Abstract
We have developed an enzyme-nanoparticle sensor array where the sensitivity is amplified through enzymatic catalysis. In this approach cationic gold nanoparticles are electrostatically bound to an enzyme (β-galactosidase, β-Gal), inhibiting enzyme activity. Analyte proteins release the β-Gal, restoring activity and providing an amplified readout of the binding event. Using this strategy we have been able to identify proteins in buffer at a concentration of 1 nM, substantially lower than current strategies for array-based protein sensing. Moreover, we have obtained identical sensitivity in studies where the proteins are spiked into the complex protein matrix provided by desalted human urine (~1.5 µM total protein; spiked protein concentrations were 0.067% of the overall protein concentration), demonstrating the potential of the method for diagnostic applications.
Introduction
Irregular protein concentration levels in biofluids, e.g. serum, urine, and saliva provide essential information for the early diagnosis of many pathological conditions such as hypoalbuminemia,1 cancers,2 Alzheimer’s disease,3 prostatisis,4 HIV,5 and other disease states.6 The development of strategies for monitoring protein levels remains a major issue in medical diagnostics, pathogen detection, and proteomics.7 Substantial efforts have been devoted to develop precise and efficient methods for protein sensing,8 including enzyme-labeled immunoassays,9 electrophoresis methods,10 and analytical techniques.11
The "chemical nose/tongue" approach12 presents a potential alternative to specific recognition and separations techniques. In this approach, a sensor array is generated to provide differential interaction with analytes via selective receptors, generating a stimulus response pattern that can be statistically analyzed and used for the identification of individual target analytes13,14 and also analysis of complex mixtures.15 Over the past few years, this technology has been successfully applied for protein detection using array-based approaches, including porphyrins,16 oligopeptide-functionalized resins,17 and polymers.18 In a real world example, a single functional conjugated polymer poly(thiophene) has been successfully applied as a food freshness sensor to detect biogenic amines in fish associated with food poisoning (e.g. histamine) with increasing concentrations from 22.5 µM (2.5 ppm) to 4.5 mM (500 ppm) to build a fish matrix.19 Recently, we have developed nanoparticle-GFP based “chemical nose” strategy for protein detection in biofluid that is highly sensitive (500 nM)20 as compared to other reported similar approaches (1–350 µM).16,17,18c,d We have also developed a sensor array composed of gold nanoparticles and fluorescent polymers that can identify proteins,21 bacteria,22 and cancerous cells23 through a fluorophore-displacement mechanism. This sensor array achieved detection limits of 215 nM for low Mw proteins.
The increased sensitivity required for many diagnostic uses24 presents a challenging goal for array-based sensors because the detection process generally relies on fluorescence responses that are restricted by the inherent emissivity of the fluorophores used. To overcome this limitation, we have explored the use of enzymes to provide array-based sensors with enhanced sensitivity. In this Enzyme Amplified Array Sensing (EAAS) approach, the sensitivity of the array is amplified through an enzymatic reaction. This approach couples the signal amplification process of ELISA with the versatility of the “chemical nose” approach. We report here the use of this method to sense and identify a range of biomedically relevant proteins at 1 nM in both buffer and desalted human urine.
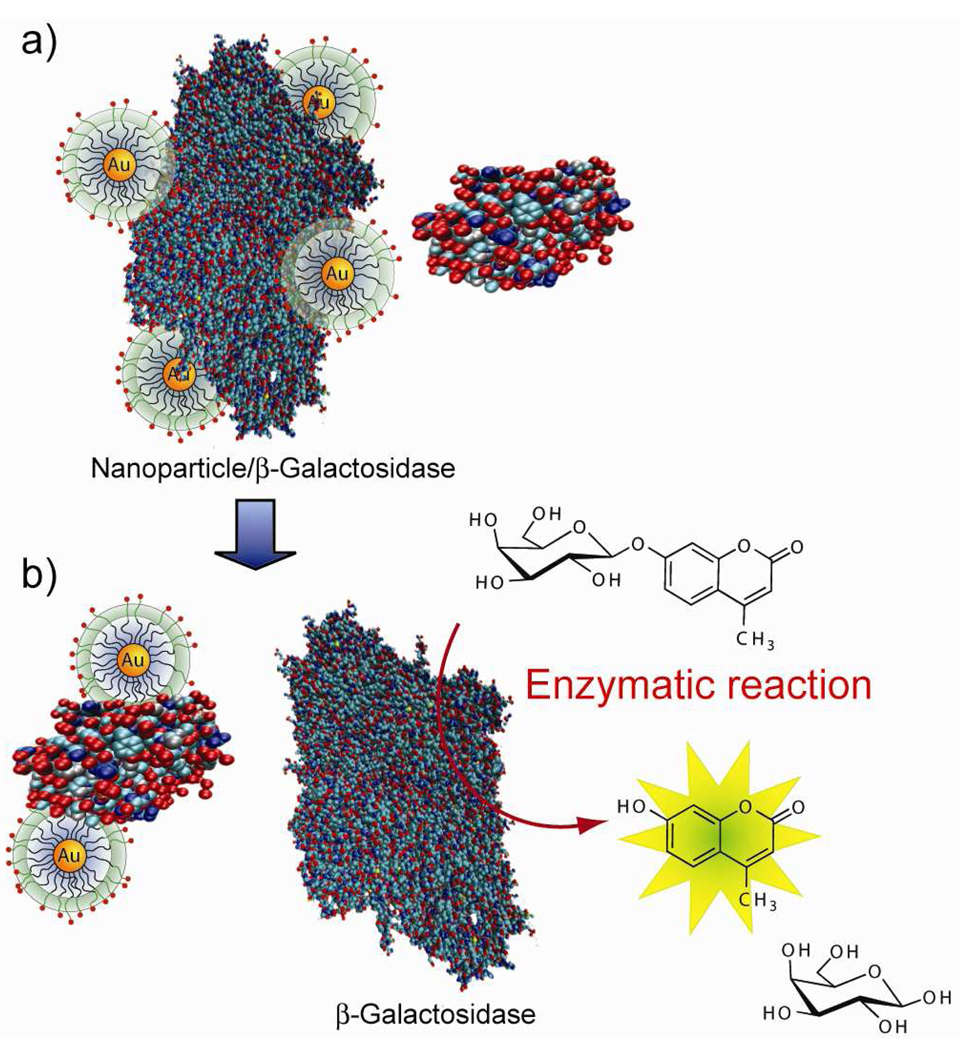
Our EAAS features three components: a) β-galactosidase (β-Gal) as the enzyme, b) 4-methylumbelliferyl-β-D-galactopyranoside (MUG) as a fluorogenic substrate to provide “turn on” sensing, and c) gold nanoparticles (AuNPs) as the receptors to provide differential protein affinity, and hence discrimination. In practice, cationic AuNPs electrostatically bind the anionic β-Gal, inhibiting the enzyme without denaturation.25 Displacement of the particle by analyte proteins restore β-Gal activity, generating a fluorescent readout signal (Figure 1) that is amplified through enzymatic catalysis.
Figure 1.
