Abstract
A sensitive, specific, and robust liquid chromatography/mass spectrometric (LC/MS) method was developed and validated that allows simultaneous analysis of arachidonic acid (AA) and its cyclooxygenase, cytochrome P450, and lipoxygenase pathway metabolites prostaglandins (PGs), dihydroxyeicosatrienoic acids (DiHETrEs), hydroxyeicosatetraenoic acids (HETEs) and epoxyeicosatrienoic acids (EETs), including PGF2α, PGE2, PGD2, PGJ2, 14,15-DiHETrE, 11,12-DiHETrE, 8,9-DiHETrE, 5,6-DiHETrE, 20-HETE, 15-HETE, 12-HETE, 9-HETE, 8-HETE, 5-HETE, 14,15-EET, 11,12-EET, 8,9-EET, and 5,6-EET in rat brain tissues. Deuterium labeled PGF2α-d4, PGD2-d4, 15(S)-HETE-d8, 14,15-EET-d8, 11,12-EET-d8, 8,9-EET-d8, and AA-d8 were used as internal standards. Solid phase extraction was used for sample preparation. A gradient LC/MS method using a C18 column and electrospray ionization source under negative ion mode was optimized for the best sensitivity and separation within 35 min. The method validation, including LC/MS instrument qualification, specificity, calibration model, accuracy, precision (without brain matrix and with brain matrix), and extraction efficiency were performed. The linear ranges of the calibration curves were 2–1000 pg for PGs, DiHETrEs, HETEs, and EETs, 10–2400 pg for PGE2 and PGD2, and 20–2000 ng for AA, respectively.
Keywords: LC/MS, Brain tissue, Arachidonic acid, Prostaglandins (PGs), Hydroxyeicosatetraenoic acids (HETEs), Dihydroxyeicosatrienoic acids (DiHETrEs), Epoxyeicosatrienoic acids (EETs)
1. Introduction
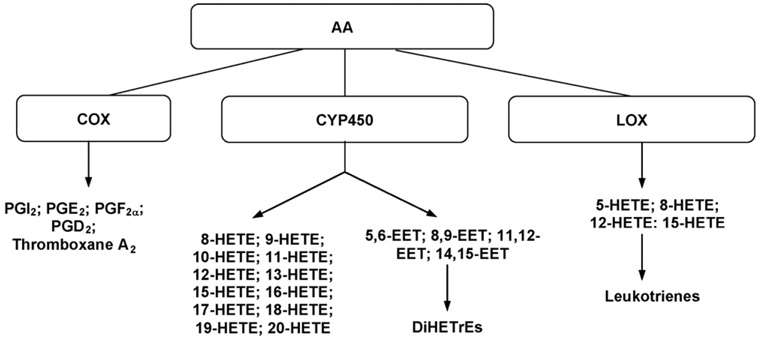
The biological oxidation products of arachidonic acid and related C20 polyene acids are summarized under the term eicosanoids that give rise to a wide variety of products of remarkable physiological activity [1]. Arachidonic acid (AA) can be metabolized to many bioactive eicosanoids (Fig. 1) [2], including oxidation to prostaglandins (PGs), hydroxyeicosatetraenoic acids (HETEs), dihydroxyeicosatrienoic acids (DiHETrEs), and epoxyeicosatrienoic acids (EETs) via cyclooxygenase (COX), cytochrome P450 (CYP450), and lipoxygenase (LOX) path-ways (Fig. 2). These endogenous eicosanoids are present at trace levels in biological fluids and tissues, including the mammalian brain. The expression of various eicosanoids is affected by injury or inflammation, in the brain and other tissues. Traumatic brain injury (TBI) is one of the leading causes of death [3]. The physical insults of TBI set into motion a cascade of biochemical events that can result in secondary brain injury that is a major contributor to the ultimate tissue loss [4–6]. One part of this response is the increased conversion of AA to PGs as a result of induction of cyclooxygenase-2 (COX2) expression after TBI [7]. COX2 is one of the isoforms of COX and is highly induced by inflammatory stimuli [8,9]. Some of the PGs (i.e. PGE2) possess pro-inflammatory properties [10–12], and many of the HETEs and EETs derived from AA via CYP450 and LOX pathways are reported to have anti-inflammatory properties [13–15]. COX2 inhibitors, are being used to treat peripheral inflammation [16], and may prove beneficial to the injured brain as well [10,17]. DFU [5,5-dimethyl-3(3-fluorophenyl)-4(4-methylsulphonyl)phenyl-2(5H)-furanone], a highly specific COX2 enzyme inhibitor, was shown to improve functional recovery and attenuate neuronal cell death and inflammation in a rat model of TBI in our lab [18,19]. To investigate the possibility that COX2 inhibitors may play a role in inhibition of pro-inflammatory eicosanoids and shunting the AA metabolism to anti-inflammatory eicosanoids in the brain after TBI, it is necessary to establish a specific, sensitive and reliable analytical method for simultaneous identification and quantification of these endogenous eicosanoid in brain samples.
