Abstract
Over the last 20 years since the discovery of the cystic fibrosis transmembrane conductance regulator (CFTR) gene, more than 1,600 different putatively pathological CFTR mutations have been identified. Until now, however, copy number mutations (CNMs) involving the CFTR gene have not been methodically analyzed, resulting almost certainly in the underascertainment of CFTR gene duplications compared with deletions. Here, high-resolution array comparative genomic hybridization (averaging one interrogating probe every 95 bp) was used to analyze the entire length of the CFTR gene (189 kb) in 233 cystic fibrosis chromosomes lacking conventional mutations. We succeeded in identifying five duplication CNMs that would otherwise have been refractory to analysis. Based upon findings from this and other studies, we propose that deletion and duplication CNMs in the human autosomal genome are likely to be generated in the proportion of approximately 2–3:1. We further postulate that intragenic gene duplication CNMs in other disease loci may have been routinely underascertained. Finally, our analysis of ±20 bp flanking each of the 40 CFTR breakpoints characterized at the DNA sequence level provide support for the emerging concept that non-B DNA conformations in combination with specific sequence motifs predispose to both recurring and nonrecurring genomic rearrangements.
Keywords: CFTR, copy number mutation, CNM, copy number variation, CNV, deletion, duplication, CGH
Introduction
Cystic fibrosis—usually described clinically as a triad—namely, chronic obstructive pulmonary disease, exocrine pancreatic insufficiency, and elevation of sodium and chloride concentrations in the sweat—is the most common life-shortening autosomal recessive disease in Caucasians. Since the positional cloning of the cystic fibrosis gene (i.e., CFTR; MIM# 602421, cystic fibrosis transmembrane conductance regulator) 20 years ago [Kerem et al., 1989; Riordan et al., 1989; Rommens et al., 1989], more than 1,600 different putatively pathological mutations have been identified (Cystic Fibrosis Mutation Database, http://www.genet.sickkids.on.ca/cftr/app). Keeping abreast of the changing landscape of molecular genetic research [Beckmann et al., 2008], we have recently analyzed CFTR copy number mutations (CNMs) in a large batch of cystic fibrosis chromosomes, by means of quantitative fluorescence multiplex PCR (QFM-PCR) [Audreézet et al., 2004; Férec et al., 2006]. (The term CNM is used, from the standpoint of pathological relevance, in contradistinction to the neutral term “copy number variation” [CNV] in accordance with our previous studies) [Chauvin et al., 2009; Masson et al., 2008]. CNV refers to a DNA segment of ≥1 kb that is present in different copy numbers with respect to a reference genome sequence [Scherer et al., 2007]. However, these earlier attempts failed to identify any CFTR duplication CNMs. With hindsight, we now know that the QFM-PCR conditions employed in these previous studies [Audrézet et al., 2004; Férec et al., 2006] were inherently biased in favor of the detection of deletions over duplications. It would appear that, under the suboptimal conditions employed, a change of copy number from two to one (in the case of heterozygous deletions) was more readily identifiable than that from two to three (in the case of heterozygous duplications). Additional caveats associated with the use of QFM-PCR are that (1) only one primer pair is employed per exon and (2) each primer pair only amplifies a short sequence tract (typically in the range of 100–200 bp), leaving the vast majority (97%) of the genomic sequence of the CFTR gene refractory to analysis [Chen et al., 2008].
In this study, we deployed high-resolution array comparative genomic hybridization (array-CGH) to overcome the aforementioned limitations of CNM detection. This technique proved itself very reliable in detecting both deletion and duplication CNMs, resulting in the identification of 8 new mutational events including five duplications. The complete ascertainment of intragenic CNMs in the CFTR gene has important implications for CNM formation and detection in other disease loci.
Materials and Methods
Cystic Fibrosis Chromosomes
A total of 233 cystic fibrosis chromosomes (162 representing available samples from among those previously reported [Audrézet et al., 2004; Férec et al., 2006], the remaining 71 being newly recruited) were analyzed in this study. All chromosomes were derived from patients diagnosed with typical cystic fibrosis but had not been found to carry any known conventional pathogenic mutations (i.e., point mutations, microinsertions and microdeletions) after all 27 exons as well as the intron/exon boundaries of the CFTR gene had been screened by denaturing gradient gel electrophoresis and/or denaturing high-performance liquid chromatography [Audrézet et al., 2004; Férec et al., 2006].
Array CGH
Array CGH was performed using a custom microarray comprising 14,000 different probes (Agilent Technologies, Santa Clara, CA). Probe distribution in the CFTR locus and the other genomic regions is provided in Supp. Table S1. Hybridizations were carried out as described previously [Chauvin et al., 2009].
Characterization of Newly Identified CNMs
QFM-PCR was used to confirm and/or narrow down the boundaries of all novel CNMs revealed by array CGH. Aberrant junctions of the newly identified CFTR CNMs were cloned by either conventional or long-range PCR. Successfully amplified patientspecific bands were then subjected to direct DNA sequencing as previously described [Audrézet et al., 2004; Férec et al., 2006].
Survey of Intragenic CNMs Reported in Several Autosomal Disease Loci
Seven autosomal loci, in which a reasonable number of intragenic CNMs have been logged in the Human Gene Mutation Database (http://www.hgmd.org), were selected for comparative analysis. All the intragenic CNMs used for the current analysis could be confidently assumed to be ≥1 kb based upon the relevant gene’s reference genomic sequence.