A schematic representation of sensors comprised of β-galactosidase (β-Gal) and cationic AuNPs. In a) supramolecular adducts of β-Gal and AuNP formed through complementary electrostatic interactions, inhibiting the enzymatic activity of β-galactosidase. As shown in b) β-galactosidase is displaced from the β-Gal/AuNP complex by protein analytes, restoring the catalytic activity of β-Gal towards the fluorogenic substrate 4-methylumbelliferyl-β-D-galactopyranoside, resulting in an amplified signal for detection.
Results and Discussion
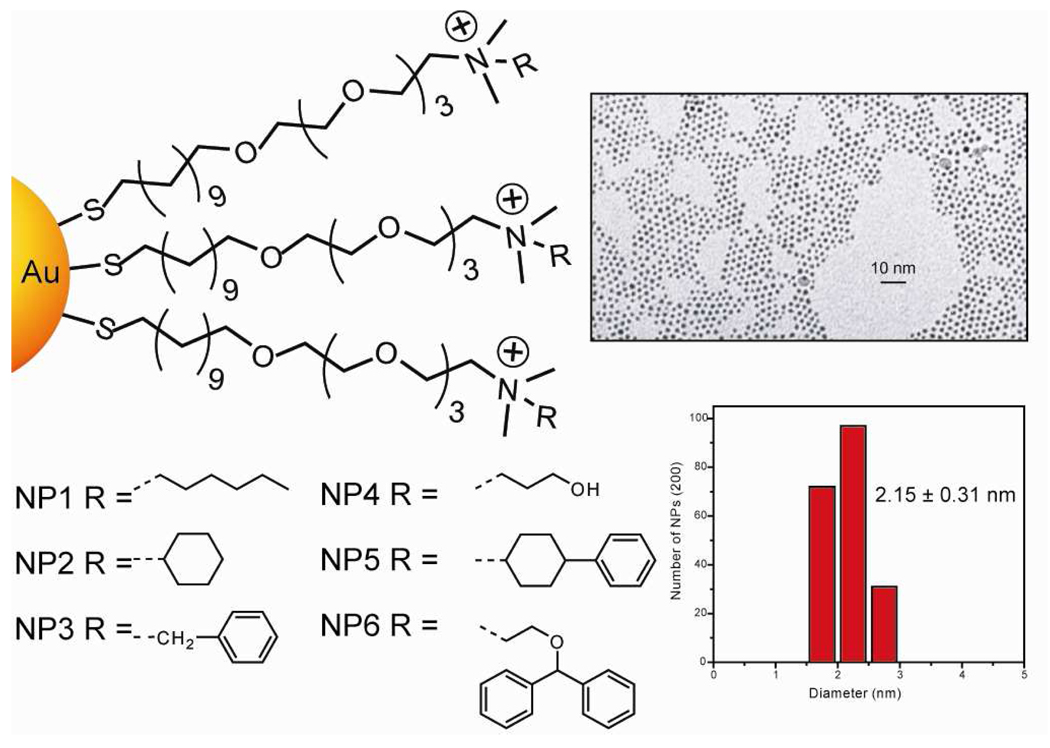
The anionic tetrameric enzyme β-Gal (17.5×13.5×9 nm, pI = 4.6, Mw = 465 kDa), was chosen as the amplifying element due its stability to a wide range of temperature, pH, and ionic strength conditions.26,27 Gold nanoparticles (~2 nm core diameter) with a positive surface charge were used to bind efficiently to the anionic β-Gal through electrostatic complementary and electrostatic charge interactions (see Figures S29, S31, and S33 for zeta potential and DLS measurements). These particles feature a large surface area with size comparable to that of proteins, allowing these systems to mimic protein-protein surface interactions,25 an excellent staring point for sensor design. These AuNPs feature a tetraethylene glycol unit in the ligand shell to minimize the denaturation of the bound enzyme/analyte protein and variable terminal functionality to generate the differential affinity required for sensing (Figure 2, Figure S29, and Table S7).28
Figure 2.
Structure features of the cationic gold nanoparticles (NP1–NP6). The transmission electron microscopy (TEM) and histogram plot show the morphology, monodispersity, and sizes of the metallic core gold nanoparticles.
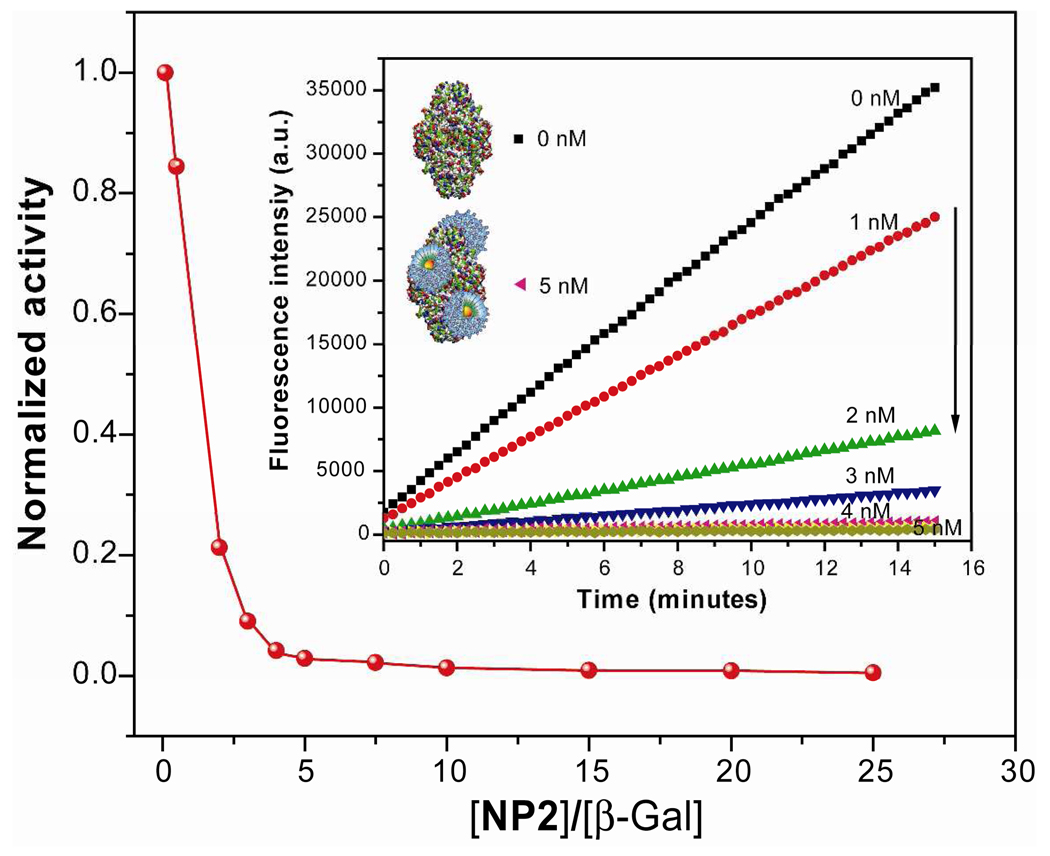
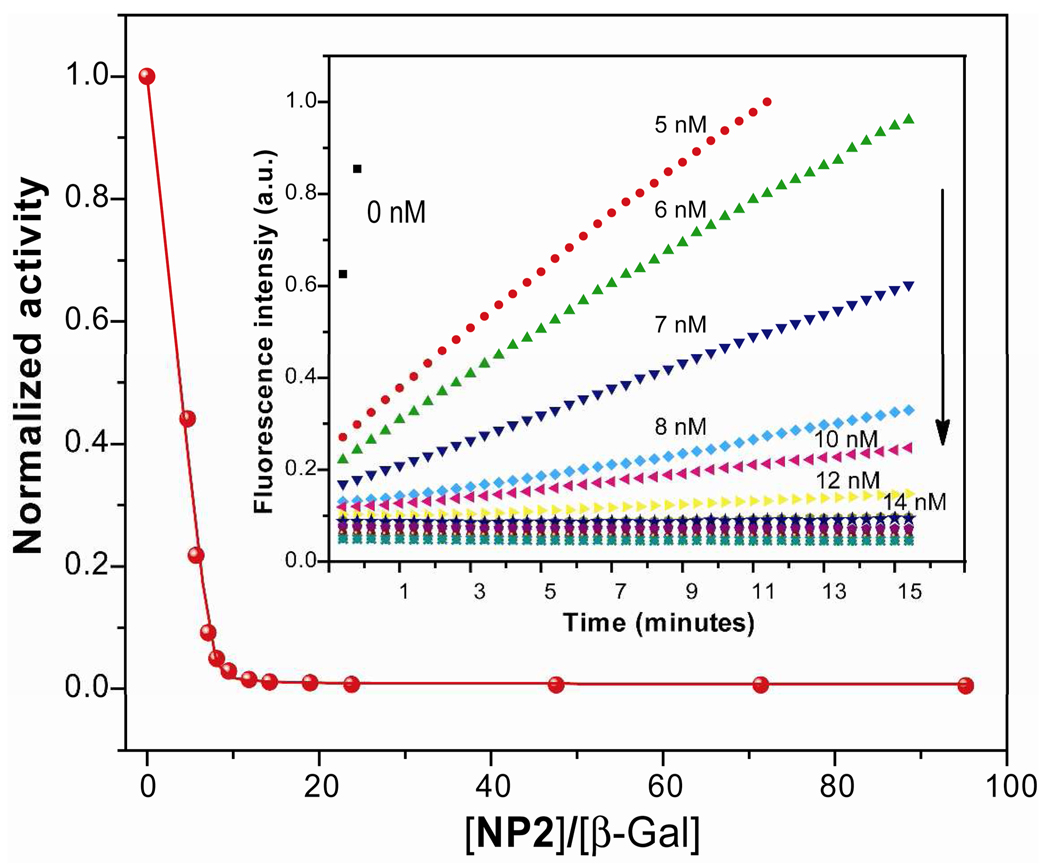
