103 copies/µl) for progression with the highest combined sensitivity (63.6%) and specificity (79.3%) was determined on the ROC curve (Fig. 1). The patients with decreased S100A8 mRNA expression had significant progression-free survival benefits compared with those with high expression (log-rank test, p<0.001) (Fig. 2). In addition to grade and tumor number, which are already well-known risk factors, S100A8 mRNA expression was a significant risk factor for progression-free survival in the univariate analysis (Table 3). Multivariate Cox regression analyses revealed that the S100A8 expression level (hazard ratio: 12.538, 95% confidence interval: 2.245-70.023, p=0.004) was significantly associated with progression in the patients with primary NMIBC (Table 3).
FIG. 1.
Receiver operating characteristic (ROC) curve generated for progression with the S100A8 mRNA expression level in non-muscle-invasive bladder cancer. AUC: area under the curve.
FIG. 2.
Time to progression in non-muscle-invasive bladder cancer with S100A8 mRNA expression levels.
TABLE 3.
Univariate and multivariate Cox regression analysis for prediction of disease progression
HR: hazards ratio, CI: confidence interval
DISCUSSION
The S100 protein family includes more than 20 entities, all of which are identified only in vertebrates, that share a common structure carrying the calcium-binding EF-hand motif [8]. S100 proteins are low molecular weight, acidic peptides of 9-13 kDa and are involved in the regulation of cell cycle progression, cell growth, differentiation, secretion, and cytoskeletal organization. Most of the S100 proteins, including S100A8, are located in a gene cluster on chromosome locus 1q21, a region in which several rearrangements that occur during tumor development have been observed [9,10]. The functions of S100 proteins in tumorigenesis and tumor progression have not been clearly elucidated, but recent reports have suggested that S100 proteins are commonly up-regulated in tumors and this is often associated with tumor progression [14]. Overexpressed S100 proteins in the colon and the breast include S100A1, S100A4, S100A6, S100A7, and S100B [19]. A number of reports have also described how S100 proteins are involved in the progression of various cancers [20,21].
Few studies have been conducted on S100A8 expression in BCs. The expression of S100A8 was ranked as 1 of 100 genes that would suggest an aggressive phenotype among tumors with protein expression [22]. Tolson et al showed S100A8 to be highly expressed in tumor cells in contrast with normal urothelium in MIBC and suggested S100A8 as a potential diagnostic and prognostic maker of BC [13]. Yao et al proved that S100A8 is significantly overexpressed in BC of rats, mice, and humans [12].
In this report, we investigated the mRNA expression levels of S100A8 in human primary NMIBC tissue. Our data showed that overexpression of S100A8 protein in primary NMIBC was strongly associated with progression in patients with NMIBC. However, overexpression of S100A8 was not associated with tumor recurrence. These results imply that S100A8 may contribute to the generation of certain aspects of the aggressive phenotype rather than simply promoting cell proliferation in NMIBC. The overexpression of S100A8 and S100A9 has also been shown to be associated with poor pathological parameters in invasive ductal carcinoma of the breast [23]. Yong and Moon revealed a functional contribution of S100A8 proteins to processes required for malignant progression including invasion in human gastric cancer cells [11]. Hermani et al suggested that S100A8 may contribute to development and progression or extension of the prostate carcinomas [24]. These results collectively suggest that induction of S100A8 expression may represent aggressive features, such as progression and invasion, in human solid cancers.
There are several possible reasons for the close association of the S100A8 protein with BC and its progression. First, chromosome 1q has been known to exhibit chromosomal rearrangements in various human tumors, including 1q21-24 in BC [25]. As mentioned previously, most S100 proteins, including S100A8, are clustered at the chromosome locus 1q21. Second, the chromosomal region 1q21-q22 contains a high density of CpG islands. Hypermethylation of CpG islands has been shown to be a common mechanism for the inactivation of tumor suppressor genes and is also found in BC [26,27].
There were some limitations in our study. We did not evaluate the protein level of S100A8, such as by Western blot or immunohistochemical staining. We think that further protein study of S100A8 will be needed to confirm its function as a more reliable prognostic marker. In our study, T stage did not predict progression. These discrepancies may reflect differences in the distribution of our patient population. In our population group, the number of T1 patients was larger than the number of Ta patients. Somehow, conventional histopathologic evaluation, encompassing cancer grade and stage, is inadequate to accurately predict the behavior of most bladder cancers, since there were heterogeneous population groups enrolled in studies [28,29]. Therefore, there has been considerable interest in identifying biological indicators of individual tumor aggressiveness. Our finding that the expression of S100A8 was a reliable prognostic indicator of progression for NMIBC, independent of traditional pathologic prognostic parameters, may be an important and valuable contribution to the management of BC.