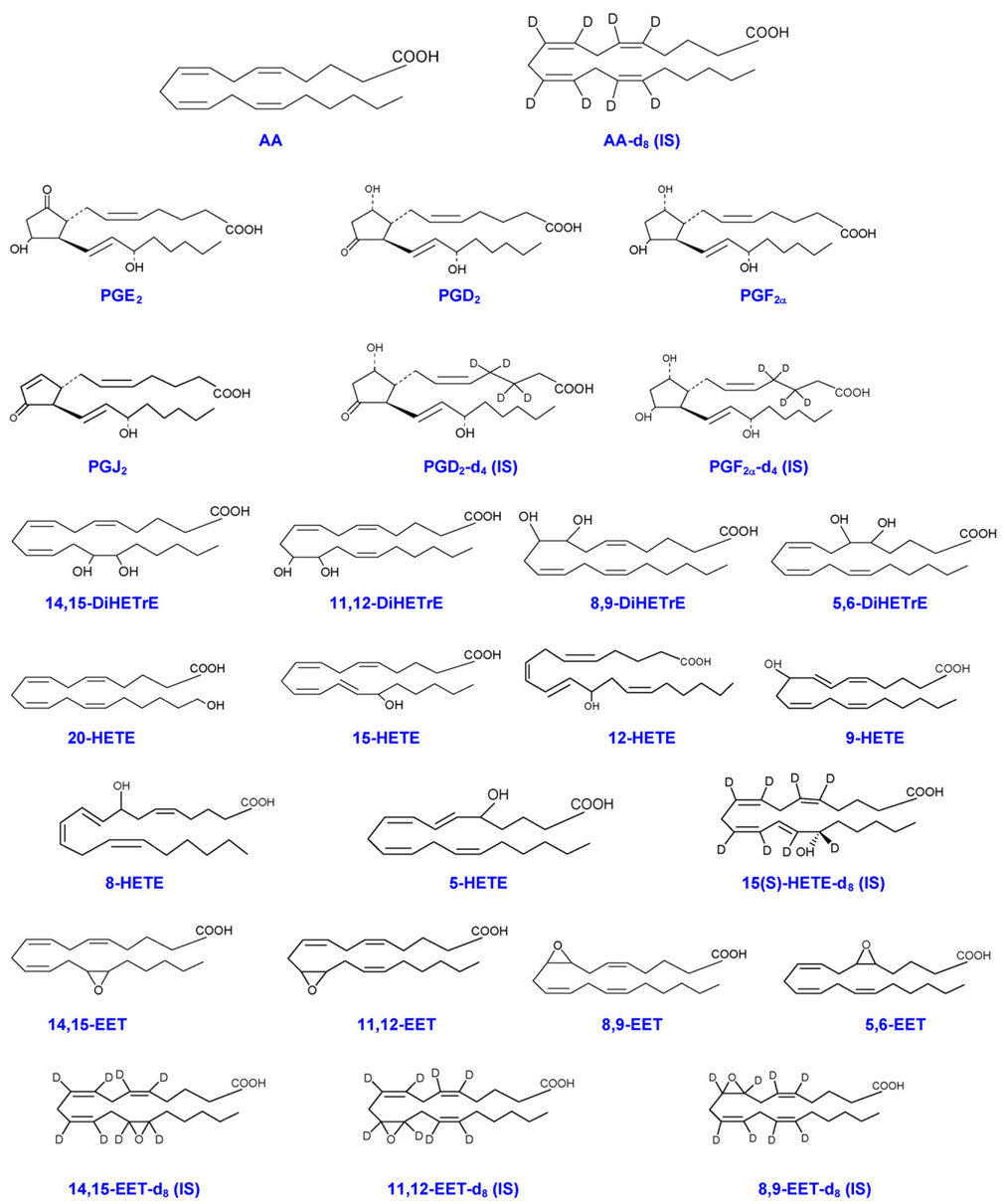
Fig. 1.
Structures of AA, PGs, DiHETrEs, HETEs, EETs, and internal standards (IS).
Fig. 2.
Selective pathways for the metabolism of arachidonic acid (AA).
Currently, GC/MS, GC/MS/MS [20–24], capillary electrophoresis/ UV [25], and HPLC/fluorescence [26–28], MS/MS, liquid chromatography/mass spectrometric (LC/MS) and LC/MS/MS methods, have been used to measure some of these eicosanoids at low levels. GC/MS and GC/MS/MS with negative ion chemical ionization has been the most commonly used technique and can give specific mass information of the peaks. However, it is not suitable for labile compounds (i.e. EETs), and needs tedious steps including TLC purification and derivatization before analysis. The need for highly sensitive and low cost methods for analyzing labile bioactive eicosanoids has initially driven the development of derivatized fluorescent HPLC method [28] to simultaneously analyze PGs, DiHETrEs, HETEs and EETs in brain tissues at pg levels in our lab. However the separation was long and the high background from tissue matrix was seen throughout the chromatogram even using solid phase extraction clean-up procedure. Recently, LC/MS and LC/MS/MS were widely used in bioanalytical work since they are powerful analytical techniques that combine the resolving power of liquid chromatography with detection specificity of mass spectrometry.
Although a few LC/MS or LC/MS/MS (either with chromatographic separation or flow injection analysis) methods have been used for identification and quantification of either single eicosanoid [29], or various PGs [30], or PGs together with HETEs [31–36], or various DiHETrEs [37], or HETEs together with EETs [38,39] or HETEs, EETs, together with DiHETrEs [40] in various biological matrices in a single analysis, we could not identify a published mass spectrometric-based method for the simultaneous identification and quantification of PGs, DiHETrEs, HETEs, EETs, and parent compound AA in biological matrices, including brain tissue. In addition, the method validation data were not available in most of these methods. In this paper, we report a sensitive, specific, robust and validated LC/MS method for simultaneously analyzing parent compound AA and its COX, CYP450 and LOX pathway metabolites PGs, DiHETrEs, HETEs and EETs, including PGF2α, PGE2, PGD2, PGJ2,14,15-DiHETrE, 11,12-DiHETrE, 8,9-DiHETrE, 5,6-DiHETrE, 20-HETE, 15-HETE, 12-HETE, 9-HETE, 8-HETE, 5-HETE, 14,15-EET, 11,12-EET, 8,9-EET, and 5,6-EET in rat cortical brain tissue.