As a starting point, we focused on the optimization of the binding ratio between AuNPs (NP1–NP6) and β-Gal through inhibition activities in phosphate buffer. We conducted an activity assay of β-Gal-catalyzed hydrolysis at various concentrations of nanoparticles (see Figure S26). Typically, a concentration of 0.5 nM of β-Gal in phosphate buffer solution (5 mM, pH = 7.4) was incubated with various concentration of NP1–NP6 for 30 minutes and 1 mM of the fluorogenic substrate (MUG, λexc = 455 nm) was added to AuNP-enzyme complexes for the inhibition and enzyme-substrate reaction studies. As a control, the enzyme inhibition was also studied using neutral tetra(ethylene glycol) functionalized nanoparticles. The normalized first-order rate of fluorogenic substrate hydrolysis was plotted versus the ratio of nanoparticles to β-Gal, and showed a tendency to decrease upon addition of nanoparticles, as shown for NP2 in Figure 3. This result clearly indicates that activity of β-Gal is inhibited by nanoparticle binding. This inhibition of β-Gal activity depends on subtle structural changes of peripheral ligands on the AuNPs with the linear end group (NP1) exhibiting less suppression than a branched isomeric structure (NP2). Control experiments with non-charged NPs (NPTEG) were carried out and no interaction or inhibition of the enzyme was observed (see Fig. S35).
Figure 3.
Normalized inhibition activity of β-Gal (0.5 nM) against 1 mM substrate MUG upon addition of cationic NP2 in 5 mM phosphate buffer. The inset shows the kinetics of the fluorescence spectra before and after addition of NP2. The arrow in the inset indicates the direction of activity (0 nM indicates free enzyme and 5 nM indicates inhibited enzyme with NPs).
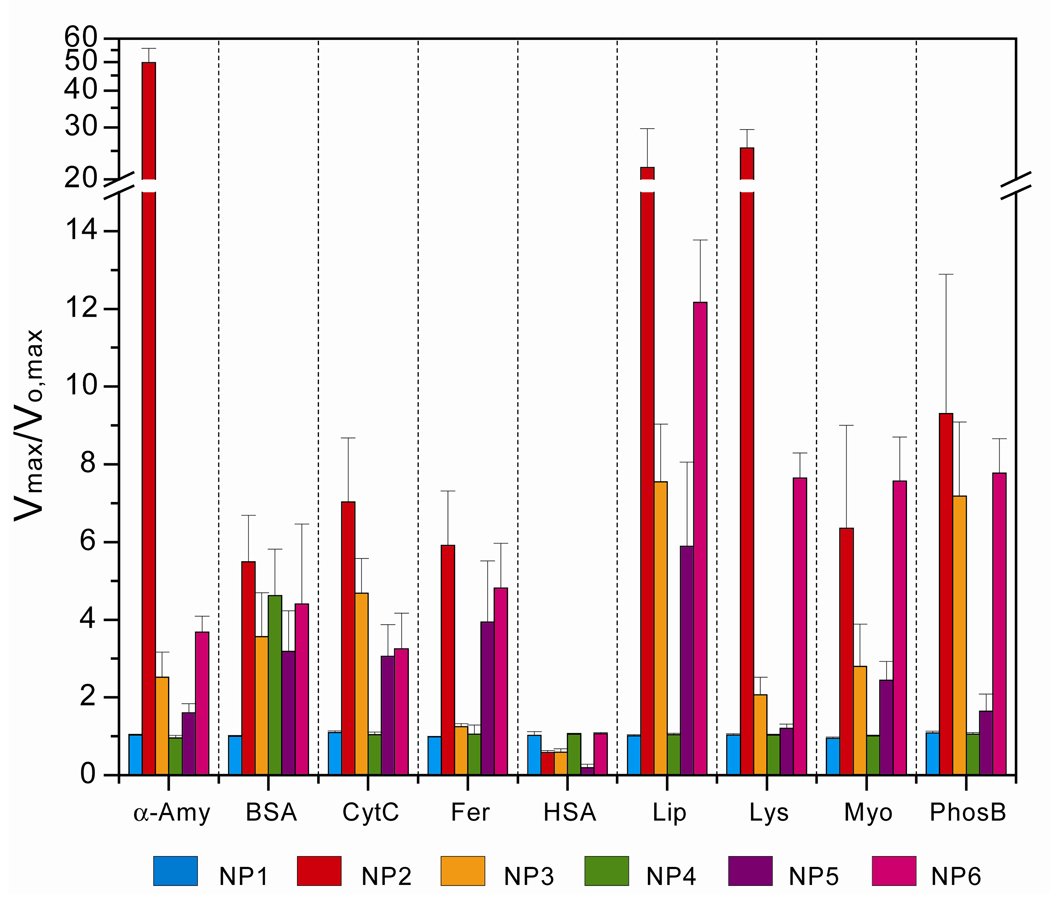
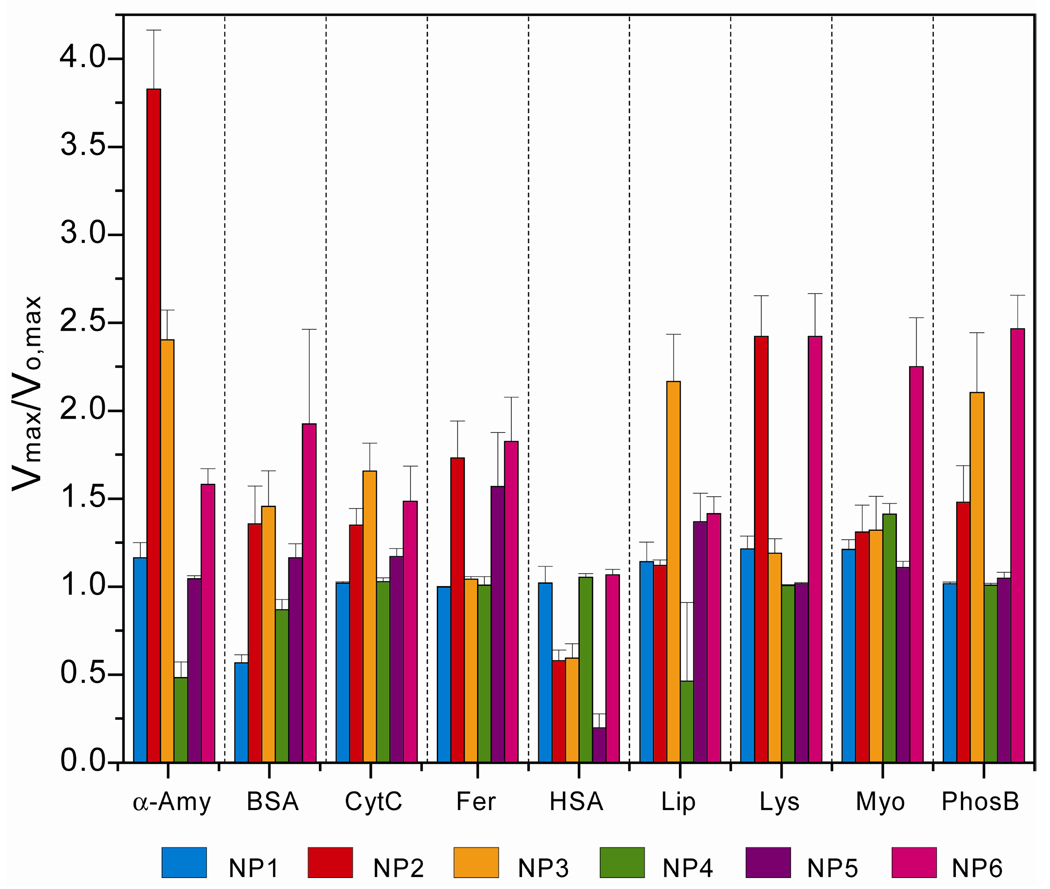
A total of nine proteins of various sizes, surface charges, molecular weights, and isoelectric points were chosen to test generality and limitations of our sensor, (Table 1, see Table S6 for zeta potentials, rh values and extinction coefficients at 280 nm of the analyte proteins). Fluorogenic substrate hydrolysis for the β-Gal/AuNP conjugates against individual proteins in buffer is summarized in Figure 4. The individual target proteins generated distinguishable and highly reproducible rates of the fluorogenesis, indicating the potential for protein discrimination.