Bioinformatic Analysis
A total of ±20 bp flanking each of the 40 CFTR gene breakpoints characterized at the DNA level were manually extracted. These 40 sequence tracts were then screened for the presence of direct, inverted, and symmetric repeats ≥6 bp, capable of non-B DNA structure formation and hence potentially involved in the induction of DNA breakage as described in [Chuzhanova et al., 2009]. The overrepresentation of a specific type of repeat (direct, inverted and symmetric) in the vicinity of the deletion/ duplication breakpoints was assessed using z-score statistics [Marino-Ramirez et al., 2004] by comparison with matching controls randomly selected from the 189 kb-long genomic sequence of the CFTR gene. The same 40 sequence tracts (each 40 bp in length) were also screened for the presence of DNA sequence motifs of length ≥5 bp and their complements (known to be associated with site-specific cleavage/recombination, highfrequency mutation and gene rearrangement as described in Chuzhanova et al. [2009]), CpG-dinucleotide frequency and R-, Y-, and RY-tract coverage.
The order statistics, r-scans [Karlin and Macken, 1991] as described in Bacolla et al. [2004] were used to detect clustering of breakpoints along the CFTR gene. In addition, the region of the CFTR gene spanning a breakpoint cluster region and randomly generated controls matching the length of the cluster found, were compared with respect to the frequency of occurrence of non-B DNA forming sequences and sequence coverage by R-, Y-, and RY-tracts.
For each of the above analyses, the statistical significance of the findings was assessed either by means of a chi-square test or by comparison with 1,000 control datasets using z-score statistics as described previously [Chuzhanova et al., 2009]. z-Scores for all sequences comprising the case dataset that exceeded the 99th, 99.9th, or 99.99th percentile of the maximum z-score found for the corresponding 1,000 control datasets were deemed to be statistically significant with P-values 0.01, 0.001, or 0.0001, respectively.
Results and Discussion
Development and Application of Array CGH to CNM Detection in the CFTR Gene
CNMs in the CFTR gene have been extensively studied over the past 5 years by means of quantitative PCR techniques [Audrézet et al., 2004; Bombieri et al., 2005; Chevalier-Porst et al., 2005; Férec et al., 2006; Hantash et al., 2006; Loumi et al., 2008; Niel et al., 2004, 2006; Paracchini et al., 2008; Schneider et al., 2007; Schrijver et al., 2008; Taulan et al., 2009]. An array CGH method has recently been developed that has been used to screen CNMs in eight human disease genes including CFTR [Saillour et al., 2008]. However, using this method, deletions were identified in only four of the five control samples with known CFTR CNMs. We are unaware of any further reports of this method being used for CFTR CNM detection.
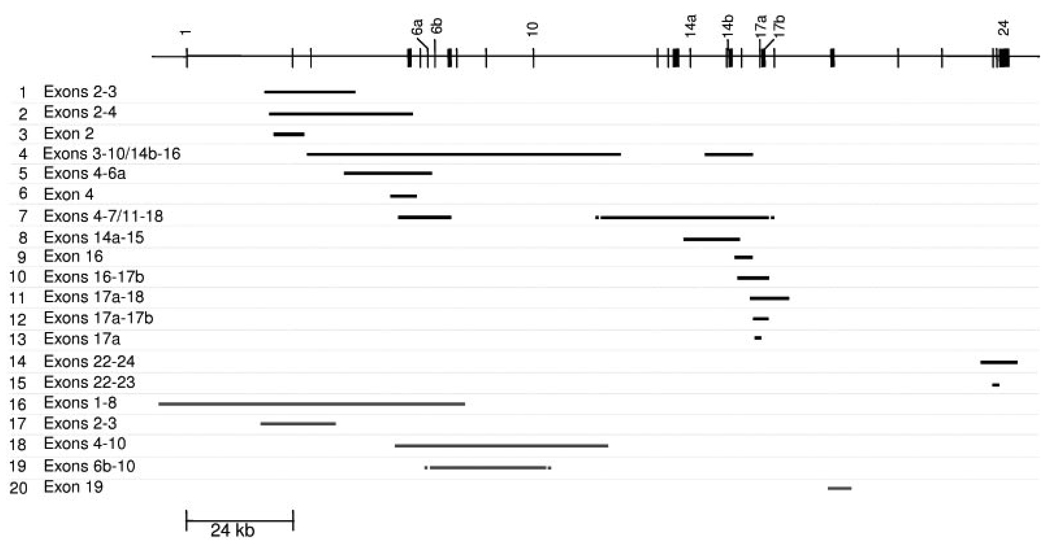
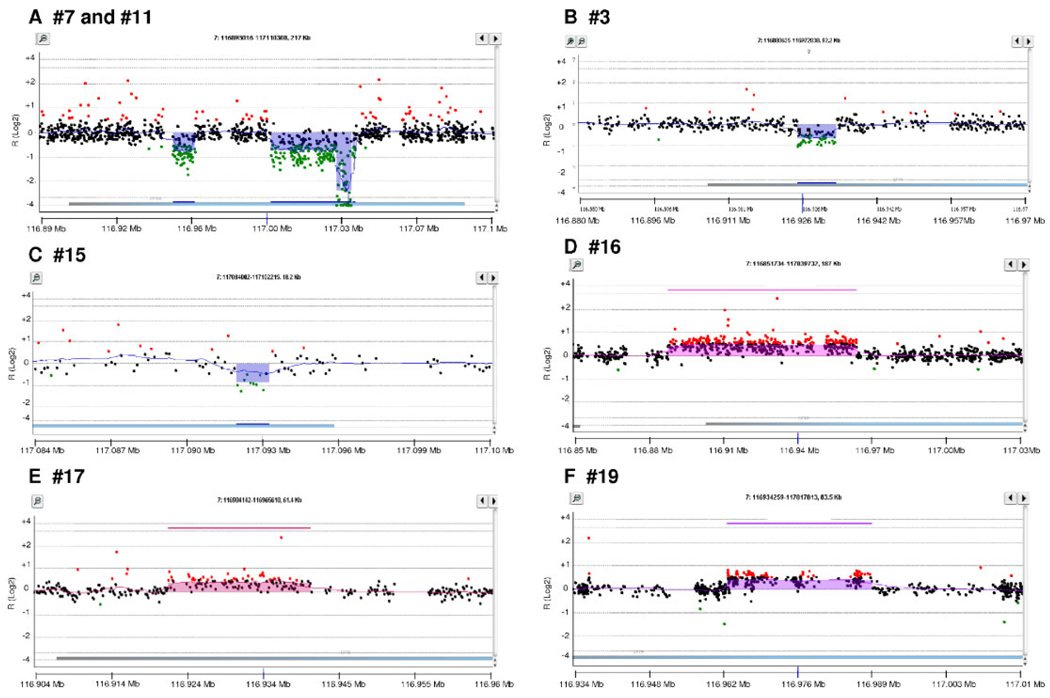