CONCLUSIONS
S100A8 mRNA levels were significantly higher in patients with progression than in those without progression. The patients with decreased S100A8 mRNA expression had significant progression-free survival benefits compared with those with high expression. The expression of S100A8 was an independent prognostic parameter for tumor progression. Thus, expression levels of S100A8 might be a useful prognostic marker for disease progression of NMIBC.
Footnotes
This work was supported by the research grant of the Chungbuk National University in 2009.
References
- 1.The statistics report: the incidence of cancer on 2003-2005 and the survival rate on 1993-2005. National Cancer Center; 2009. [accessed Apr 1, 2009]. http://www.ncc.re.kr. [Google Scholar]
- 2.Messing EM. Urothelial tumors of the bladder. In: Wein AJ, Kavoussi LR, Novick AC, Partin AW, Peters CA, editors. Campbell-Walsh urology. 9th ed. Philadelphia: Saunders; 2007. pp. 2426–2427. [Google Scholar]
- 3.Wu TT, Chen JH, Lee YH, Huang JK. The role of bcl-2, p53, and ki-67 index in predicting tumor recurrence for low grade superficial transitional cell bladder carcinoma. J Urol. 2000;163:758–760. doi: 10.1016/s0022-5347(05)67798-1. [DOI] [PubMed] [Google Scholar]
- 4.Babjuk M, Oosterlinck W, Sylvester R, Kaasinen E, Böhle A, Palou-Redorta J. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder. Eur Urol. 2008;54:303–314. doi: 10.1016/j.eururo.2008.04.051. [DOI] [PubMed] [Google Scholar]
- 5.Quan C, Park MS, Jo SW, Lee SC, Kim WJ. Effects of transforming growth factor-beta1 and its receptor on the development, recurrence and progression of human bladder cancer. Korean J Urol. 2006;47:426–435. [Google Scholar]
- 6.Ha YS, Yun SJ, Kim YJ, Lee SC, Kim WJ. Utility of Smo as a prognostic marker for human bladder tumors. Korean J Urol. 2007;48:997–1003. [Google Scholar]
- 7.Schäfer BW, Heizmann CW. The S100 family of EF-hand calcium-binding proteins: functions and pathology. Trends Biochem Sci. 1996;21:134–140. doi: 10.1016/s0968-0004(96)80167-8. [DOI] [PubMed] [Google Scholar]
- 8.Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature) Biochem Biophys Res Commun. 2004;322:1111–1122. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- 9.Schäfer BW, Wicki R, Engelkamp D, Mattei MG, Heizmann CW. Isolation of a YAC clone covering a cluster of nine S100 genes on human chromosome 1q21: rationale for a new nomenclature of the S100 calcium-binding protein family. Genomics. 1995;25:638–643. doi: 10.1016/0888-7543(95)80005-7. [DOI] [PubMed] [Google Scholar]
- 10.Schutte BC, Carpten JD, Forus A, Gregory SG, Horii A, White PS. Report and abstracts of the sixth international workshop on human chromosome 1 mapping 2000. Iowa City, Iowa, USA. 30 September-3 October 2000. Cytogenet Cell Genet. 2001;92:23–41. doi: 10.1159/000056867. [DOI] [PubMed] [Google Scholar]
- 11.Yong HY, Moon A. Roles of calcium-binding proteins, S100A8 and S100A9, in invasive phenotype of human gastric cancer cells. Arch Pharm Res. 2007;30:75–81. doi: 10.1007/BF02977781. [DOI] [PubMed] [Google Scholar]
- 12.Yao R, Lopez-Beltran A, Maclennan GT, Montironi R, Eble JN, Cheng L. Expression of S100 protein family members in the pathogenesis of bladder tumors. Anticancer Res. 2007;27:3051–3058. [PubMed] [Google Scholar]
- 13.Tolson JP, Flad T, Gnau V, Dihazi H, Hennenlotter J, Beck A, et al. Differential detection of S100A8 in transitional cell carcinoma of the bladder by pair wise tissue proteomic and immunohistochemical analysis. Proteomics. 2006;6:697–708. doi: 10.1002/pmic.200500033. [DOI] [PubMed] [Google Scholar]