Table 1.
Physical properties of the proteins used as sensing targets in phosphate buffer solution at pH 7.4.18a
| Protein‡ | Mw (kDa) | pI |
|---|---|---|
| α-Amylase (α-Am) | 50.0 | 5.0 |
| Bovine serum albumin (BSA) | 66.3 | 4.8 |
| Cytochrome c (CytC) | 12.3 | 10.7 |
| Ferritin (Fer) | 750.0 | 4.5 |
| Human serum albumin (HSA) | 69.4 | 5.2 |
| Lipase (Lip) | 58.0 | 5.6 |
| Lysozyme (Lys) | 14.4 | 11.0 |
| Myoglobin (Myo) | 17.0 | 7.2 |
| Alkaline phosphatase (PhosB) | 140.0 | 5.7 |
Proteins in italics are commonly found in human urine.
Figure 4.
Fluorescence response patterns ratio of β-Gal and six AuNP adducts against various target proteins. Each value represents an average of six parallel measurements with standard deviation.
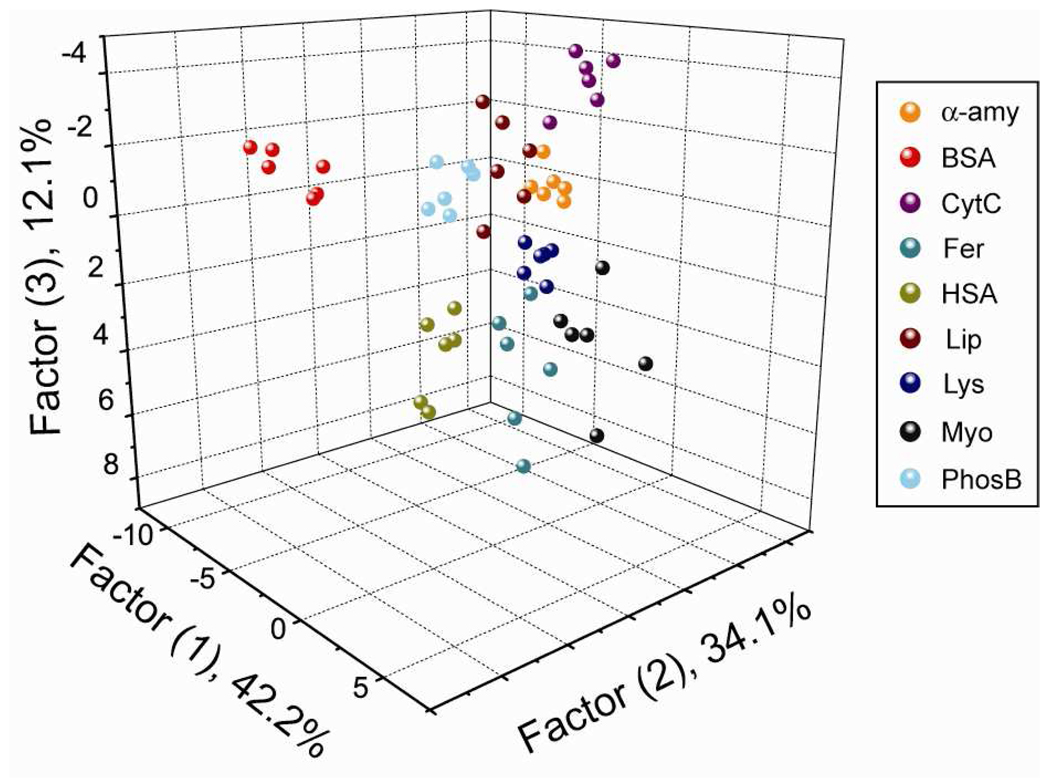
All proteins were tested using a fluorescence displacement assay of six β-Gal/AuNP assemblies array for six replicate measurements, providing a data set as a 6 × 6 × 9 matrix. The resulting data was analyzed through linear discrimination analysis (LDA) using SYSTAT software (version 11)29 and transformed into five canonical factors. This statistical analysis method is used to recognize the linear combination of features that differentiate two or more classes of objects or events. The five canonical factors contain 42.2%, 34.1%, 12.1%, 5.8%, and 4.9% of the variation, respectively. The canonical score plot of the first three factors is presented in Figure 5, where each dot represents the fluorescence response pattern of a single protein target to the β-Gal/AuNP sensor array. The canonical plot reveals nine distinct clusters corresponding to individual target proteins that give rise to a 100% classification accuracy obtained from a jackknifed matrix in LDA. This result demonstrates that the β-Gal/AuNP sensor array is sensitive enough to differentiate target proteins in the 1 nM range, significantly more sensitive than prior methods (1–350 µM),16,17,18c,d including our previous fluorescent polymer-nanoparticle conjugates system (215 nM for the low Mw proteins).21
Figure 5.