2. Experimental
2.1. Chemicals and materials
(±)14,15-Dihydroxy-5Z,8Z,11Z-eicosatrienoic acid (14,15-DiHETrE), (±)11,12-dihydroxy-5Z,8Z,14Z-eicosatrienoic acid (11,12-DiHETrE), (±)8,9-dihydroxy-5Z,11Z,14Z-eicosatrienoic acid (8,9-DiHETrE), (±)5,6-dihydroxy-8Z,11Z,14Z-eicosatrienoic acid (5,6-DiHETrE), 20-hydroxy-5Z,8Z,11Z, 14Z-eicosatetraenoic acid (20-HETE), (±)15-hydroxy-5Z,8Z, 11Z,13E-eicosatetraenoic acid (15-HETE), (±)12-hydroxy-5Z,8Z,10E,14Z-eicosatetraenoic acid (12-HETE), (±)9-hydroxy- 5Z,7E,11Z,14Z-eicosatetraenoic acid (9-HETE), (±)8-hydroxy-5Z,9E,11Z,14Z-eicosatetraenoic acid (8-HETE), (±) 5-hydroxy-6E,8E,11Z,14Z-eicosatetraenoic acid (5-HETE), (±)11(12)epoxy-5Z,8Z,14Z-eicosatrienoic acid (11,12-EET), (±)5(6)epoxy-8Z,11Z,14Z-eicosatrienoic acid (5,6-EET) and 15S-hydroxy-5Z,8Z,11Z,13E-eicosatetraenoic-5,6,8,9,11,12, 14,15-d8 acid (15(S)-HETE-d8), 9α,15S-dihydroxy-11-oxoprosta-5Z,13E-dien-1-oic-3,3,4,4-d4 acid (PGD2-d4), 9a, 1α, 15S-trihydroxy-prosta-5Z,13E-dien-1-oic-3,3,4,4-d4 acid (PGF2α-d4) were purchased from Cayman Chemical Company (Ann Arbor, MI, USA). 9α, 11α,15S-trihydroxy-prosta-5Z,13E-dien-1-oic acid (PGF2α), 9-oxo-11α, 15S-dihydroxy-prosta-5Z,13E-dien-1-oic acid (PGE2), 9α, 15S-dihydroxy-11-oxo-prosta-5Z,13E-dien-1-oic acid (PGD2), 11-oxo-15S-hydroxy-prosta-5Z,9,13E-trien-1-oic acid (PGJ2), (±)14(15)epoxy-5Z,8Z,11Z-eicosatrienoic acid (14,15-EET), (±)8(9)epoxy- 5Z,11Z,14Z-eicosatrienoic acid (8,9-EET), (±) 14(15)epoxy-5Z,8Z,11Z-eicosatrienoic-5,6,8,9,11,12,14,15-d8 acid(14,15-EET-d8), (±)11(12)epoxy-5Z,8Z,14Z-eicosatrienoic-5,6,8,9,11,12,14,15-d8 acid(11,12-EET-d8), (±)8(9)epoxy-5Z,11Z,14Z-eicosatrienoic-5,6,8,9,11,12,14,15-d8 acid(8,9-EET-d8), 5Z,8Z,11Z,14Z-eicosatetraenoic acid (arachidonic acid) and 5Z,8Z,11Z,14Z-eicosatetraenoic-5,6,8,9,11,12,14,15-d8 acid (AA-d8) were obtained from BIOMOL Research Laboratories Inc. (Plymouth Meeting, PA, USA). Deuterium labeled PGF2α-d4, PGD2-d4,15(S)-HETE-d8, 14,15-EET-d8, 11,12-EET-d8, 8,9-EET-d8, and AA-d8 were used as internal standards for quantitation. HPLC grade acetonitrile, water, methanol, and A.C.S. reagent grade formic acid (96%) were obtained from Sigma–Aldrich (Steinheim, Germany). Solid phase extraction (SPE) cartridges Oasis®HLB (30 mg, 30 µm) were purchased from Waters Corporation (Maliford, MA, USA).
2.2. Biological samples
Male Sprague–Dawley rats, each weighing 300–400 g, were studied. A lateral cortical impact model of traumatic brain injury (TBI) was utilized (this model generates moderate levels of head injury including the temporary loss of strength and coordination in the limbs contra-lateral to the injury and significant retrograde amnesia and learning deficits). Animals were preanesthetized using 2% isoflurane, then given oxygen with 0.75% isoflurane through a facemask on a sterotactic frame. The cranium was exposed and a lateral craniectomy was made over the somatosensory cortex using a 6mm trephine. The exposed dura was subjected to a 5mm diameter piston impact (4 m/s velocity, 3.0mm depth, 100 ms duration). Animals were sacrificed by decapitation at 6 h, 24 h, and 72 h post-injury. Brains were rapidly removed, snap frozen on dry ice and stored at −80 °C. Thick sections (300 µm) were cut on a cryostat at −8 °C. The study included one negative control group (Sham), one positive control group (injured), and one post-treatment group (DFU): Sham group, surgery, no injury, and treated with vehicle; injured group, treated with vehicle and injured; and DFU group, treated with DFU (10 mg/kg i.p.) 30 min after injury, then twice daily. DFU was suspended in saline by mixing vigorously for 30 s at room temperature with a rotor-stator device.
2.3. Sample preparation
20–40 mg cortex brain tissue was dissected from the section inside a cryostat at −15°C, then weighed, and transferred to a 2 ml polypropylene tube surrounded with crush ice. Following addition of 200 µl of methanol and 2 µl of formic acid, the cortex brain tissue was homogenized using a micro ultrasonic cell disrupter at 30 amplitudes (2 mm probe, highest power 100 amplitudes, Kontes). The homogenates were centrifuged at 14,000 rpm for 10 min at 0 °C. 5% of the supernatant was transferred to a fresh tube for protein assay. 95% of the supernatant was used for LC/MS analysis and was transferred to a fresh tube followed by dilution with 1.8 ml of water. The diluted supernatant was loaded onto an Oasis®HLB SPE cartridge that was pre-cleaned, conditioned, and equilibrated with 1 ml of methanol, 1 ml of ethylacetate, 1 ml of methanol and 2 ml of water by low feeding speed in sequence. Additional 200 µl of 10% methanol was used to rinse the tube and was load onto the cartridge. The cartridge was washed with 3 ml of water, and 1 ml of 10% methanol. The cartridge was then dried under vacuum for 20 min. The analytes were collected in a 2 ml polypropylene tube by elution with 0.5 ml of acetonetrile followed by 1.5 ml of ethylacetate. Followed by addition of 100 pg of PGDF2α-d4, PGD2-d4, 15(S)-HETE-d8, 8,9-EET-d8, and 11,12-EET-d8, 14,15-EET-d8 and 40 ng of AA-d8 (5 µl of internal standard working solution IV—preparation is described in Section 2.5). The solvent was then evaporated under a gentle stream of argon (5.0 grade). The residue was reconstituted with 20 µl of methanol, vortexed briefly and transferred to an autosampler vial insert for LC/MS analysis.