- 14.Salama I, Malone PS, Mihaimeed F, Jones JL. A review of the S100 proteins in cancer. Eur J Surg Oncol. 2008;34:357–364. doi: 10.1016/j.ejso.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 15.Borden LS, Jr, Clark PE, Hall MC. Bladder cancer. Curr Opin Oncol. 2003;15:227–233. doi: 10.1097/00001622-200305000-00009. [DOI] [PubMed] [Google Scholar]
- 16.Greene FL. The American Joint Committee on Cancer: updating the strategies in cancer staging. Bull Am Coll Surg. 2002;87:13–15. [PubMed] [Google Scholar]
- 17.Hall MC, Chang SS, Dalbagni G, Pruthi RS, Seigne JD, Skinner EC, et al. Guideline for the management of nonmuscle invasive bladder cancer (stages Ta, T1, and Tis): 2007 update. J Urol. 2007;178:2314–2330. doi: 10.1016/j.juro.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Bland JM, Altman DG. Transformations, means, and confidence intervals. BMJ. 1996;312:1079. doi: 10.1136/bmj.312.7038.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsieh HL, Schäfer BW, Sasaki N, Heizmann CW. Expression analysis of S100 proteins and RAGE in human tumors using tissue microarrays. Biochem Biophys Res Commun. 2003;307:375–381. doi: 10.1016/s0006-291x(03)01190-2. [DOI] [PubMed] [Google Scholar]
- 20.Cross SS, Hamdy FC, Deloulme JC, Rehman I. Expression of S100 proteins in normal human tissues and common cancers using tissue microarrays: S100A6, S100A8, S100A9 and S100A11 are all overexpressed in common cancers. Histopathology. 2005;46:256–269. doi: 10.1111/j.1365-2559.2005.02097.x. [DOI] [PubMed] [Google Scholar]
- 21.Rafii S, Lyden D. S100 chemokines mediate bookmarking of premetastatic niches. Nat Cell Biol. 2006;8:1321–1323. doi: 10.1038/ncb1206-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dyrskjot L, Kruhoffer M, Thykjaer T, Marcussen N, Jensen JL, Moller K, et al. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64:4040–4048. doi: 10.1158/0008-5472.CAN-03-3620. [DOI] [PubMed] [Google Scholar]
- 23.Arai K, Takano S, Teratani T, Ito Y, Yamada T, Nozawa R. S100A8 and S100A9 overexpression is associated with poor pathological parameters in invasive ductal carcinoma of the breast. Curr Cancer Drug Targets. 2008;8:243–252. doi: 10.2174/156800908784533445. [DOI] [PubMed] [Google Scholar]
- 24.Hermani A, Hess J, De Servi B, Medunjanin S, Grobholz R, Trojan L, et al. Calcium-binding proteins S100A8 and S100A9 as novel diagnostic markers in human prostate cancer. Clin Cancer Res. 2005;11:5146–5152. doi: 10.1158/1078-0432.CCR-05-0352. [DOI] [PubMed] [Google Scholar]
- 25.Qin SL, Chen XJ, Xu X, Shou JZ, Bi XG, Ji L, et al. Detection of chromosomal alterations in bladder transitional cell carcinomas from Northern China by comparative genomic hybridization. Cancer Lett. 2006;238:230–239. doi: 10.1016/j.canlet.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 26.Kim EJ, Kim WJ. The causal relationship between RUNX3 and bladder tumor. Korean J Urol. 2005;46:1192–1198. [Google Scholar]
- 27.Kim EJ, Kim YJ, Jeong P, Ha YS, Bae SC, Kim WJ. Methylation of the RUNX3 promoter as a potential prognostic marker for bladder tumor. J Urol. 2008;180:1141–1145. doi: 10.1016/j.juro.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Sylvester RJ, van der Meijden AP, Oosterlinck W, Witjes JA, Bouffioux C, Denis L, et al. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: a combined analysis of 2596 patients from seven EORTC trials. Eur Urol. 2006;49:466–475. doi: 10.1016/j.eururo.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 29.Millan-Rodriguez F, Chechile-Toniolo G, Salvador-Bayarri J, Palou J, Vicente-Rodriguez J. Multivariate analysis of the prognostic factors of primary superficial bladder cancer. J Urol. 2000;163:73–78. doi: 10.1016/s0022-5347(05)67975-x. [DOI] [PubMed] [Google Scholar]