Canonical score plot of the first three factors of fluorescence response patterns obtained through β-Gal/AuNP sensor array against nine target proteins in 1 nM concentration.
The high sensitivity of our β-Gal/AuNP sensor array can be attributed to signal amplification through the enzyme-substrate reaction of β-Gal. Significantly, the same training matrix analyzed using only one nanoparticle structure gives rise to classification accuracies of 33%, 44%, 37%, 31%, 44%, and 35% for NP1 to NP6 respectively, indicating almost equal ability of each particle to discriminate between protein targets (Table S3).
To investigate the robustness identification accuracy of the β-Gal/AuNP sensor array, we prepared sixty unknown protein samples at 1 nM randomly chosen from the training set for identification. The fluorescence response patterns obtained for each unknown against the sensor array were analyzed through LDA analysis. The resulting patterns were classified through the canonical score plot by the first two factors of simplified fluorescence patterns based on the Mahalanobis distances of unknowns to the centroid of the respective protein clusters in the canonical score plot. An identification accuracy of 92% (55 correct out 60) demonstrates reproducibility of our enzyme-nanoparticle sensor system for identification.
Sensing of proteins in real world biofluids such as protein in human urine provides a far more demanding test than sensing in simple buffer solutions. The overall protein content (>1.5 µM, 0.150 g/L) and the multianalyte nature of the human urine (>1500 proteins as competing biomolecules) generate a complex matrix that is challenging for sensor design.30 An additional complication is variation in ionic strength, an issue that is addressed biomedically through desalting using spin column chromatography. We employed this technique in our studies but we are aware that as with current analytical methods this desalting adds an additional step to the analytical procedure. (see Figures S27 and S28). 30,31
The complexation between β-Gal and cationic AuNPs in desalted human urine (Bioreclamation Inc.) was determined by the hydrolysis of MUG by β-Gal in the presence of various concentrations of AuNPs (Figure 6, additional information see Figures S34 and S35). In this experiment, β-Gal was dissolved in desalted human urine (~1.5 µM, see SI for details) buffered to pH = 7.4 using 5 mM phosphate buffer. This solution was then equilibrated with a stoichiometric amount of nanoparticles for 15 min. Then, an excess amount of the MUG solution (1 mM) was added to initiate the enzymatic reaction. The activity of β-Gal was directly correlated with the AuNPs concentration, indicating that the activity of β-Gal is inhibited by AuNPs complex formation.
Figure 6.
Normalized inhibition activity of β-Gal (0.5 nM) against 1 mM substrate MUG upon addition of cationic NP2 in the presence of desalted human urinary proteins. The inset shows the kinetics of the fluorescence spectra before and after addition of NP2. The arrow in the inset indicates the direction of activity.
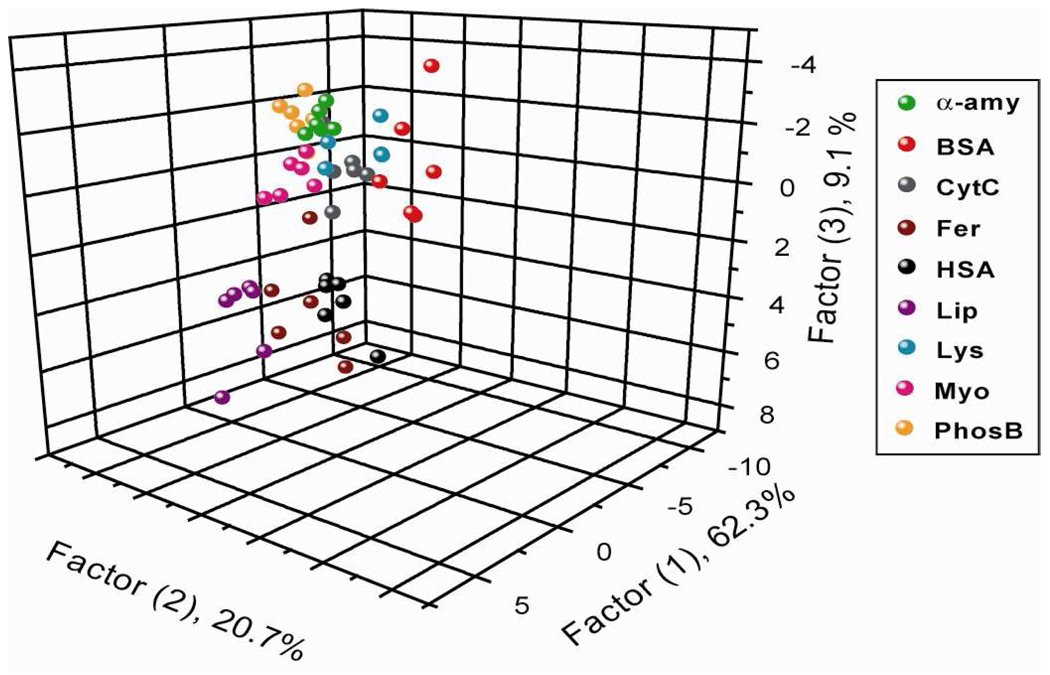
Using the optimized conditions (vide supra), it was established that 1 nM concentration of spiked proteins was required for reproducible differentiation of the target analytes (Table 1). As before, we created a training matrix (six β-Gal/AuNP adducts × nine proteins × six replicates) with β-Gal/AuNP adduct and each of the proteins. Each protein in the human urine protein solution generated a distinct fluorescence response. The rates of fluorogenic substrate hydrolysis for β-Gal/AuNP pair in the presence of individual protein analytes are summarized in Figure 7, showing that NP1 and NP4 exhibit stronger affinity for β-Gal than for other proteins, producing smaller hydrolysis rates and less fluorescence response. As before, this fluorescent response pattern was subjected to further LDA analysis producing a 6 × 6 × 9 matrix. This matrix was transformed into five canonical factors. The five canonical factors contain 62.3%, 20.7%, 9.1%, 4.3%, and 0.9% of the variation, respectively.