2.4. Protein assay
The protein amounts in methanol extractions were measured using Bio-Rad® Protein Microtiter Plate Protocol (CA, USA). Dye reagent (Coomassie® Brilliant Blue G-250 dye) was prepared by diluting 2 parts of Dye Reagent Concentrate with 7 parts of water. 0.5 mg/ml of bovine serum albumin solution was prepared as protein standard solution. For standard curve, 1 µl, 2 µl, 4 µl, and 8 µl of the protein standard solution with corresponding 19 µl, 18 µl, 16 µl, and 12 µl of water were added into separate microtiter plate wells in duplicate followed by addition of 4 µl of methanol and 200 µl of diluted dye reagent to each well. For brain samples, 4 µl of each methanol extraction, 20 µl of water and 200 µl of diluted dye reagent were added into separate wells. Solutions were mixed using a microplate mixer and incubated at room temperature for 15 min. The optical density was then read at 620 nm using a Vmax Kinetic Microplate Reader (Molecular Devices). The protein amount in each sample was calculated using standard curve.
2.5. Preparation of stock solutions, calibration standards and quality control samples
The stock solution I consisting of PGF2α, PGJ2,14,15-DiHETrE, 11,12-DiHETrE, 8,9-DiHETrE, 5,6-DiHETrE, 20-HETE, 15-HETE, 12-HETE, 8-HETE, 5-HETE, 14,15-EET, 11,12-EET, 8,9-EET, and 5,6-EET (200 pg/µl) was made in methanol. A series of corresponding working solutions I at 0.4 pg/µl, 2 pg/µl, 6 pg/µl, 10 pg/µl, 20 pg/µl, 40 pg/µl, 80 pg/µl, 100 pg/µl, 160 pg/µl, and 200 pg/µl were prepared by diluting the stock solution I with methanol. The stock solution II consisting of PGE2 and PGD2 (600 pg/µl) was made in methanol. A series of corresponding working solutions II at 2 pg/µl, 6 pg/µl, 12 pg/µl, 30 pg/µl, 60 pg/µl, 120 pg/µl, 160 pg/µl, 240 pg/µl, 480 pg/µl, and 600 pg/µl were prepared by diluting the stock solution II with methanol. The stock solution III consisting of AA (400 ng/µl) was made in methanol. A series of corresponding working solutions III at 4 ng/µl, 8 ng/µl, 20 ng/µl, 40 ng/µl, 80 ng/µl, 120 ng/µl, 160 ng/µl, 240 ng/µl, 320 ng/µl, and 400 ng/µl were prepared by diluting the stock solution III with methanol. The internal standard stock solution IV consisting of PGDF2α-d4, PGD2-d4, 15(s)-HETE-d8, 8,9-EET-d8, 1 1,12-EET-d8, 14,15-EET-d8, and AA-d8 (500 pg/µl except for AA-d8 at 200 ng/µl) was made in methanol. The corresponding working solution IV (20 pg/µl except for AA-d8 at 8 ng/µl) was prepared by diluting the stock solution IV with methanol. All of the solutions were stored in 2 ml polypropylene tubes at ≤−76 °C.
Calibration standards and quality control (QC) samples were prepared by spiking analyte-free extraction solvent (195 µl of methanol with 2 µl of formic acid) with a series of 5 µl of working solutions I, II, and III. The resultant nominal spiked amounts of calibration standards for PGF2α, PGJ2, 14,15-DiHETrE, 11,12-DiHETrE, 8,9-DiHETrE, 5,6-DiHETrE, 20-HETE, 15-HETE, 12-HETE, 8-HETE, 5-HETE, 14,15-EET, 11,12-EET, 8,9-EET, and 5,6-EET were 2 pg, 10 pg, 30 pg, 50 pg, 100 pg, 200 pg, 400 pg, 500 pg, 800 pg, and 1000 pg; for PGE2 and PGD2 were 10 pg, 30 pg, 60 pg, 150 pg, 300 pg, 600 pg, 800 pg, 1200 pg, and 2400 pg; for AA were 20 ng, 40 ng, 100 ng, 200 ng, 400 ng, 600 ng, 800 ng, 1200 ng, 1600 ng, and 2000 ng. The resultant nominal spiked amounts of QC samples for PGF2α, PGE2, PGD2, PGJ2, 14,15-DiHETrE, 11,12-DiHETrE, 8,9-DiHETrE, 5,6-DiHETrE, 20-HETE, 15-HETE, 12-HETE, 8-HETE, 5-HETE, 14,15-EET, 11,12-EET, 8,9-EET, and 5,6-EET were 50 pg, 200 pg, and 800 pg; for AA were 40 ng, 400 ng, and 1600 ng. All calibration standards and QC samples were subjected to solid phase extraction procedure described in Section 2.3. The samples were then stored at ≤−76 °C until LC/MS analysis. A blank sample (analyte-free extraction solvent) and a zero sample (analyte-free extraction solvent spiked with internal standards) were also prepared by being taken through the solid phase extraction procedure described in Section 2.3.