For this study, we designed an array comprising 14,000 probes, of which 2,000 were located within the bounds of the 189 kb CFTR locus (Supp. Table S1). The probe density within the CFTR locus—one probe every 95 bp—is probably among the highest for any studied disease locus so far reported. To validate this method, we first analyzed samples that had previously been found to carry a CFTR deletion CNM by QFM-PCR [Audrézet et al., 2004; Férec et al., 2006]. All 12 known deletions were accurately identified by the newly established array CGH technique. We then analyzed the remaining patient samples and identified eight new mutational events, of which five were duplications. Of these five duplications, only one (#19) was identified in the newly recruited samples. In other words, four of the duplication CNMs had gone undetected in our previous QFM-PCR analysis. Parental samples were only available for eight of the 20 CNMs detected; however, in all eight cases, the CNM was found to be inherited. In short, a total of 15 intragenic deletion CNMs and 5 intragenic duplication CNMs were identified in our well-defined set of 233 cystic fibrosis chromosomes (Table 1 and Fig. 1). Some of these mutational events are illustrated in Figure 2.
Table 1.
Cystic Fibrosis-Causing CNMs in the CFTR Gene
| CNM # | Type of CNM | Exon(s) involved | Length (kb) | Starting position | Ending position | Referencea |
|---|---|---|---|---|---|---|
| 1 | Deletion | 2–3 | 21.1 | 116925603 | 116946682 | [Dork et al., 2000] |
| 2 | Deletion | 2–4 | 33.1 | 116925701 | 116958805 | [Férec et al., 2006] |
| 3 | Deletion (ins182) | 2 | 8.1 | 116925732 | 116933839 | [Férec et al., 2006] |
| 4–1b | Deletion (ins39) | 3–10 | 70.5 | 116936033 | 117006501 | [Niel et al., 2006) |
| 4–2b | Deletion (ins16) | 14b–16 | 12.3 | 117024636 | 117036970 | [Niel et al., 2006] |
| 5 | Deletion (insACCTCG) | 4–6a | 18.7 | 116944415 | 116963066 | [Audrézet et al., 2004] |
| 6 | Deletion (ins41) | 4 | 8.2 | 116952251 | 116960415 | [Audrézet et al., 2004] |
| 7–1b | Deletionc | 4–7 | 10.6 | 116957148 | 116967747 | [Morral et al., 1993]; this study |
| 7–2b | Deletion | 11–18 | ~38.2 | (117003504–117003905)d | (117042124–117042947)d | [Morral et al., 1993] |
| 8 | Deletion | 14a–15 | 10.9 | 117021454 | 117032400 | This study |
| 9 | Deletion | 16 | 4.4 | 117032020 | 117036447 | This study |
| 10 | Deletion | 16–17b | 7.2 | 117032158 | 117039358 | This study |
| 11 | Deletion | 17a–18 | 8.9 | 117035216 | 117044114 | [Lerer et al., 1999] |
| 12 | Deletion | 17a–17b | 2.5 | 117036832 | 117039346 | [Niel et al., 2004] |
| 13 | Deletion (ins5bp) | 17a | 1.0 | 117036901 | 117037905 | [Férec et al., 2006] |
| 14 | Deletion (insTAACT) | 22–24 | 9.5 | 117088088 | 117097541 | [Chevalier-Porst et al., 2005] |
| 15 | Deletion | 22–23 | 1.5 | 117091900 | 117093431 | [Audrézet et al., 2004] |
| 16 | Duplication (insAAAT) | 1–8 | 78.9 | 116891547 | 116970474 | This study |
| 17 | Duplication | 2–3 | 20.5 | 116922009 | 116942545 | This study |
| 18 | Duplication | 4–10 | 49.7 | 116955282 | 117004935 | This study |
| 19 | Duplication | 6b–10 | ~26.8 | (116962800–116963124)d | (116989889–116990039)d | This study |
| 20 | Duplication (insCA) | 19 | 5.3 | 117054760 | 117060092 | This study |
See also Figure 1.
First report to describe the relevant CNM.
Deletions are linked in cis.
First described in Morral et al. [1993] but not fully characterized at this time.
Not yet characterized at the DNA sequence level; breakpoint region defined by array CGH analysis.
Figure 1.
Overview of cystic fibrosis-causing CNMs reported in this study. Upper panel: Genomic structure of the CFTR gene (scaled). Vertical bars represent the 27 exons. Lower panel: black horizontal lines indicate intragenic deletions whereas gray horizontal lines indicate intragenic duplications. Breakpoint ends not yet fully characterized are indicated by dots. The CNMs are arranged in the same order as in Table 1.
