Nielsen and Beaumont (2009) extol the virtues of a coalescent-based, likelihood approach to phylogeographic inference. In contrast, they compare the inferences drawn from nested-clade, phylogeographic analysis (NCPA) as similar to an “interest in newspaper horoscopes”. I fully agree with Nielsen and Beaumont (2009) on the desirability of a coalescent-based, likelihood approach to phylogeographic inference. Where I disagree with them is in their description of the inference structure of NCPA, which is both inaccurate and incomplete.
NCPA is a coalescent-based method of statistical phylogeographic inference that tests hypotheses about the association of clades (branches) of haplotype trees with spatial information. The fundamental units of analysis in NCPA are haplotype trees. Coalescent theory deals with the genealogical history of a sample of homologous DNA regions. In the absence of complete pedigree data, much of this genealogical history is unobservable. However, some of the DNA replication events that make up this history are marked by mutations, which make that particular part of the genealogical history potentially observable. When there is little to no recombination in a DNA region, many mutations can mark different portions of the common genealogical history. When the DNA replication events that are not marked by mutations are collapsed together because they are not distinguishable, the resulting observable genealogical history is called a haplotype tree. A haplotype tree is therefore the observable portion of a realized coalescent process for a DNA region with little to no recombination (for a more extended discussion on the relationship between coalescence, gene genealogies, and haplotype trees, see Chapter 5 in Templeton (2006)). These observed portions of realized coalescent processes are the core units of analysis in NCPA. Limiting inference to observable genetic variants in NCPA is not different from the coalescent simulation approach because the simulations ultimately have to be related to the observed genetic data for estimation or hypothesis testing. At this point in the simulation analysis, the genetic data are always collapsed into the observable allelic or haplotype categories. Hence, both NCPA and the methods favored by Nielsen and Beaumont (2009) are coalescent based; NCPA uses realized coalescent processes and Nielsen and Beaumont use simulated coalescent processes to gauge the fit to observable patterns of genetic variation.
Because haplotype trees are valid only in DNA regions with little to no recombination, a statistical analysis of recombination should be executed for the genes or genomic regions chosen for study (Templeton et al. 2000a). This step can be skipped when dealing with DNA regions with no recombination, such as mtDNA or Y-DNA. If recombination is common and uniform in a genomic region, that region is inappropriate for NCPA and should be excluded. If recombination is common but concentrated into one or more hotspots, the region can be divided into subregions of little to no recombination that are appropriate for NCPA (Templeton et al. 2000b). Once DNA regions with little to no recombination have been identified, the haplotype trees are estimated via statistical parsimony (Templeton et al. 1992). Statistical parsimony is a Bayesian procedure based on an explicit coalescent model (Crandall & Templeton 1993; Templeton et al. 1992) that estimates a 95% confidence set of haplotype trees, thereby quantifying the error in tree estimation.
Coalescent theory predicts much variation in realized haplotype trees in terms of tree topology and coalescent times even for a given demographic history. NCPA takes that coalescent variation into account by converting the haplotype tree into a nested design and basing all statistical inference on local contrasts defined by the nested design. One benefit of using such local contrasts is that much of the tree estimation error has no impact on the nested design. When tree estimation error does affect the nested design, additional nesting rules are used to incorporate this ambiguity directly into the design (Templeton et al. 1992), or the NCPA can be repeated over all possible nested designs in the 95% confidence set (Brisson et al. 2005). In either case, the only phylogeographic inferences that are retained are those that are robust to the underlying uncertainty in tree estimation. The more important benefit of using only local contrasts within the tree is that inference in NCPA does not depend upon the overall haplotype tree topology. As a consequence, inference in NCPA is robust to the evolutionary stochasticity of tree topology that is inherent in the coalescent process. Indeed, this robustness to tree topology was the primary reason for extending nested clade analysis, originally developed to test for genotype/phenotype associations at candidate loci (Templeton et al. 1987), to perform statistical phylogeographic inference (Templeton 1993). Accordingly, NCPA has never equated a haplotype tree to a population tree. Indeed, the very fist null hypothesis that is tested with NCPA is the null hypothesis that there is no association between geography and the haplotype tree; i.e., there is no population tree at all. Even when this null hypothesis is rejected, NCPA can still lead to the inference that no population tree exists. For example, the NCPA of human populations leads to the inference that there was sufficient gene flow and admixture such that there is no population tree of humans but rather an interconnected genetic trellis (Templeton 2002, 2005; Templeton 2007a; Templeton 2007b). Another example illustrating robustness to haplotype tree topology is shown in Figure 1 from Templeton (2009a) that portrays five different haplotype trees from elephants based on data from Roca et al. (2005). All five topologies are different with respect to the populations of Asian elephant, African savanna elephant, and African forest elephant, due either to lineage sorting and/or limited introgression. Only one of these five haplotype trees has a topology that corresponds to the accepted population tree; yet, all five of these different tree topologies yield the same NCPA inference of a fragmentation event between forest and savanna elephants in Africa. Moreover, the null hypothesis that all five of these inferences are detecting the same phylogeographic event is accepted (the NCPA log-likelihood ratio test is 1.497 with 4 degrees of freedom, yielding a probability tail value of 0.8272). As these examples clearly show, NCPA inferences incorporate the coalescent randomness of haplotype tree topology through the nesting design and do not depend upon the exact haplotype tree topology.