2.6. Liquid chromatography/mass spectrometry
The Agilent 1100 Series Liquid Chromatograph/Mass Selective Detector (LC/MSD) system was used for LC separation and detection. The HPLC system consisted of a binary pump, a thermostated autosampler and a column thermostat. A gradient chromatographic separation was performed on a HPLC Symmetry® C18 column (4.6 mm × 250 mm, 5 µm) (Waters Corporation, USA) at 40 °C. Mobile phase A consisted of 0.1% formic acid in water, and mobile phase B consisted of 0.1% formic acid in acetonitrile. A flow rate 1 ml/min was used to deliver the mobile phase A and B gradient: 60–80% B in 30 min; 80–85% B in 5 min; 85–100% in 1 min; 100% B for 9 min. The autosampler was set at 4 °C and the injection volume was 10 µl. The HPLC column effluent was pumped to a SL model quadrupole mass spectrometer with atmospheric electrospray ionization source. The detection was under the selected ion monitoring (SIM) negative mode. Ions with m/z 353 (PGF2α), m/z 351 (PGD2 and PGE2), m/z 333 (PGJ2), m/z 355 (PGD2-d4), and m/z 357 (PGF2α-d4) were monitored from 0 min to 7 min using gain 2; ions with m/z 337 (DiHETrEs), m/z 319 (HETEs and EETs), and m/z 327 (HETE-d8 and EET-d8) were monitored from 7 min to 23 min using gain 2; ions with m/z 303 (AA) and m/z 311 (AA-d8) were monitored from 23 min to 45 min using gain 1. The following mass detector parameters were used: capillary voltage = 3000 V; gas temperature = 350 °C; drying gas flow rate = 11 l/min; nebulizer pressure = 40 psig; fragmentor voltage = 120 V from 0 min to 7 min, 110 V from 7 min to 23 min, and 130 V from 23 min to 45 min. The data acquisitions were accomplished using LC/MSD ChemStation Rev. A.09.03 (Agilent Technologies).
2.7. Method validation
The method validation including LC/MS instrument qualification, selectivity, calibration model, accuracy, precision (with brain matrix and without brain matrix), extraction efficiency, and stability of samples sitting in autosampler were performed to establish our method is suitable for quantifying endogenous eicosanoids including PGs, DiHETrEs, HETEs, EETs, and AA in brain tissue. The guiding principles [41–57] for validation of bioanalytical methods were used as references to design the experiments.
The operational qualification and performance verification tests were performed for LC/MSD with electrospray ionization source according to HP 1100 Series Qualification Workbook. Caffeine water solutions ranged from 0.5 µg/ml to 50 µg/ml were used. The tests included flow accuracy/precision, injector precision/carry over, response linearity, and gradient composition. The selectivity of the method was evaluated by comparing calibration standard to the blank and zero samples. The intra- and inter-day accuracy and precision were determined using independent QC samples at two levels. The deviation (%) of the mean from the nominal spiked amount served as the measure of accuracy. The % R.S.D. of the mean served as the measure of precision. The intra-day precision with brain matrix was determined at three levels using independent samples prepared from brain homogenates spiked with known amounts of PGs, DiHETrEs, HETEs, EETs, AA and internal standards. The response ratios of individual eicosanoid to internal standard were calculated, and the % R.S.D. of the ratios was used to measure the precision with brain matrix. The extraction efficiencies (%) were evaluated at three levels.
At each level, three independently prepared samples were used for determination. The response of the sample spiked with standards before sample preparation (normalized with internal standards) was compared to the response of the sample spiked with standard after sample preparation (normalized with internal standards), and the ratio (%) was used to measure extraction efficiency. The stability of samples after sitting in autosampler tray was tested using brain homogenate (160 mg brain tissue) spiked with 2400 pg eicosanoid standards.
3. Results and discussion
The overall method development included four stages:
An LC/MS method for 19 eicosanoid standards including PGs, HETEs, DiHETrEs, EETs and AA, and 7 deuterated internal standards was developed and optimized with the aim of resolution and sensitivity (Section 3.1).
The method was fine-tuned by coupling the LC/MS conditions with the sample preparation procedure, the sensitivity and linearity of standards, internal and external standardization, concentration range, and the method validation design were studied at pre-validation stage (Section 3.2).
Method validation was performed (Section 3.3).
The method was applied to analyze PGs, DiHETrEs, HETEs, EETs, and AA in cortical brain tissue (Section 3.4).
3.1. Development of liquid chromatography/mass spectrometry method
For initial the HPLC method development with the aim of resolution and sensitivity; electrospray ionization with negative ion scan mode with default parameters was used, because PGs, HETEs, DiHETrEs, EETs, and AA are readily ionized to form carboxylate anions, and that is what electrospray ionization favored. To simultaneously separate 19 eicosanoids (PGs, DiHETrEs, HETEs, EETs, and AA) and 7 deuterated internal standards, we used a Symmetry® C18 column (4.6mm × 250 mm, 5 µm) that was used in our lab for separating 14 fluorescent labeling PGs, DiHETrEs, HETEs, EETs, and AA simultaneously [28]. A flow rate of 1 ml/min was used throughout the method development. Simple and volatile aqueous acetonitrile mobile phase with 0.1% formic acid was used. Gradient conditions was used and optimized to obtain the best separation less than 40 min. The elution sequence was identified by running individually standard, and was PGF2α-d4, followed by PGF2α, PGE2, PGD2-d4, PGD2, PGJ2, 14,15-DiHETrE, 11,12-DiHETrE, 8,9-DiHETrE, 20- HETE, 5,6-DiHETrE, 15(S)-HETE-d8, 15-HETE, 12-HETE, 9-HETE, 8-HETE, 5-HETE, 14,15-EET-d8, 14,15-EET, 11,12-EET-d8, 11,12-EET, 8,9-EET-d8, 5,6-EET, AA-d8, and AA. 12-HETE and 9-HETE that have the same molecular weight were co-eluted and were quantified as a group in our method. Interestingly, 20-HETE was eluted before 5,6-DiHETrE. For the purpose of obtaining higher sensitivity, a Symmetry® C18 column (2.1 mm × 150 mm, 3.5 µm) was further compared. However, by using this narrow-bore column, a better sensitivity was not achieved and some of the regioisomers that have the same molecular weigh were only partially separated.