Figure 2.
Examples of CFTR CNMs identified by array-CGH. Green plots indicate deletions, red plots duplications. CNM numbers are in accord with those described in Table 1 and Figure 1. Note that in A, the carrier is a compound heterozygote of CNMs 7 and 11.
One obvious advantage of the current array CGH method over QFM-PCR (and other PCR-based techniques) is that whenever a CNM was identified, its breakpoints could usually be narrowed down to rather precise genomic regions. This greatly facilitated subsequent efforts to clone the breakpoints involved. Thus, of the eight newly identified CNMs, only one (#19) still remains to be characterized at the DNA sequence level. In addition, we fully characterized the first component of the double deletion CNM (#7), first described by Morral et al. [1993], but also identified among our collected samples. characterization of the breakpoints of CNM #19 and the second component of CNM #7 are under way.
Given the extremely high density of probes spanning the entire 189 kb CFTR locus (Supp. Table S1) and the considerable number of cystic fibrosis chromosomes analyzed, this study has provided, some 20 years after the identification of the CFTR gene [Kerem et al., 1989; Riordan et al., 1989; Rommens et al., 1989], the first comprehensive overview of cystic fibrosis-causing intragenic CNMs at the CFTR locus. It did not escape our attention that all the identified mutational events affected at least one exon, a finding that should be interpreted in the context of clinical selection. Deletions or duplications that are confined to single introns and which do not involve any splice sites should, at least in principle, give rise to less severe functional impairment of the affected allele. In this regard, it is important to point out that ~5% of normal CFTR function is enough to confer pancreatic sufficiency [Chen and Férec, 2009]. In other words, deletions or duplications that involve only single introns, with the exception of those that disrupt splice donor or acceptor sites, are unlikely to result in a functional impairment severe enough to cause exocrine pancreatic insufficiency, one of the diagnostic hallmarks of cystic fibrosis.
Implications for CNM Generation and Detection in Other Autosomal Loci
Given the high sensitivity of array CGH in detecting both deletion and duplication CNMs, we can be fairly confident that all intragenic CNMs present in the 233 cystic fibrosis chromosomes were accurately and reliably ascertained. These data may thus shed new light on the basic issue of the relative frequencies of initial occurrence of deletion and duplication CNMs in vivo. To properly address this question, a clear distinction should be made between two specific categories of CNM from the standpoint of the underlying generational mechanism. The first CNM category results from nonallelic homologous recombination (NAHR) between very similar duplicated sequences during meiosis [Lupski and Stankiewicz, 2005]. Recent analysis of de novo meiotic deletions and duplications in three autosomal NAHR hotspots using a sperm-based assay demonstrated quite similar ratios (i.e., 2.10, 2.14, and 2.43) of deletions to duplications [Turner et al., 2008]. The higher rate of deletion over duplication is readily explicable in terms of intrachromatidal NAHR occurring more frequently than either interchromatidal or interchromosomal NAHR; the former type of NAHR generates only deletions,whereas the latter two types generate reciprocal deletions and duplications [Turner et al., 2008]. Consistent with this explanation, the chromosome Y-located AZFa-HERV hotspot, which cannot on the basis of its chromosomal location undergo interchromosomal NAHR, exhibits the highest reported ratio (4.11) of deletions:duplications [Turner et al., 2008]. It is for this reason that, in the following analysis, we focus exclusively upon autosomal disease loci. The second CNM category comprises those deletions and duplications that originate in unique (or quasi-unique) genomic regions through either nonhomologous end joining (NHEJ) or alternatively diverse replication-based mechanisms [Bauters et al., 2008; Chauvin et al., 2009; Chen et al., 2005a,b,c; Hastings et al., 2009a,b; Lee et al., 2007; Sheen et al., 2007; Zhang et al., 2009]. These generative mechanisms, originating in unique genomic regions, are thought to occur mainly during the premeiotic division of germ cells [Chauvin et al., 2009; Zhang et al., 2009]. Inspection of breakpoint-spanning regions revealed that all the CNMs listed in Table 1 fall into the second CNM category. In this regard, it is pertinent to point out that no low copy repeats (stretches of duplicated DNA >1 kb in size that share >90% sequence similarity) were found to be present either within the 189-kb CFTR locus itself or within its ±50 kb-flanking sequences.
Unlike in vitro assays, clinically observed findings are often confounded by many diverse factors, most notably clinical selection. This is perhaps best exemplified by the recurrent reciprocal 1.4-Mb deletions and duplications at the CMT1A-REP NAHR hotspot (17p11.2) associated with quite distinct clinical phenotypes (viz. deletion/hereditary neuropathy with liability to pressure palsies [HNPP]; duplication/Charcot-Marie-Tooth disease type 1A [CMT1A], respectively). Although a sperm-based assay has demonstrated that the ratio of deletions to duplications at this hotspot should be of the order of 2:1 [Turner et al., 2008], the actual ratio of HNPP to CMT1A coming to clinical attention is ~1:4 [Turner et al., 2008]. The underascertainment of HNPP has been attributed to the relatively mild and variable clinical phenotype associated with the CMT1A-REP deletion [Turner et al., 2008]. By contrast, in the case of the CFTR gene, the mutational events to be considered are both nonrecurrent and intragenic. Given the complex multiple domain structure of CFTR and the wide distribution of pathogenic missense mutations along its coding sequence, large intragenic deletions and duplications are unlikely to differ in terms of their functional effects. Another issue is that duplications, once they have arisen, could be more unstable than deletions (indeed, duplications can revert either to wild type or to deletions [Gitschier, 1988]), and this possibility is not easily refuted. However, duplicated (or even triplicated) sequences appear to be stably transmissible from one generation to another both in the context of evolution [Redon et al., 2006] and inherited disease [Le Maréchal et al., 2006; Padiath et al., 2006]. Hence, taking all the above into consideration, it seems not unreasonable to conclude that the ratio of disease-causing intragenic CFTR deletions to duplications (3:1) obtained in this study may approximate to the actual relative occurrence of de novo deletion CNMs to duplication CNMs at this locus. However, we should like to emphasize two points here. First, a deletion hotspot appears to exist around CFTR exons 16–18 (Fig. 1), a region that has previously been shown to contain sequences of relatively low complexity [Férec et al., 2006]. It remains to be seen whether duplication breakpoints will be discovered within this deletion hotspot in the future. Second, we performed an ANOVA test to compare the mean length of the CFTR deletions with that of the duplications, but although the mean length of duplications (36.25 kb) exceeded that of deletions (15.69 kb) this difference was only of borderline significance (one-way ANOVA, P = 0.06).