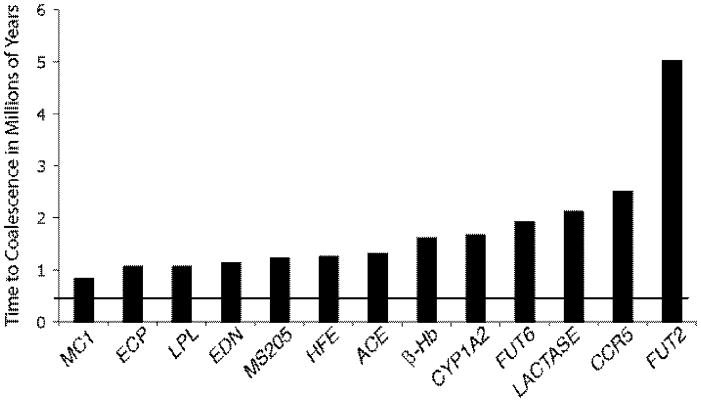
Figure 1.
The times to coalescence to a common ancestor for thirteen autosomal loci using a 6 million year divergence for humans and chimpanzees (Takahata et al. 2001; Templeton 2007b). The thick line shows the average autosomal coalescence time predicted from the simulation assumptions given in Fagundes et al. (2007).
When the null hypothesis of no geographical association is rejected, an inference key based upon predictions from coalescent theory is used to arrive at biological interpretations. This key does not depend upon an overall phylogeographic model, but rather focuses upon localized components of the species’ phylogeography. The validity of this key has been established through the use of 150 positive controls (Templeton 2004b). False positive error rates can be adjusted for single locus NCPA using the independence of the nested tests, and this adjustment has been show to work well, albeit conservatively, with actual data sets (Templeton 2009b). However, reducing false positive error rates in this manner also reduces power, so a cross-validation procedure was developed to reduce both false positives and false negatives simultaneously (Templeton 2002). This procedure requires NCPA upon two or more loci or DNA regions. To reduce type I errors (false positives), all inferences are discarded that are not cross-validated by two or more genomic regions with respect to type of inference (e.g., range expansion, gene flow with isolation by distance, etc.) and geographical location (Templeton 2002). The inferences cross-validated by type and location are next tested for concordance in time. This is accomplished by using a likelihood function that explicitly incorporates the randomness associated with the coalescent and mutational processes (Templeton 2004a; Templeton 2004b; Templeton 2009a). In the probability model used, the variance in time is proportional to its mean squared, a result that follows directly from coalescent theory and reflects the evolutionary stochasticity of the coalescent process. The variance is also inversely proportional to the number of accumulated mutations since the inferred event or process. This variance component stems from the Poisson nature of the molecular clock. Hence, both coalescent stochasticity and mutational randomness are explicitly incorporated into the NCPA likelihood function. The effectiveness of cross-validation in eliminating false positives and negatives is shown by the coalescent simulations of Knowles and Maddison (2002) when applied to multilocus NCPA (Templeton 2009b). NCPA reconstructs the simulated phylogeographic scenario of Knowles and Maddison with 100% accuracy, making not a single type I or type II error (Templeton 2009b). This result is particularly remarkable because the scenario simulated by Knowles and Maddison (2002) was an extremely difficult one, as pointed out by Nielsen and Beaumont (2009); one for which the simulation approach performed poorly (Knowles & Maddison 2002). Thus, the performance of the current version of NCPA in simulation studies is excellent (Templeton 2009b). Any one truly concerned about false positives would never use the weak inference structure advocated by Nielsen and Beaumont (2009) in which the false positive rate is both unknown and unknowable (Templeton 2009a). The ability to measure and correct for false positives is a major statistical advantage of NCPA over the coalescent simulation methods.