Mass spectrometric detection conditions were then optimized based on the resulting HPLC conditions. HPLC runs on both atmospheric pressure chemical ionization and electrospray ionization with either positive or negative ion scan mode were further compared. The electrospray ionization with negative ion mode provided the best detection. The most intense pseudo molecular ion of PGs, DiHETrEs, HETEs, EETs, AA, and deuterated internal standards was [M–H]− and its m/z was used for selected ion monitoring (SIM) detection. The representative mass spectra of PGs, DiHETrEs, HETEs, EETs, and AA are shown in Fig. 3. For better selectivity and sensitivity, the time program was used for mass detection. Since the levels of endogenous PGs, DiHETrEs, HETEs, and EETs were very low, to improve the sensitivity, extra effect was devoted to optimize spray chamber parameters including fragmentor voltage, capillary voltage, drying gas flow, nebulizer gas pressure and gain. For determining the best setting of the spray chamber for analysis, representative PGF2α, PGD2, 5,6-DiHETrE, 14,15-DiHETrE, 5-HETE, 15-HETE, 20-HETE, 5,6-EET, 14,15-EET, and AA (4 ng/µl in methanol) were chosen and flow injection analyses (FIA) (with 1 µl of injection volume) were performed while the mobile phase and flow rate were kept the same as corresponding HPLC conditions. By changing the gain from 1 to 2, peak responses were doubled. However, by changing the gain from 2 to 3, peak responses only increased by 38% while noise responses increased accordingly at the same time. Therefore, a gain of 2 was used for detecting trace PGs, DiHETrEs, HETEs and EETs, and a gain of 1 was used for detecting AA with relatively high level in brain tissue. Fragmentor voltage, capillary voltage, drying gas flow, and nebulizer gas pressure were then optimized in sequence. As a result, the responses of eicosanoids increased by 15% using the optimum setting of spay chamber parameters in Table 1. Once developed, the LC/MS was further adjusted in terms of specificity by coupling with sample preparation procedure. The representative LC/MS chromatogram for separation 19 eicosanoids and 7 deuterium labeled internal standards is shown in Fig. 4.
Fig. 3.
Scan mass spectra of representative PGs, DiHETrEs, HETEs, EETs, and AA in mobile phase under negative electrospray ionization mode.
Table 1.
Optimum settings of spay chamber parameters
| Operating parameter | Range | Incremental step | Optimum condition |
|---|---|---|---|
| Fragmentor voltage (V) | 50–150 | 10 | 110–130 |
| Capillary voltage (V) | 2500–5000 | 500 | 3000 |
| Drying gas flow (l/min) | 8–13 | 1 | 11 |
| Nebulizer gas pressure (psig) | 20–60 | 5 | 40 |
| Gain | 1–3 | 1 | 1–2 |
Fig. 4.
LC/MS chromatogram for separation 19 eicosanoids and 7 deuterium labeled internal standards.
3.2. Pre-validation
Before validation of the method, the method was fine-tuned by coupling the LC/MS conditions with the sample preparation procedure modified from the one developed in our lab [28]. Each step was optimized relative the others for the purpose of easy operation and robustness. The ruggedness of the method was tested in different labs. The sensitivity, linearity, concentration range, recovery, and stability of the working solution were checked.
3.2.1. Sensitivity and quantification linearity of standards
To obtain information about the sensitivity, and assist with troubleshooting in later stages of the method development, the limit of detection (LOD), limit of quantification (LOQ), and quantification linearity of standards were tested on individually without performing sample preparation and without the biological matrix, i.e. standard solutions were used. Primary stock solutions (1 µg/ml) of individual PGD2, 11,12-DiHETrE, 12-HETE, 5,6-EET, or AA were prepared in methanol, respectively. Serial dilutions of these stock solutions with methanol were performed for LOD and LOQ experiments. The on-column LOD (S/N > 4) and LOQ (S/N > 10) were estimated to be 5 × 10−4 pg and 5 × 10−3 pg individually using FIA with 5 µl injection. The linearity of PGs, DiHETrEs, HETEs, EETs, and AA within the range of 1–4500 pg was evaluated using developed LC/MS conditions, and correlation coefficients (R2) were all higher than 0.999.
3.2.2. Evaluation internal and external standardization approaches
Although deuterated internal standards were designed to be used for quantification initially, to determine the final procedure for application, some studies were still necessary. First, five independent standards were prepared by spiking extraction solvent with different amount of standards and fix amount of internal standards (1200 pg except AA is 120 ng) followed by being taken through SPE procedure. The external calibration curves and internal calibration curves of PGs, DiHETrEs, HETEs, EETs, and AA were constructed, and all provided very good linearity, R2 > 0.99 and low values of the associated residuals. No significant differences were noted in the residuals between the internal standard and external standard analyses. To find out the concentration range for our application, the representative real brain samples occurred in TBI model were analyzed and compared to the curves, including samples from uninjured brain, injured brain, injured brain treated with COX2 inhibitor. The results were used to define the validation ranges for PGs, DiHETrEs, HETEs, EETs, and AA at later stage. Second, the inter-day reproducibility of the instrument response was evaluated in continuous 3 days using a methanol solution consisted of 19 eicosanoid standards and 7 deuterated internal standards (200 ng/ml). The % R.S.D. of absolute peak responses ranged from 7% to 18%. However, the % R.S.D. of the response ratios of standards to internal standards ranged from 0.28% to 2.09% while the absolute peak responses in the second day was higher than the first day and lower than the third day. This indicated that internal standardization was still necessary in our method to minimize the variance introduced by different day-to-day LC/MS instrument responses. Therefore, the deuterated internal standards were spiked into the eluent from SPE cartridge in our final procedure. In addition to above evaluation, the reproducibility of recoveries and cleanness of the samples were checked at this stage.
3.2.3. Method validation design
The guiding principles [41–57] for validation of bioanalytical methods were used as references to design the experiments. Since PGs, DiHETrEs, HETEs, EETs, and AA are endogenous compounds, the analyte-free brain matrix is not available for matrix-based method validation and there is no other proxy matrix available to mimic the complicated solid brain tissue. Meanwhile, standard addition methods are not suitable in our application because more errors will be introduced due to endogenous trace levels of these eicosanoids in brain. Therefore, preparation of calibration standards by spiking analyte-free extraction solvent with a series of working solutions followed by being taken through the whole procedure is the best choice for our application. In addition, a clean-up SPE procedure was used to reduce the matrix effect. To evaluate the systematic errors arising from not using matrix-matched calibration curve [47,53], the standard addition curves prepared in brain homogenate were compared to the calibration curves prepared in analyte-free extraction solvent. The relative differences between slopes were less than 8%, and the intercepts of standard addition curves were higher corresponding to endogenous presenting of these eicosanoids in brain. Furthermore, the quantitative changes of PGs, DiHETrEs, HETEs, EETs, and AA were studies in our application, errors introduced by matrix effect were significantly minimized due to background subtraction between samples of uninjured brain, injured brain and injured brain treated with COX2 inhibitor. For these reasons, specificity, calibration model study, accuracy, precision, extraction efficiency were studied using samples prepared by spiking analyte-free extraction solvent solvent with standards followed by being taken through SPE procedure, the precision using samples prepared by spiking brain homogenate with standards was evaluated as well.