Figure 7.
Fluorescence response patterns ratio of β-Gal and six AuNP adducts against various target proteins. Each value represents an average of six parallel measurements.
The canonical score plot of the first three factors is presented in Figure 8, where each dot represents the fluorescence response pattern of a single protein target to the β-Gal/AuNP sensor array. The canonical plot reveals nine distinct clusters corresponding to individual target proteins, giving rise to a 100% classification accuracy based on the jackknifed matrix in LDA. This result demonstrates that the β-Gal/AuNP sensor array is sensitive enough to differentiate each of the target proteins at 1 nM in the biofluid matrix (0.067% of the total protein content in urine), comparable with the preliminary study carried out in buffer. This sensitivity is improved as described before 4-2×102-fold in comparison with simple fluorophore displacement21 and it is also comparable with the preliminary study carried out in buffer. The particles in this study are well suited for differentiation: the same training matrix analyzed using a single nanoparticle gives rise to classification accuracies of 33%, 52%, 41%, 43%, 48%, and 31% from NP1 to NP6, respectively (Table S10). This indicates almost an equal contribution of each particle in the discrimination of the examined protein targets.
Figure 8.
Canonical score plot of the first three factors of fluorescence response patterns ratio obtained through β-Gal/AuNP sensor array against nine target proteins at 1 nM in desalted human urine (~1.5 µM total protein content).
The accuracy of the β-Gal/AuNP sensor array was validated by identifying unknown proteins in the competitive environment of desalted human urine protein solution. Sixty unknown protein solutions spiked at 1 nM were chosen arbitrarily from the training set. The fluorescence response patterns were newly analyzed through LDA analysis, and further classified by the Mahalanobis distances of unknowns to the centroid of the respective protein clusters in the canonical score plot. This process identified 55 out of 60 unknowns correctly, corresponding to a 92% identification accuracy, demonstrating both the feasibility and reproducibility of our enzyme-nanoparticle sensor system.
Conclusions
In this study, we have demonstrated that the use of enzymatic amplification dramatically increases the sensitivity of the array-based sensing of proteins. Using this EAAS method, we rapidly and reproducibly sensed proteins at concentrations of 1 nM in both phosphate buffer and desalted human urine. These studies demonstrate that sensing can be achieved with high sensitivity in a complex biomatrix, providing an important first step for the creation of array-based biosensors for real-word diagnostic applications. In our ongoing studies, we are exploiting both new alternative approaches for protein detection and new data analysis strategies to apply this methodology to more complex matrices featuring a large diversity of target analytes.
Supplementary Material
Acknowledgement
This work was supported by the NSF Center for Hierarchical Manufacturing at the University of Massachusetts Nanoscale Science and Engineering Center (NSEC, DMI-0531171), the NSF (V.M.R., CHE-0808945), and the NIH (GM077173). U.H.F.B thanks the Department of Energy Grant for generous financial support (DE-FG02-04ER46141).
Footnotes
Supporting Information Available. Experimental section, synthesis of ligands, synthesis of gold nanoparticles, 13C NMR and 1H NMR spectra, fluorescence titration curves, training matrix, protein purification, gel electrophoresis, zeta potential, and dynamic light scattering. These materials are available free of charge via the Internet at http://pubs.acs.org.
Contributor Information
Uwe H. F. Bunz, Email: uwe.bunz@chemistry.gatech.edu.
Vincent M. Rotello, Email: rotello@chem.umass.edu.
References
- 1.a) Abdelhafiz AH, Myint MP, Tayek JA, Wheeldon NM. Clin. Thera. 2009;21:1534. doi: 10.1016/j.clinthera.2009.07.015. [DOI] [PubMed] [Google Scholar]; b) Galic G, Tomic M, Galesic K, Kvesic A, Soljic M, Mozetic V, Loncar Z, Maricic A, Martinovic Z. Coll. Antropol. 2009;33:559. [PubMed] [Google Scholar]; c) Zismam DA, Kawut SM, Lederer DJ, Belperio JA, Lynch JP, Schwarz MI, Tayek JA, Reuben DB, Karlamangla AS. Chest. 2009;135:929. doi: 10.1378/chest.08-0754. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Abdelhafiz AH, Wheeldon NM. Clin. Thera. 2004;26:1470. doi: 10.1016/j.clinthera.2004.09.002. [DOI] [PubMed] [Google Scholar]; e) Abdelhafiz AH, Wheeldon NM. Am. J. Geriatr. Pharmacother. 2008;6:1. doi: 10.1016/j.amjopharm.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 2.a) Hogdall EVS, Christensen L, Kjaer SK, Blaakaer J, Kjaerbye-Thygesen A, Gayther S, Jacobs IJ, Hogdall CK. Gynecol.Oncol. 2007;104:508. doi: 10.1016/j.ygyno.2006.09.028. [DOI] [PubMed] [Google Scholar]; b) Daniels MJ, Wang Y, Lee M-Y, Venkitaraman AR. Science. 2004;306:876. doi: 10.1126/science.1102574. [DOI] [PubMed] [Google Scholar]; c) Ross JS, Fletcher JA. Stem Cells. 1998;16:413. doi: 10.1002/stem.160413. [DOI] [PubMed] [Google Scholar]