The inferences cross-validated by type and location are formally rephrased as the null hypothesis that a single phylogeographic event or process occurred in the cross-validated locations. Different likelihood ratio tests are used for events (e.g., range expansions, fragmentation events) and recurrent processes (e.g., gene flow with isolation by distance). The only inferences that are retained are those that pass a formal likelihood ratio test with the support of two or more loci or genomic regions, a point totally ignored by Nielsen and Beaumont (2009). It is important to note that just as inference in NCPA is robust to tree topology, it is likewise robust to differences in coalescence times for the cross-validating loci. For example, in the NCPA of human evolution, a recent out-of-Africa range expansion into Eurasia was cross-validated by five DNA regions that differed greatly in their coalescence times: mtDNA with an estimated coalescent time of 240,000 years ago (using a 6 million year calibration point for the divergence of humans and chimpanzees), Y-DNA at 230,000 years ago, RRM2P4 at 1,714,000 years ago, HS571B2 at 710,000 years ago, and HFE at 1,270,000 years ago (Templeton 2005, 2007b). NCPA extracted the same inference of an out-of-Africa expansion into Eurasia around 100,000 years ago from tall five of these DNA regions that varied greatly in tree topology, overall branch length, and coalescent time (the NCPA log-likelihood ratio test had a value of 0.698 with 4 degrees of freedom, yielding a probability value of 0.952 for the null hypothesis of homogeneity). As this and the previous examples of likelihood ratio tests patently reveal, NCPA is not only a coalescent-based method of statistical phylogeography, but ever since 2002 it has also been a coalescent-based likelihood method as well.
Despite the fact that the inference key with cross-validation has been shown to work extremely well with both actual and simulated data sets (Templeton 2009b), Nielsen and Beaumont (2009) likened the inference key of NCPA to a horoscope. Science is a human enterprise, so there is always the danger of a user interpreting ambiguous questions in a manner that reflects a subjective bias. However the purpose of the key was and is to increase objectivity. Users are not asked to interpret some overall phylogeographic pattern, but instead are asked highly directed questions on specific, local aspects of their data set and output. The key is driven by predictions from coalescent and statistical sampling theory and uses uniform, a priori criteria for all interpretation. Far from being ambiguous, answering the questions can be and has been automated (Zhang et al. 2006). This does not mean that subjective bias and over-interpretation are impossible, but the inference key is an important tool to reduce both.
In contrast, Nielsen and Beaumont (2009) argue for an alternative in which phylogeographic inference is based on computer simulations of complex phylogeographic scenarios with multiple parameters. This procedure is thoroughly riddled with subjectivity and ambiguities. First, there is ambiguity and subjectivity in deciding upon which models are included and which are excluded. The ambiguity in this inference step is illustrated well by the simulations given by Fagundes et al.(2007) on three models of human evolution, a paper that is held out as an exemplar of how to perform phylogeographic analyses with coalescent simulations (Beaumont & Panchal 2008). One of the models Fagundes et al. include is the “multiregional model.” This model, first proposed by Weidenreich (Weidenreich 1946), posits that hominin populations throughout Africa and Eurasia were interconnected by gene flow constrained by isolation by distance throughout the Pleistocene, resulting in a population trellis and not a population tree (Templeton 2007a). In contrast to Weidenreich’s trellis, Fagundes et al. (2007) present a tree-like structure with weak gene flow in their version of the “multiregional” model. Moreover, they simulate direct gene flow between sub-Saharan African populations and far East-Asian populations rather than an isolation-by-distance model. There has been no major model of human evolution that has posited such direct genetic interchange during the Pleistocene (Templeton 2007a), yet this radical and unprecedented model is simply invoked without any discussion or rationale for why the authors feel that simulating direct interchange between populations separated by over 10,000 kilometers is reasonable for Pleistocene hominins. Moreover, the original multiregional model with isolation-by-distance is excluded from their interpretation space. Such an exclusion has non-trivial consequences. For example, Fagundes et al. (2007) conclude that the out-of-Africa replacement model is the best of the three models that they simulated, whereas Eswaran et al. (2005) used simulations to show that the out-of-Africa replacement model is definitely refuted in comparison to a model with isolation-by-distance. This discrepancy illustrates the extreme ambiguity of inference via computer simulations; conclusions are strongly shaped not just by the models that are simulated but also by the models that are not simulated. NCPA does not pre-specify models and is based on falsification (strong inference), so it does not suffer from this profound inference defect of being affected by what is not tested.