3.3. Method validation
3.3.1. LC/MS instrument qualification
The operational qualification and performance verification tests were performed for LC/MSD with electrospray ionization source according to HP 1100 Series Qualification Workbook. The results of standard verification tests including flow accuracy/precision, injector precision/carry over, response linearity, and gradient composition all passed the acceptance limits defined by Agilent Technologies, and indicated that the LC/MSD instrument performed as intended throughout all anticipated operating ranges.
3.3.2. Stability of processed brain samples after sitting in autosampler tray
The stability of processed samples after storage in autosampler tray (4 °C) was tested using samples prepared by spiking brain homogenate with representative PGs, DiHETrEs, HETEs, EET, and AA. The results are given in Table 2. These results indicated that reconstituted samples were stable for at least 14 h. Therefore, more samples can be run using sequence without the presence of analysts.
Table 2.
Stability of processed brain samples after sitting in autosampler tray
| Stability (h) | AA (%) | PGE2 (%) | 14,15-DiHETrE (%) | 20-HETE (%) | 8,9-EET (%) |
|---|---|---|---|---|---|
| 0 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| 2 | 98.74 | 105.27 | 97.56 | 99.17 | 106.06 |
| 4 | 99.74 | 99.57 | 102.74 | 101.91 | 96.40 |
| 6 | 99.34 | 93.72 | 101.51 | 100.24 | 99.62 |
| 8 | 100.72 | 100.31 | 104.40 | 102.01 | 100.99 |
| 10 | 99.82 | 99.55 | 99.24 | 100.53 | 97.70 |
| 12 | 99.51 | 97.86 | 97.91 | 100.38 | 95.40 |
| 14 | 97.41 | 100.66 | 98.01 | 100.98 | 99.97 |
3.3.3. Selectivity
The selectivity of the method was evaluated by comparing calibration standard, blank, and zero samples. No false positive peaks were introduced by internal standards. This is demonstrated by absence of internal standard peaks observed in the extracted ion chromatograms of blank samples. In addition, no other interferences were observed except that two peaks being introduced by SPE procedure were co-eluted with PGF2α and 15-HETE.
3.3.4. Calibration model
The internal standardization approach was used: PGD2-d4 was used for PGs; 15(S)-HETE-d8 was used for DiHETrEs and HETEs; 14,15-EET-d8 was used for 14,15-EET; 1 1,12-EET-d8 was used for 11,12-EET and 5,6-EET; 8,9-EET-d8 was used for 8,9-EET; AA-d8 was used for AA. Concentration ranges for PGs, DiHETrEs, HETE, EETs, and AA were investigated at pre-validation stage and were chosen on the basis of real brain samples in TBI model. PGF2α, PGJ2, DiHETrEs, HETEs, and EETs ranged from 2 pg to 1000 pg. PGE2 and PGD2 ranged from 10 pg to 2400 pg. AA ranged from 20 ng to 2000 ng. Over 80% of 10 calibration standards whose deviations of back-calculated amount from nominal spiked amount fell within 15% were used to construct the calibration curves. The deviation at the lower end of the range was relatively large using simple least-squares linear regression because of the concentration ranges over 3 orders of magnitude. This phenomenon is common in bioanalytical analysis using LC/MS based methods [48]. As such, a weighted least-squares linear regression was used [58,59] in our method. Different weighting factors including linear (1/X and 1/Y normalized to the smallest amount) and quadratic (1/X2 and 1/Y2 normalized to the smallest amount) were compared using Agilent ChemStation software Rev.A.09.03. The weighting factor 1/Y2 (normalized to the smallest amount) provided the best fits of calibration curves (Table 3). The deviations (%) of back-calculated amounts from nominal spiked amounts ranged from −14.05% to 13.63% for all levels and correlation coefficients (R2) were over 0.99. The lower limit of quantification (LLOQ) was the lowest level on the calibration curve with the deviations (%) ranged from −8.08% to 13.63%, and its response was five times greater than the blank response.
Table 3.
Calibration curve parameters of PGs, HETEs, EETs, and AA
| Equation | Correlation coefficient(R2) | SIM (m/z) | Range | |
|---|---|---|---|---|
| AA | Y = 607.165X + 40.318* | 0.996 | 303 | 20–2000 ng |
| PGE2 | Y = 1.042X + 0.098 | 0.995 | 351 | 10–2400 pg |
| PGD2 | Y = 1.487X + 0.218 | 0.999 | 351 | 10–2400 pg |
| PGJ2 | Y = 2.518X + 0.117 | 0.999 | 333 | 2–1000 pg |
| 14,15-DiHETrE | Y = 5.284X + 0.096 | 0.997 | 337 | 2–1000 pg |
| 11,12-DiHETrE | Y = 1.931X + 0.107 | 0.997 | 337 | 2–1000 pg |
| 8,9-DiHETrE | Y = 1.402X + 0.181 | 0.997 | 337 | 2–1000 pg |
| 5,6-DiHETrE | Y = 1.562X + 0.109 | 0.997 | 337 | 2–1000 pg |
| 20-HETE | Y = 1.089X − 0.010 | 0.998 | 319 | 2–1000 pg |
| 12-HETE | Y = 1.464X + 0.103 | 0.995 | 319 | 2–1000 pg |
| 8-HETE | Y = 1.380X + 0.061 | 0.998 | 319 | 2–1000 pg |
| 5-HETE | Y = 1.267X + 0.202 | 0.992 | 319 | 2–1000 pg |
| 14,15-EET | Y = 1.621X + 0.670 | 0.994 | 319 | 2–1000 pg |
| 11,12-EET | Y = 0.613X + 0.088 | 0.998 | 319 | 2–1000 pg |
| 8,9-EET | Y = 1.431X + 0.167 | 0.997 | 319 | 2–1000 pg |
| 5,6-EET | Y = 1.368X + 0.296 | 0.995 | 319 | 2–1000 pg |
Y = area ratio; X = amount ratio.