To the best of our knowledge, of the studies that have analyzed ≥50 well-defined disease chromosomes for intragenic CNMs at a given autosomal locus, only one has yielded a comparable deletion:duplication ratio, viz. 2.4 in an analysis of 53 patients with isolated lissencephaly (all patients were previously found to be negative for microdeletions in the 17p13.3 region by FISH and were also negative for conventional mutations upon sequencing the LIS1 gene) using the Lissencephaly P061 MLPA (multiplex ligation-dependent probe amplification) kit; this analysis revealed 12 intragenic deletions and 5 intragenic duplications in the 91 kb LIS1 gene [Haverfield et al., 2009]. Intriguingly, the intragenic deletion:duplication ratios for the CFTR and LIS1 loci are consistent with those reported for the aforementioned autosomal NAHR-derived de novo deletions:duplications [Turner et al., 2008]. The similarity of these ratios, despite the widely different contexts and means of detection, could imply the operation of a common biological mechanism underlying the generation of deletion and duplication CNMs. Indeed, intrachromatidal events, irrespective of whether they originate via NHEJ or replicationbased mechanisms, should occur more frequently than either interchromatidal or interchromosomal events; the former type of event can only generate deletions, whereas the latter two types should generate deletions and duplications in equal proportions.
We would therefore tentatively propose that a deletion:duplication ratio of between 2 and 3 is likely to represent the best estimate of the relative occurrence of deletion and duplication CNMs in the human autosomal genome. This conclusion could have important implications for the detection of intragenic CNMs in clinical diagnosis. Using data from the Human Gene Mutation Database [Stenson et al., 2009], we collated intragenic CNMs from seven additional autosomal loci for which a respectable number of deletions/duplications have been characterized at the kilobase level. These loci consistently displayed rather higher ratios of deletion CNMs to duplication CNMs (Supp. Table S2), ranging from 5.7 to 25.0, suggesting that intragenic duplications may have been significantly underascertained at these loci.
Sequence Features that may have Predisposed to Nonrecurring CFTR CNMs
It is increasingly recognized that local DNA sequence features, either in the form of recombination-promoting motifs or non-B DNA structures, predispose to the generation of genomic rearrangements [Chuzhanova et al., 2009; Wells, 2007]. The most persuasive evidence has come, however, from meta-analyses of mutational events derived from many different genes [Bacolla et al., 2004; Chuzhanova et al., 2009]. In the context of a given locus, it has often been long sequence tracts (usually in the range of >1 kb) that have been used for analysis because most of the breakpoints have not been precisely characterized at the DNA sequence level. In this study cohort, a total of 40 breakpoints were fully characterized (see Table 1 and Fig. 1). This enabled us to perform the most detailed CFTR breakpoint analysis performed to date, involving the analysis of ±20 bp DNA sequence flanking each breakpoint for the presence of known recombination-associated motifs and non-B DNA-forming sequences.
Sequences capable of non-B DNA slipped structure, cruciform, and triplex formation were found to be significantly over-represented (z-score statistics, P = 0.001) in the vicinity of the CFTR breakpoints compared to 1,000 matching control datasets randomly selected from the genomic CFTR gene (Supp. Table S3). This provides further evidence for the potential involvement of sequences capable of non-B DNA-formation in double-strand break formation.
One specific motif, the “super hotspot” CCAAR, found in the vicinity of 10 CFTR breakpoints, was significantly overrepresented at the 1% level (z-score statistics, P<0.01). Both short (5 bp) and long (10 bp) RY-tracts were also overrepresented in the vicinity of the CFTR breakpoints at the 1% level. In addition, alternating purine-pyrimidine tracts of >5 bp, representing ~13% of the total length of the CFTR locus, were overrepresented within sequences flanking the CFTR breakpoints by comparison with 1,000 random controls (Pearson’s χ2 test, P = 3.1 × 10−6). The combined coverage of flanking sequences by R- (9%), Y- (9%), and RY-tracts (13%) of >5 bp was significantly higher than that evident in matched controls (Pearson’s chi-square test, P = 0.0001). The overrepresentation of these tracts suggests that that they may have been responsible for promoting deletion/ duplication mutagenesis.
The region spanning CFTR exons 16, 17a, and 17b was found to harbour a cluster of 11 breakpoints (P = 0.01). Comparison of this 7,339-bp region with 1,000 randomly selected matching controls revealed a significant difference in terms of sequence coverage by R- and Y-tracts; the R-tracts were overrepresented (P = 0.003), whereas Y-tracts were underrepresented (P = 6.9 × 10−5) within the breakpoint cluster region. The only other notable feature found to characterize this breakpoint cluster was its enrichment in inverted repeats capable of cruciform (non-B) structure formation (P = 0.001).