Second, once the models to be simulated have been decided upon, the actual simulation is subject to extreme ambiguity and subjectivity in the process of parameterization. For example, Fagundes et al. (Table 7 of the supplementary material, 2007) make assumptions about 23 parameters in their simulated models of human evolution. Little to no information exists about most of these parameter values, such as the number of Homo erectus individuals originally coming out of Africa about 1.9 million years ago to colonize Eurasia. Hence, the parameter ranges and distributions are only guesses based upon the subjective opinion of the investigators. In some cases, information does exist about parameter values but is ignored. For example, Fagundes et al. (2007) assume small inbreeding effective sizes for archaic African and Eurasian populations, which results in small predicted average coalescent times for autosomal loci (the data source in that paper), as shown in Figure 1. Figure 1 also shows the coalescent times estimated for several autosomal loci using the method of Takahata et al. (2001). As can be seen, the observed coalescent times are highly discrepant with the assumptions made by Fagundes et al. (2007). Many of the co-authors of Fagundes et al. (2007) have a prior publication record of supporting the idea that there was no admixture or genetic exchange between Africans and archaic Eurasians, and these small, highly unrealistic, effective sizes and their guess about a small number of Homo erectus leaving Africa are necessary assumptions to ensure that the computer simulations will yield an outcome that does not favor admixture or genetic interchange (Templeton 2009a). Hence, an assumption based only on imagination and an assumption that seriously violates known data are crucial for the central conclusion of Fagundes et al. that there was replacement and no admixture. In contrast, the simulations of Eswaran et al. (2005) indicate that 80% of the loci in humans were affected by admixture. These two conclusions, both based on simulations, could not be more discrepant. The only common thread in the phylogeographic inferences emerging from these two sets of simulations is that each group concluded that their simulations supported the model that they had supported in prior publications, even though those models were diametric opposites. This is an example of the extreme subjectivity of inference from computer simulations.
Third, the interpretation of why a complex model is rejected is highly ambiguous and subjective with computer simulations. For example, Fagundes et al. (2007) interpreted their simulations as rejecting admixture on the basis of the poor fit of their “assimilation” model that included admixture. However, their assimilation model also assumed a long period of total genetic isolation between archaic Eurasians and Africans during the Pleistocene and unrealistically low inbreeding effective sizes (Figure 1). Thus, the rejection of their assimilation model could be due to the unrealistic inbreeding effective sizes, could be due to the assumed extended period of Pleistocene isolation, and/or could be due to the admixture component. These components are confounded because the estimate of the admixture rate depends directly upon the assumptions of isolation and low inbreeding effective size (Templeton 2009a). Consequently, there is no objective basis for Fagundes et al. (2007) to interpret the rejection of their complex assimilation model as a rejection of the single component of admixture; yet, that is what they do. As Figure 1 shows, the assumptions about effective sizes are clearly wrong, and the flexibility of the maximum-likelihood testing framework of NCPA shows that the hypothesis of Pleistocene isolation is also wrong (the NCPA likelihood ratio test value is 30.02 with 18 degrees of freedom, yielding a probability level of 0.0094, Templeton 2009a), as is the null hypothesis of no admixture (probability < 10−17) (Templeton 2005; Templeton 2007a). Hence, the only part of their assimilation model that is supported by rigorous statistical hypothesis testing is the existence of admixture, the exact opposite of their subjective interpretation. Simulations of complex phylogeographic scenarios result in the confoundment of several biological factors and in a high level of interpretative ambiguity. Focusing upon only one aspect of a complex model as the cause of rejecting the entire model merely reflects the subjective bias of the investigators. In contrast, NCPA tests individual features of complex models, so objective interpretations for rejecting a null hypothesis are straightforward (Templeton 2009a).
As a result of its greater objectivity and lack of pre-specified models, NCPA can discover phylogeographic features that were not anticipated by anyone, whereas the simulation approach is completely dependent on the prior beliefs of the investigators. For example, the NCPA of human evolution revealed an unanticipated expansion of humans out of Africa around 700,000 years ago which was not a part of any prior model of human evolution at the time (Templeton 2002, 2005). Once articulated, this inference was strongly corroborated by fossil, archaeological and paleo-climatic data (Templeton 2007a; Templeton 2007b). The discovery of unanticipated results obviously cannot be attributed to subjective bias; and such a discovery is impossible for the simulation procedure because all models have to be specified a priori.