- 3.a) Couderc R. Ann. Biol. Clin. 2000;58:581. [PubMed] [Google Scholar]; b) Yaffe K, Barrett-Connor E, Lin F, Grady D. Arch. Neurol. 2002;59:378. doi: 10.1001/archneur.59.3.378. [DOI] [PubMed] [Google Scholar]; c) Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, Rossi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ, Hulette C, Crain B, Goldgaber D, Roses AD. Neurology. 1993;43:1467. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]; d) Padmanabhan J, Levy M, Dickson DW, Potter H. Brain. 2006;129:3020. doi: 10.1093/brain/awl255. [DOI] [PubMed] [Google Scholar]; e) Hampel H, Burger K, Teipel SJ, Bokde AL, Zetterberg H, Blennow K. Alzheimers Dement. 2008;4:38. doi: 10.1016/j.jalz.2007.08.006. [DOI] [PubMed] [Google Scholar]; f) Andreasson U, Portelius E, Andersson ME, Blennow K, Zetterberg H. Biomarkers in Medicine. 2007;1:59. doi: 10.2217/17520363.1.1.59. [DOI] [PubMed] [Google Scholar]
- 4.a) Tricoli JV, Schoenfeldt M, Conley B. Clin. Cancer Res. 2004;10:3943. doi: 10.1158/1078-0432.CCR-03-0200. [DOI] [PubMed] [Google Scholar]; b) Bai VU, Kaseb A, Tejwani S, Divine GW, Barrack ER, Menon M, Pardee AB, Reddy GPV. Proc. Nat. Acad. Sci. 2007;104:2343. doi: 10.1073/pnas.0610504104. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Gutman AB, Gutman EB. J. Clin. Investig. 1938;17:473. doi: 10.1172/JCI100974. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Brawer MK, Chetner MP, Beatie J, Buchner DM, Vessella RL, Lange PHJ. Urol. 1992;147:841. doi: 10.1016/s0022-5347(17)37401-3. [DOI] [PubMed] [Google Scholar]; e) Deftos LJ, Abrahamsson PA. Urol. 1998;51:141. doi: 10.1016/s0090-4295(98)00062-4. [DOI] [PubMed] [Google Scholar]; f) Pisitkun T, Johnstone R, Knepper MA. Mol. Cell. Proteomics. 2006;5:1760. doi: 10.1074/mcp.R600004-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.a) Payne BAI, Price DA, Schmid ML, Ong E, Snow MH. J. Infect. 2007;54:e195. [Google Scholar]; b) Nielsen SL, Andersen PL, Koch C, Jensenius JC, Thiel S. Clin. Exp. Immunol. 1995;100:219. doi: 10.1111/j.1365-2249.1995.tb03656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Cantry O, Moja P, Quesnel A, Pozzetto B, Lucht FR, Genin C. Clin. Expe. Immunol. 1997;109:47. doi: 10.1046/j.1365-2249.1997.4261320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Hardy J, Selkoe DJ. Science. 2002;297:353. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]; b) Pulido R, van Huijsduijnen RH. FEBS J. 275:848. doi: 10.1111/j.1742-4658.2008.06250.x. [DOI] [PubMed] [Google Scholar]; c) Masson J-F, Battaglia TM, Khairallah P, Beaudoin S, Booksh KS. Anal. Chem. 2007;79:612. doi: 10.1021/ac061089f. [DOI] [PubMed] [Google Scholar]
- 7.Kodadek T. Chem. Biol. 2001;8:105. doi: 10.1016/s1074-5521(00)90067-x. [DOI] [PubMed] [Google Scholar]
- 8.a) Soukka T, Paukkunen J, Harma H, Lonnberg S, Lindroos H, Lovgren T. Clin. Chem. 2001;47:1269. [PubMed] [Google Scholar]; b) Acharya G, Chang C-L, Doorneweerd DD, Vlashi E, Henne WA, Hartmann LC, Low PS, Savran CA. J. Am. Chem. Soc. 2007;129:15824. doi: 10.1021/ja073094m. [DOI] [PubMed] [Google Scholar]; c) Muller UR. Mol. Biosyst. 2006;2:470. doi: 10.1039/b608442g. [DOI] [PubMed] [Google Scholar]
- 9.a) Engvall E, Perlmann P. J. Immunol. 1972;109:129. [PubMed] [Google Scholar]; b) Schuurs AHWM, Van Weemen BK. J. Immunoassay. 1980;1:229. doi: 10.1080/01971528008055786. [DOI] [PubMed] [Google Scholar]; c) Haab BB. Curr. Opin. Biotechnol. 2006;17:415. doi: 10.1016/j.copbio.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 10.McPherson RA, Pincus MR. Henry’s Clinical Diagnosis and Management by Laboratory Methods. Ch 19. Saunders–Elsevier; 2007. [Google Scholar]
- 11.a) Li JN, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Clin. Chem. 2002;48:1296. [PubMed] [Google Scholar]; b) Baggerly KA, Morris JS, Coombes KR. Bioinformatics. 2004;20:777. doi: 10.1093/bioinformatics/btg484. [DOI] [PubMed] [Google Scholar]
- 12.a) Goodey A, Lavigne JJ, Savoy SM, Rodriguez MD, Curey T, Tsao A, Simmons G, Wright J, Yoo SJ, Sohn Y, Ansyln EV, Shear JB, Neikirk DP, McDevitt JT. J. Am. Chem. Soc. 2001;123:2559. doi: 10.1021/ja003341l. [DOI] [PubMed] [Google Scholar]; b) Albert KJ, Lewis NS, Schauer CL, Sotzing GA, Stitzel SE, Vaid TP, Walt DR. Chem. Rev. 2000;100:2595. doi: 10.1021/cr980102w. [DOI] [PubMed] [Google Scholar]
- 13.a) Wiskur SL, Floriano PN, Anslyn EV, McDevitt JT. Angew. Chem. Int. Ed. 2003;42:2070. doi: 10.1002/anie.200351058. [DOI] [PubMed] [Google Scholar]; b) Lavigne JJ, Savoy SM, Clevenger MB, Ritchie JE, McDoniel B, Yoo SJ, Anslyn EV, McDevitt JT, Shear JB, Neikirk DP. J. Am. Chem. Soc. 1998;120:6429. [Google Scholar]; c) Curey TE, Goodey A, Tsao A, Lavigne JJ, Sohn Y, McDevitt JT, Anslyn EV, Neikirk D, Shear JB. Anal. Biochem. 2001;293:178. doi: 10.1006/abio.2001.5114. [DOI] [PubMed] [Google Scholar]; d) McCleskey SC, Griffin MJ, Schneider SE, McDevitt JT, Anslyn EV. J. Am. Chem. Soc. 2003;125:1114. doi: 10.1021/ja021230b. [DOI] [PubMed] [Google Scholar]
- 14.a) Folmer-Andersen JF, Kitamura M, Anslyn EV. J. Am. Chem. Soc. 2006;128:5652. doi: 10.1021/ja061313i. [DOI] [PubMed] [Google Scholar]; b) Buryak A, Severin KA. J. Am. Chem. Soc. 2005;127:3700. doi: 10.1021/ja042363v. [DOI] [PubMed] [Google Scholar]; c) Greene NT, Shimizu KD. J. Am. Chem. Soc. 2005;127:5695. doi: 10.1021/ja0468022. [DOI] [PubMed] [Google Scholar]; d) Wright AT, Anslyn EV. Chem. Soc. Rev. 2006;35:14. doi: 10.1039/b505518k. [DOI] [PubMed] [Google Scholar]; e) Lee JW, Lee J-S, Chang Y-T. Angew. Chem. Int. Ed. 2006;45:6485. doi: 10.1002/anie.200602055. [DOI] [PubMed] [Google Scholar]; f) Rakow NA, Suslick KS. Nature. 2000;406:710. doi: 10.1038/35021028. [DOI] [PubMed] [Google Scholar]