Conclusions and Perspective
This study represents the first to have successfully used highresolution array CGH to detect CNMs in the CFTR gene. We have demonstrated that this method is both extremely reliable and efficient at detecting both intragenic deletions and duplications. Owing to the extremely high probe density employed, the breakpoints of the newly identified events were immediately and very precisely localized, greatly facilitating the task of breakpoint characterization at the DNA sequence level. The extremely high probe density should also serve to avoid the pitfall of false negative results that are sometimes encountered with quantitative PCR-based methods owing to the presence of polymorphisms located in primer binding sites. Given its overwhelming superiority over PCR-based methods such as QFM-PCR and MLPA, high-resolution array CGH is expected to find wide application in both research and diagnostic laboratories over the coming years. In this regard, we would like to emphasize that the extensive heterogeneity of large CFTR rearrangements occurring in unique or quasi-unique genomic sequences would make specific genotyping assays impractical. Thus, as we previously proposed [Férec et al., 2006], the ideal strategy for CFTR mutation detection should include the following sequential steps: (1) genotyping the 20–30 most common CFTR gene mutations by commercially available kits; if negative, proceeding (2) to complete screening of the 27 CFTR exons by means of rapid screening techniques such as denaturing high-performance liquid chromatography [Le Maréchal et al., 2001] and high-resolution melting analysis [Audrézet et al., 2008]; if still negative, then (3) array CGH should be employed.
The comprehensive nature of this study on the CFTR gene led us to propose a general value for the ratio of occurrence of deletion and duplication CNVs in the human autosomal genome. As high-resolution array CGH becomes widely accepted in diagnostic laboratories, an increasing number of disease loci will be methodically analyzed for CNMs. This should eventually allow us to assess the validity of our postulate that intragenic gene duplications have hitherto been routinely underascertained. Finally, our analysis of ±20 bp flanking each of the 40 CFTR breakpoints lends support to the emerging concept that non-B DNA conformations and/or certain sequence motifs predispose to both recurrent and nonrecurrent genomic rearrangements [Chuzhanova et al., 2009; Wells, 2007].
Supplementary Material
Acknowledgments
This work was supported by the INSERM (Institut National de la Santé et de la Recherche Médicale), the VLM (Vaincre La Mucoviscidose), and the Association de Transfusion Sanguine et de Biogénétique Gaetan Saleun, France; and VZFNM00064203 and Health-F5-2009-2231434 Techgene (to M.M.).
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Audrézet MP, Chen JM, Raguénès O, Chuzhanova N, Giteau K, Le Maréchal C, Quéré I, Cooper DN, Férec C. Genomic rearrangements in the CFTR gene: extensive allelic heterogeneity and diverse mutational mechanisms. Hum Mutat. 2004;23:343–357. doi: 10.1002/humu.20009. [DOI] [PubMed] [Google Scholar]
- Audrézet MP, Dabricot A, Le Maréchal C, Férec C. Validation of high-resolution DNA melting analysis for mutation scanning of the cystic fibrosis transmembrane conductance regulator (CFTR) gene. J Mol Diagn. 2008;10:424–434. doi: 10.2353/jmoldx.2008.080056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacolla A, Jaworski A, Larson JE, Jakupciak JP, Chuzhanova N, Abeysinghe SS, O’Connell CD, Cooper DN, Wells RD. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc Natl Acad Sci USA. 2004;101:14162–14167. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauters M, Van Esch H, Friez MJ, Boespflug-Tanguy O, Zenker M, Vianna-Morgante AM, Rosenberg C, Ignatius J, Raynaud M, Hollanders K, Govaerts K, Vandenreijt K, Niel F, Blanc P, Stevenson RE, Fryns JP, Marynen P, Schwartz CE, Froyen G. Nonrecurrent MECP2 duplications mediated by genomic architecture-driven DNA breaks and break-induced replication repair. Genome Res. 2008;18:847–858. doi: 10.1101/gr.075903.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann JS, Sharp AJ, Antonarakis SE. CNVs and genetic medicine (excitement and consequences of a rediscovery) Cytogenet Genome Res. 2008;123:7–16. doi: 10.1159/000184687. [DOI] [PubMed] [Google Scholar]
- Bombieri C, Bonizzato A, Castellani C, Assael BM, Pignatti PF. Frequency of large CFTR gene rearrangements in Italian CF patients. Eur J Hum Genet. 2005;13:687–689. doi: 10.1038/sj.ejhg.5201387. [DOI] [PubMed] [Google Scholar]
- Chauvin A, Chen JM, Quemener S, Masson E, Kehrer-Sawatzki H, Ohmle B, Cooper DN, Le Maréchal C, Férec C. Elucidation of the complex structure and origin of the human trypsinogen locus triplication. Hum Mol Genet. 2009;18:3605–3614. doi: 10.1093/hmg/ddp308. [DOI] [PubMed] [Google Scholar]
- Chen JM, Chuzhanova N, Stenson PD, Férec C, Cooper DN. Meta-analysis of gross insertions causing human genetic disease: novel mutational mechanisms and the role of replication slippage. Hum Mutat. 2005a;25:207–221. doi: 10.1002/humu.20133. [DOI] [PubMed] [Google Scholar]