I completely agree with Nielsen and Beaumont (2009) about the dangers of subjectivity and over-interpretation of ambiguous results. NCPA fortunately protects against this by using a highly focused inference key that deals with local, specific issues, and by objectively testing specific components of complex phylogeographic models with likelihood ratio tests. In contrast, the simulation approach is highly ambiguous and subjective at all stages of inference: model inclusion and exclusion, model parameterization, and model interpretation.
Both NCPA and many of the coalescent simulation approaches make use of likelihood functions, but there are major differences. First, the likelihoods used in simulations are very specific to a particular simulation and do not constitute a general-use phylogeographic tool. In contrast, the likelihood-ratio testing framework of NCPA is both general and flexible, so additional specific phylogeographic hypotheses can be tested besides just temporal concordance, such as replacement events, gene flow versus fragmentation in a specific time period and location, coevolution of hosts and parasites, etc. (Templeton 2004a; Templeton 2004b; Templeton 2005; Templeton 2007a; Templeton 2007b; Templeton 2009a). Second, the NCPA likelihoods are based on probability distributions that are conditioned upon the observed number of accumulated substitutions (Tajima 1983). This conditional probability allows the coalescent to be parameterized more simply and, specifically, eliminates two parameters, the mutation rate μ and the inbreeding effective size Nef. All remaining parameters are easily estimable with standard maximum likelihood procedures. As a result, NCPA can take advantage of the parametric power of log-likelihood ratio tests while retaining a predominantly non-parametric approach to phylogeographic inference. In contrast, the likelihoods associated with complex simulated models contain many, non-independent parameters that necessitate either additional assumptions about these parameters and/or approximations or local solutions that are not guaranteed to be optimal under either maximum likelihood or Bayesian criteria (Templeton 2009a).
The third difference is that the likelihood ratios used to test hypotheses in NCPA all have well-defined degrees of freedom (the difference between the dimensionality of the data versus the dimensionality of the model). In contrast, the non-independent parameters in complex simulation models make dimensionality difficult to calculate, and typically the dimensionality of the models is simply ignored (e.g., Fagundes et al. 2007). In testing complex phylogeographic models via computer simulation, the basic output is some measure of goodness of fit of the simulations of the pre-specified models to the observed genetic data. In order to translate a goodness of fit statistic of a complex model into a probability statement, it is essential to define the dimensionality of both the model and the data set for both maximum likelihood (Fisher 1922) and for Bayesian (Schwarz 1978) inference. By not specifying the dimensionalities of the models, the “tests” of phylogeographic models through simulation are devoid of statistical meaning (Templeton 2009a). In contrast, NCPA builds up complex models from simple components, so the likelihood ratio tests associated with NCPA all have well defined dimensionality, allowing them to be transformed to probability values using well established maximum-likelihood statistical techniques (Fisher 1922).
Until the dimensionality problem has been addressed, it is mathematically impossible for coalescent simulation procedures to assign a true probability measure to alternative hypotheses (Templeton 2009a). Simulations are still valuable for investigating feasibility and concordance in a non-statistical fashion and for parameter estimation (Templeton 2009a), but until true probability measures can be assigned to hypotheses, simulation procedures are inappropriate for testing complex phylogeographic hypotheses. A further complication is that some simulation procedures, such as Approximate Bayesian Computation, ignore important sources of sampling error, which also invalidates them for statistical inference (Templeton 2009a). Fortunately, there is a coalescent-based, maximum-likelihood inference method for testing phylogeographic hypotheses. It is called nested-clade, phylogeographic analysis.
Acknowledgments
This work was supported in part by NIH grants P50-GM65509 and 2RO1-GM02871924. The comments of Dr. Nolan Kane and anonymous reviewers on earlier versions are greatly appreciated.