- 15.a) Rakow NA, Suslick KS. Nature. 2000;406:710. doi: 10.1038/35021028. [DOI] [PubMed] [Google Scholar]; b) Krantz-Rulcker C, Stenberg M, Winquist F, Lundstrom I. Anal. Chim. Acta. 2001;426:217. [Google Scholar]; c) Lewis NS. Acc. Chem. Res. 2004;37:663. doi: 10.1021/ar030120m. [DOI] [PubMed] [Google Scholar]; d) Collins BE, Wright AT, Anslyn EV. Top. Curr. Chem. 2007;277:181. [Google Scholar]; e) Buryak A, Pozdnoukhov A, Severin K. Chem. Commun. 2007:2366. doi: 10.1039/b705250b. [DOI] [PubMed] [Google Scholar]; f) Wright AT, Edwards NY, Anslyn EV, McDevitt JT. Angew. Chem. Int. Ed. 2007;46:8212. doi: 10.1002/anie.200701236. [DOI] [PubMed] [Google Scholar]; g) Stephenson CJ, Shimizu KD. Polym. Int. 2007;56:482. [Google Scholar]
- 16.a) Baldini L, Wilson AJ, Hong J, Hamilton AD. J. Am. Chem. Soc. 2004;126:5656. doi: 10.1021/ja039562j. [DOI] [PubMed] [Google Scholar]; b) Zhou H, Baldini L, Hong J, Wilson AJ, Hamilton AD. J. Am. Chem. Soc. 2006;128:2421. doi: 10.1021/ja056833c. [DOI] [PubMed] [Google Scholar]
- 17.Wright AT, Griffin MJ, Zhong ZL, McCleskey SC, Anslyn EV, McDevitt JT. Ang. Chem. Int. Ed. 2005;44:6375. doi: 10.1002/anie.200501137. [DOI] [PubMed] [Google Scholar]
- 18.a) Miranda OR, You C-C, Phillips R, Kim IB, Ghosh PS, Bunz UHF, Rotello VM. J. Am. Chem. Soc. 2007;129:9856. doi: 10.1021/ja0737927. [DOI] [PubMed] [Google Scholar]; b) Sandanaraj BS, Demont R, Aathimanikandan SV, Savariar EN, Thayumanavan S. J. Am. Chem. Soc. 2006;128:10686. doi: 10.1021/ja063544v. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Sandanaraj SB, Demont R, Thayumanavan S. J. Am. Chem. Soc. 2007;129:3506. doi: 10.1021/ja070229f. [DOI] [PubMed] [Google Scholar]
- 19.Maynor MS, Nelson TL, O'Sullivan C, Lavigne JJ. Org. Lett. 2007;9:3217. doi: 10.1021/ol071065a. [DOI] [PubMed] [Google Scholar]
- 20.De M, Rana S, Akpinar H, Miranda OR, Arivizo R, Bunz UHF, Rotello VM. Nat. Chem. 2009;1:461. doi: 10.1038/nchem.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.You C-C, Miranda OR, Gider B, Ghosh PS, Kim IB, Erdogan B, Krovi SA, Bunz UHF, Rotello VM. Nature Nanotech. 2007;2:318. doi: 10.1038/nnano.2007.99. [DOI] [PubMed] [Google Scholar]
- 22.Phillips RL, Miranda OR, You CC, Rotello VM, Bunz UHF. Angew. Chem. Int. Ed. 2008;47:2590. doi: 10.1002/anie.200703369. [DOI] [PubMed] [Google Scholar]
- 23.Bajaj A, Miranda OR, Kim I-B, Phillips RL, Jerry DJ, Bunz UHF, Rotello VM. Proc. Natl. Acad. Sci. 2009;106:10912. doi: 10.1073/pnas.0900975106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giljohann DA, Mirkin CA. Nature. 2009;462:461. doi: 10.1038/nature08605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.a) De M, You C-C, Srivastava S, Rotello VM. J. Am. Chem. Soc. 2007;129:10147. doi: 10.1021/ja071642q. [DOI] [PubMed] [Google Scholar]; b) Hong R, Fisher NO, Verma A, Goodman CM, Emrick T, Rotello VM. J. Am. Chem. Soc. 2004;126:739. doi: 10.1021/ja037470o. [DOI] [PubMed] [Google Scholar]; c) Jones S, Thornton JM. Proc. Natl. Acad. Sci. 1996;93:13. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) W. Stites E. Chem. Rev. 1997;97:1233. doi: 10.1021/cr960387h. [DOI] [PubMed] [Google Scholar]; e) Conte LL, Chothia C, Janin J. J. Mol. Biol. 1999;285:2177. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]; f) De M, Miranda OR, Rana S, Rotello VM. Chem. Commun. 2009;16:2157. doi: 10.1039/b900552h. [DOI] [PubMed] [Google Scholar]
- 26.a) Jacobson RH, Zhang XJ, Dubose RF, Matthews BW. Nature. 1994;369:761. doi: 10.1038/369761a0. [DOI] [PubMed] [Google Scholar]; b) Craven GR, Steers E, Anfinsen CB. J. Biol. Chem. 1965;240:2468. [PubMed] [Google Scholar]; c) Verma A, Simard JM, Worrall JWE, Rotello VM. J. Am. Chem. Soc. 2004;126:13987. doi: 10.1021/ja046572r. [DOI] [PubMed] [Google Scholar]; d) Wallenfels K, Weil R. Enzymes. (3rd Ed.) 1972;7:617. [Google Scholar]
- 27.a) Fowler AV, Zabin I. J. Biol. Chem. 1978;253:5521. [PubMed] [Google Scholar]; b) Fowler AV, Zabin I. J. Biol. Chem. 1970;245:5032. [PubMed] [Google Scholar]; c) Wallenfels K, Weil R. Enzymes. (3rd Ed.) 1972;7:617. [Google Scholar]
- 28.a) Hong R, Fischer NO, Verma A, Goodman CM, Emrick T, Rotello VM. J. Am. Chem. Soc. 2004;126:739. doi: 10.1021/ja037470o. [DOI] [PubMed] [Google Scholar]; b) Nikolic MS, Krack M, Aleksandrovic V, Kornowski A, Förster S, Weller H. Angew. Chem. 2006;118:6727. doi: 10.1002/anie.200602209. [DOI] [PubMed] [Google Scholar]
- 29.SYSTAT11.0. Richmond, CA94804, USA: SystatSoftware; 2004. [Google Scholar]
- 30.Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. Genome Biol. 2006;7(9):R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Li S-J, Peng M, Li H, Liu B-S, Wang C, Wu J-R, Li Y-X, Zeng R. Nucl. Acids Res. 2009;37:D907. doi: 10.1093/nar/gkn849. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Fliser D, Novak J, Thongboonkerd V, Argilés A, Jankowski V, Girolami MA, Jankowski J, Mischa H. J. Am. Soc. Nephrol. 2007;18:1057. doi: 10.1681/ASN.2006090956. [DOI] [PubMed] [Google Scholar]
- 31.a) Magistroni R, Ligabue G, Lupo V, Furci F, Leonelli M, Manganelli L, Masellis M, Gatti V, Cavazzini F, Tizzanini W, Albertazzi A. Nephrol. Dial. Transplan. 2009;24:1672. doi: 10.1093/ndt/gfp020. [DOI] [PubMed] [Google Scholar]; b) Roden AC, Lockington KS, Trostrud LJ, Katzmann JA. Am. J. Clin. Pathol. 2008;130:141. doi: 10.1309/6K33KTFA7A5VUQ1T. [DOI] [PubMed] [Google Scholar]; c) Nedelkolv D, Nelson RW. Am. J. Kidney Dis. 2001;38:481. doi: 10.1053/ajkd.2001.26831. [DOI] [PubMed] [Google Scholar]; d) Bottini P, Almerinda M, Alves R, Garlipp CR. Am. J. Kidney Dis. 2002;39:E2. doi: 10.1053/ajkd.2002.29920. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.