- Chen JM, Chuzhanova N, Stenson PD, Férec C, Cooper DN. Complex gene rearrangements caused by serial replication slippage. Hum Mutat. 2005b;26:125–134. doi: 10.1002/humu.20202. [DOI] [PubMed] [Google Scholar]
- Chen JM, Chuzhanova N, Stenson PD, Férec C, Cooper DN. Intrachromo-somal serial replication slippage in trans gives rise to diverse genomic rearrangements involving inversions. Hum Mutat. 2005c;26:362–373. doi: 10.1002/humu.20230. [DOI] [PubMed] [Google Scholar]
- Chen JM, Férec C. Chronic pancreatitis: genetics and pathogenesis. Annu Rev Genomics Hum Genet. 2009;10:63–87. doi: 10.1146/annurev-genom-082908-150009. [DOI] [PubMed] [Google Scholar]
- Chen JM, Masson E, Macek M, Jr, Raguénès O, Piskackova T, Fercot B, Fila L, Cooper DN, Audrézet MP, Férec C. Detection of two Alu insertions in the CFTR gene. J Cyst Fibros. 2008;7:37–43. doi: 10.1016/j.jcf.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Chevalier-Porst F, Souche G, Bozon D. Identification and characterization of three large deletions and a deletion/polymorphism in the CFTR gene. Hum Mutat. 2005;25:504. doi: 10.1002/humu.9335. [DOI] [PubMed] [Google Scholar]
- Chuzhanova N, Chen JM, Bacolla A, Patrinos GP, Férec C, Wells RD, Cooper DN. Gene conversion causing human inherited disease: evidence for involvement of non-B-DNA-forming sequences and recombination-promoting motifs in DNA breakage and repair. Hum Mutat. 2009;30:1189–1198. doi: 10.1002/humu.21020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dork T, Macek M, Jr, Mekus F, Tummler B, Tzountzouris J, Casals T, Krebsova A, Koudova M, Sakmaryova I, Macek M, Sr, Vavrova V, Zemkova D, Ginter E, Petrova NV, Ivaschenko T, Baranov V, Witt M, Pogorzelski A, Bal J, Zekanowsky C, Wagner K, Stuhrmann M, Bauer I, Seydewitz HH, Neumann T, Jakubiczka S. Characterization of a novel 21-kb deletion, CFTRdele2,3(21 kb), in the CFTR gene: a cystic fibrosis mutation of Slavic origin common in Central and East Europe. Hum Genet. 2000;106:259–268. doi: 10.1007/s004390000246. [DOI] [PubMed] [Google Scholar]
- Ellsworth RE, Jamison DC, Touchman JW, Chissoe SL, Braden Maduro VV, Bouffard GG, Dietrich NL, Beckstrom-Sternberg SM, Iyer LM, Weintraub LA, Cotton M, Courtney L, Edwards J, Maupin R, Ozersky P, Rohlfing T, Wohldmann P, Miner T, Kemp K, Kramer J, Korf I, Pepin K, Antonacci-Fulton L, Fulton RS, Minx P, Hillier LW, Wilson RK, Waterston RH, Miller W, Green ED. Comparative genomic sequence analysis of the human and mouse cystic fibrosis transmembrane conductance regulator genes. Proc Natl Acad Sci USA. 2000;97:1172–1177. doi: 10.1073/pnas.97.3.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Férec C, Casals T, Chuzhanova N, Macek M, Jr, Bienvenu T, Holubova A, King C, McDevitt T, Castellani C, Farrell PM, Sheridan M, Pantaleo SJ, Loumi O, Messaoud T, Cuppens H, Torricelli F, Cutting GR, Williamson R, Ramos MJ, Pignatti PF, Raguénès O, Cooper DN, Audrézet MP, Chen JM. Gross genomic rearrangements involving deletions in the CFTR gene: characterization of six new events from a large cohort of hitherto unidentified cystic fibrosis chromosomes and meta-analysis of the underlying mechanisms. Eur J Hum Genet. 2006;14:567–576. doi: 10.1038/sj.ejhg.5201590. [DOI] [PubMed] [Google Scholar]
- Gitschier J. Maternal duplication associated with gene deletion in sporadic hemophilia. Am J Hum Genet. 1988;43:274–279. [PMC free article] [PubMed] [Google Scholar]
- Hantash FM, Redman JB, Starn K, Anderson B, Buller A, McGinniss MJ, Quan F, Peng M, Sun W, Strom CM. Novel and recurrent rearrangements in the CFTR gene: clinical and laboratory implications for cystic fibrosis screening. Hum Genet. 2006;119:126–136. doi: 10.1007/s00439-005-0082-0. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009a;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009b;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverfield EV, Whited AJ, Petras KS, Dobyns WB, Das S. Intragenic deletions and duplications of the LIS1 and DCX genes: a major disease-causing mechanism in lissencephaly and subcortical band heterotopia. Eur J Hum Genet. 2009;17:911–918. doi: 10.1038/ejhg.2008.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S, Macken C. Some statistical problems in the assessment of inhomogenesis of DNA sequence data. J Am Statist Assoc. 1991;86:27–35. [Google Scholar]
- Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, Buchwald M, Tsui LC. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- Le Maréchal C, Audrézet MP, Quéré I, Raguénès O, Langonné S, Férec C. Complete and rapid scanning of the cystic fibrosis transmembrane conductance regulator (CFTR) gene by denaturing high-performance liquid chromatography (D-HPLC): major implications for genetic counselling. Hum Genet. 2001;108:290–298. doi: 10.1007/s004390100490. [DOI] [PubMed] [Google Scholar]