References
- Beaumont MA, Panchal M. On the validity of nested clade phylogeographical analysis. Molecular Ecology. 2008;17:2563–2565. doi: 10.1111/j.1365-294X.2008.03786.x. [DOI] [PubMed] [Google Scholar]
- Brisson JA, De Toni DC, Duncan I, Templeton AR. Abdominal pigmentation variation in Drosophila polymorpha: Geographic variation in the trait, and underlying phylogeography. Evolution. 2005;59:1046–1059. [PubMed] [Google Scholar]
- Crandall KA, Templeton AR. Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics. 1993;134:959–969. doi: 10.1093/genetics/134.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaran V, Harpending H, Rogers AR. Genomics refutes an exclusively African origin of humans. Journal of Human Evolution. 2005;49:1–18. doi: 10.1016/j.jhevol.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Fagundes NJR, Ray N, Beaumont M, et al. Statistical evaluation of alternative models of human evolution. Proceedings of the National Academy of Sciences. 2007;104:17614–17619. doi: 10.1073/pnas.0708280104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. On the mathematical foundations of theoretical statistics. Philosophical Transactions of the Royal Society A. 1922;22:309–368. [Google Scholar]
- Knowles LL, Maddison WP. Statistical phylogeography. Molecular Ecology. 2002;11:2623–2635. doi: 10.1046/j.1365-294x.2002.01637.x. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Beaumont MA. Statistical inferences in phylogeography. Molecular Ecology. 2009;18:1034–1047. doi: 10.1111/j.1365-294X.2008.04059.x. [DOI] [PubMed] [Google Scholar]
- Roca AL, Georgiadis N, O’Brien SJ. Cytonuclear genomic dissociation in African elephant species. Nature Genetics. 2005;37:96–100. doi: 10.1038/ng1485. [DOI] [PubMed] [Google Scholar]
- Schwarz G. Estimating the Dimension of a Model. The Annals of Statistics. 1978;6:461–464. [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N, Lee S-H, Satta Y. Testing Multiregionality of Modern Human Origins. Mol Biol Evol. 2001;18:172–183. doi: 10.1093/oxfordjournals.molbev.a003791. [DOI] [PubMed] [Google Scholar]
- Templeton AR. The “Eve” hypothesis: a genetic critique and reanalysis. Amer Anthropol. 1993;95:51–72. [Google Scholar]
- Templeton AR. Out of Africa again and again. Nature. 2002;416:45–51. doi: 10.1038/416045a. [DOI] [PubMed] [Google Scholar]
- Templeton AR. A maximum likelihood framework for cross validation of phylogeographic hypotheses. In: Wasser SP, editor. Evolutionary Theory and Processes: Modern Horizons. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2004a. pp. 209–230. [Google Scholar]
- Templeton AR. Statistical phylogeography: methods of evaluating and minimizing inference errors. Molecular Ecology. 2004b;13:789–809. doi: 10.1046/j.1365-294x.2003.02041.x. [DOI] [PubMed] [Google Scholar]
- Templeton AR. Haplotype trees and modern human origins. Yearbook of Physical Anthropology. 2005;48:33–59. doi: 10.1002/ajpa.20351. [DOI] [PubMed] [Google Scholar]
- Templeton AR. Population Genetics and Microevolutionary Theory. John Wiley & Sons; Hoboken, New Jersey: 2006. [Google Scholar]
- Templeton AR. Perspective: Genetics and recent human evolution. Evolution. 2007a;61:1507–1519. doi: 10.1111/j.1558-5646.2007.00164.x. [DOI] [PubMed] [Google Scholar]
- Templeton AR. Population biology and population genetics of Pleistocene Hominins. In: Henke W, Tattersall I, editors. Handbook of Palaeoanthropology. Springer-Verlag; Berlin: 2007b. pp. 1825–1859. [Google Scholar]
- Templeton AR. Statistical hypothesis testing in intraspecific phylogeography: nested clade phylogeographical analysis vs. approximate Bayesian computation. Molecular Ecology. 2009a;18:319–331. doi: 10.1111/j.1365-294X.2008.04026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR. Why does a method that fails continue to be used: the answer. Evolution. 2009b;63:807–812. doi: 10.1111/j.1558-5646.2008.00600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Boerwinkle E, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. I. Basic theory and an analysis of Alcohol Dehydrogenase activity in Drosophila. Genetics. 1987;117:343–351. doi: 10.1093/genetics/117.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Clark AG, Weiss KM, et al. Recombinational and mutational hotspots within the human Lipoprotein Lipase gene. American Journal of Human Genetics. 2000a;66:69–83. doi: 10.1086/302699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Weiss KM, Nickerson DA, Boerwinkle E, Sing CF. Cladistic structure within the human Lipoprotein lipase gene and its implications for phenotypic association studies. Genetics. 2000b;156:1259–1275. doi: 10.1093/genetics/156.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidenreich F. Apes, Giants, and Man. University of Chicago Press; Chicago: 1946. [Google Scholar]
- Zhang AB, Tan S, Sota T. AUTOINFER 1.0: a computer program to infer biogeographical events automatically. Molecular Ecology Notes. 2006;6:597–599. [Google Scholar]