- Le Maréchal C, Masson E, Chen JM, Morel F, Ruszniewski P, Levy P, Férec C. Hereditary pancreatitis caused by triplication of the trypsinogen locus. Nat Genet. 2006;38:1372–1374. doi: 10.1038/ng1904. [DOI] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Lerer I, Laufer-Cahana A, Rivlin JR, Augarten A, Abeliovich D. A large deletion mutation in the CFTR gene (3120+1Kbdel8.6Kb): a founder mutation in the Palestinian Arabs. Mutation in brief no. 231. Online. Hum Mutat. 1999;13:337. doi: 10.1002/(SICI)1098-1004(1999)13:4<337::AID-HUMU13>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Loumi O, Férec C, Mercier B, Creff J, Fercot B, Denine R, Grangaud JP. CFTR mutations in the Algerian population. J Cyst Fibros. 2008;7:54–59. doi: 10.1016/j.jcf.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino-Ramirez L, Spouge JL, Kanga GC, Landsman D. Statistical analysis of over-represented words in human promoter sequences. Nucleic Acids Res. 2004;32:949–958. doi: 10.1093/nar/gkh246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson E, Le Maréchal C, Chandak GR, Lamoril J, Bezieau S, Mahurkar S, Bhaskar S, Reddy DN, Chen JM, Férec C. Trypsinogen copy number mutations in patients with idiopathic chronic pancreatitis. Clin Gastroenterol Hepatol. 2008;6:82–88. doi: 10.1016/j.cgh.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Morral N, Nunes V, Casals T, Cobos N, Asensio O, Dapena J, Estivill X. Uniparental inheritance of microsatellite alleles of the cystic fibrosis gene (CFTR): identification of a 50 kilobase deletion. Hum Mol Genet. 1993;2:677–681. doi: 10.1093/hmg/2.6.677. [DOI] [PubMed] [Google Scholar]
- Niel F, Legendre M, Bienvenu T, Bieth E, Lalau G, Sermet I, Bondeux D, Boukari R, Derelle J, Levy P, Ruszniewski P, Martin J, Costa C, Goossens M, Girodon E. A new large CFTR rearrangement illustrates the importance of searching for complex alleles. Hum Mutat. 2006;27:716–717. doi: 10.1002/humu.9431. [DOI] [PubMed] [Google Scholar]
- Niel F, Martin J, Dastot-Le Moal F, Costes B, Boissier B, Delattre V, Goossens M, Girodon E. Rapid detection of CFTR gene rearrangements impacts on genetic counselling in cystic fibrosis. J Med Genet. 2004;41:e118. doi: 10.1136/jmg.2004.022400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padiath QS, Saigoh K, Schiffmann R, Asahara H, Yamada T, Koeppen A, Hogan K, Ptacek LJ, Fu YH. Lamin B1 duplications cause autosomal dominant leukodystrophy. Nat Genet. 2006;38:1114–1123. doi: 10.1038/ng1872. [DOI] [PubMed] [Google Scholar]
- Paracchini V, Seia M, Coviello D, Porcaro L, Costantino L, Capasso P, Degiorgio D, Padoan R, Corbetta C, Claut L, Costantini D, Colombo C. Molecular and clinical features associated with CFTR gene rearrangements in Italian population: identification of a new duplication and recurrent deletions. Clin Genet. 2008;73:346–352. doi: 10.1111/j.1399-0004.2007.00957.x. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL, Drumm ML, Iannuzzi MC, Collins FS, Tsui LC. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, Rozmahel R, Cole JL, Kennedy D, Hidaka N, Zsiga M, Buchwald M, Riordan JR, Tsui LC, Collins FS. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- Saillour Y, Cossee M, Leturcq F, Vasson A, Beugnet C, Poirier K, Commere V, Sublemontier S, Viel M, Letourneur F, Barbot JC, Deburgrave N, Chelly J, Bienvenu T. Detection of exonic copy-number changes using a highly efficient oligonucleotide-based comparative genomic hybridization-array method. Hum Mutat. 2008;29:1083–1090. doi: 10.1002/humu.20829. [DOI] [PubMed] [Google Scholar]
- Scherer SW, Lee C, Birney E, Altshuler DM, Eichler EE, Carter NP, Hurles ME, Feuk L. Challenges and standards in integrating surveys of structural variation. Nat Genet. 2007;39:S7–S15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Hirt C, Casaulta C, Barben J, Spinas R, Buhlmann U, Spalinger J, Schwizer B, Chevalier-Porst F, Gallati S. Large deletions in the CFTR gene: clinics and genetics in Swiss patients with CF. Clin Genet. 2007;72:30–38. doi: 10.1111/j.1399-0004.2007.00820.x. [DOI] [PubMed] [Google Scholar]
- Schrijver I, Rappahahn K, Pique L, Kharrazi M, Wong LJ. Multiplex ligation-dependent probe amplification identification of whole exon and single nucleotide deletions in the CFTR gene of Hispanic individuals with cystic fibrosis. J Mol Diagn. 2008;10:368–375. doi: 10.2353/jmoldx.2008.080004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen CR, Jewell UR, Morris CM, Brennan SO, Férec C, George PM, Smith MP, Chen JM. Double complex mutations involving F8 and FUNDC2 caused by distinct break-induced replication. Hum Mutat. 2007;28:1198–1206. doi: 10.1002/humu.20591. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, Cooper DN. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taulan M, Guittard C, Theze C, Claustres M, Georges MD. A novel double deletion underscores the importance of characterizing end points of the CFTR large rearrangements. Eur J Hum Genet. 2009;17:1683–1687. doi: 10.1038/ejhg.2009.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DJ, Miretti M, Rajan D, Fiegler H, Carter NP, Blayney ML, Beck S, Hurles ME. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat Genet. 2008;40:90–95. doi: 10.1038/ng.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RD. Non-B DNA conformations, mutagenesis and disease. Trends Biochem Sci. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, Lupski JR. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.