3.3.5. Accuracy and precision
The intra- and inter-day accuracy and precision using samples prepared by spiking analyte-free extraction solvent with standards followed by being taken through SPE procedure were determined at two levels. The results for representative PGs, DiHETrEs, HETEs, EETs, and AA are given in Table 4. The precision using samples prepared by spiking brain homogenate with PGs, DiHETrEs, HETEs, EETs, and AA was evaluated at three levels. The results are given in Table 5. Good values for the % R.S.D. for all analytes (within 15%) indicated that the procedure related to brain matrix was reproducible.
Table 4.
Accuracy and precision of representative PG, DiHETrE, HETE, and AA (n = 3)
| Nominal spiked amount | Deviation (%) | R.S.D. (%) | |||
|---|---|---|---|---|---|
| Intra-day | Inter-day | Intra-day | Inter-day | ||
| PGJ2 | 50 pg | 0.17 | 1.61 | 1.86 | 5.40 |
| 200 pg | 0.94 | −4.17 | 3.41 | 1.03 | |
| 14,15-DiHETrE | 50 pg | −10.21 | −1.21 | 8.34 | 3.79 |
| 200 pg | −3.61 | −11.36 | 5.78 | 17.60 | |
| 20-HETE | 50 pg | 0.94 | 9.50 | 1.87 | 14.02 |
| 200 pg | −7.12 | −2.18 | 0.57 | 3.29 | |
| 8,9-EET | 50 pg | 8.46 | 11.18 | 13.31 | 12.22 |
| 200 pg | 5.16 | 26.45 | 9.03 | 2.29 | |
| AA | 400 ng | 2.45 | 1.67 | 2.60 | 8.34 |
| 1600 ng | 3.17 | 5.13 | 2.30 | 3.55 | |
Table 5.
Precision of PGs, DiHETrEs, HETEs, EETs, and AA in brain matrix (n = 3)
| Nominal spiked amount (% R.S.D.) | |||
|---|---|---|---|
| 50 pg | 200 pg | 800 pg | |
| PGJ2 | 2.73 | 0.66 | 9.91 |
| PGE2 | 4.77 | 1.91 | 0.61 |
| PGD2 | 4.12 | 5.21 | 0.61 |
| 14,15-DiHETrE | 3.30 | 8.80 | 2.25 |
| 11,12-DiHETrE | 9.06 | 7.71 | 13.16 |
| 8,9-DiHETrE | 8.08 | 8.60 | 11.69 |
| 5,6-DiHETrE | 4.79 | 10.34 | 1.99 |
| 20-HETE | 2.98 | 4.11 | 0.83 |
| 12-HETE | 4.35 | 7.71 | 5.43 |
| 8-HETE | 0.99 | 8.77 | 5.86 |
| 5-HETE | 4.05 | 8.73 | 1.74 |
| 14,15-EET | 14.43 | 11.35 | 0.44 |
| 11,12-EET | 6.17 | 4.68 | 6.44 |
| 8,9-EET | 14.33 | 12.04 | 0.84 |
| 5,6-EET | 5.65 | 11.25 | 0.86 |
| Nominal spiked amount (% R.S.D.) | |||
| 40 ng | 400 ng | 1600 ng | |
| AA | 12.62 | 5.50 | 0.04 |
3.3.6. Extraction efficiency
The extraction efficiency was evaluated at three levels (three independent samples each): at 50 pg, 200 pg, and 800 pg for PGs, DiHETrEs, HETEs, and EETs; and at 40 ng, 400 ng, and 1600 ng for AA. The results for representative PG, DiHETrE, HETE, EET, and AA are given in Table 6. Good values of the % R.S.D. (1.93–14.73%) indicate reproducible extraction efficiency, and this is the most important for developing a reliable bioanalytical method.
Table 6.
Extraction efficiency of representative PG, DiHETrE, HETE, EET, and AA (n = 3)
| Level | Extraction efficiency (%) | R.S.D. (%) | |
|---|---|---|---|
| 50 pg | 93.39 | 2.79 | |
| PGJ2 | 200 pg | 98.40 | 10.84 |
| 800 pg | 99.56 | 13.46 | |
| 50 pg | 94.35 | 11.32 | |
| 14,15-DiHETrE | 200 pg | 94.97 | 5.88 |
| 800 pg | 98.16 | 13.32 | |
| 50 pg | 72.13 | 8.66 | |
| 20-HETE | 200 pg | 92.31 | 6.49 |
| 800 pg | 93.96 | 14.23 | |
| 50 pg | 77.78 | 14.73 | |
| 11,12-EET | 200 pg | 82.14 | 10.34 |
| 800 pg | 83.31 | 9.28 | |
| 40 ng | 54.84 | 13.04 | |
| AA | 400 ng | 77.67 | 8.12 |
| 1600 ng | 90.08 | 11.31 |
3.4. Analysis of cortical brain tissue samples
The method was applied to analyze rat brain samples from TBI model, including samples from uninjured brain, injured brain and injured brain treated with COX2 inhibitor. The peak retention times in extracted ion chromatograms were used to identify PGs, DiHETrEs, HETEs, EETs, and AA. In addition, the retention time of deuterium labeled internal standards were used to assist differentiation of corresponding unlabeled peaks from closely eluted peaks presenting in brain matrix. Peaks of PGF2α, PGE2, PGD2, 20-HETE, 15-HETE, 12-HETE and/or 9-HETE, 8-HETE, 5-HETE, 14,15-EET, 11,12-EET, 8,9-EET, 5,6-EET, and AA were found in all the brain samples, however, PGJ2, DiHETrEs were not detected. PGF2α and 15-HETE peaks were interfered with components introduced by SPE procedure, and 8,9-EET peak was interfered with the component existing in brain matrix. In uninjured-brain samples, the levels of these eicosanoids, for EETs ranged from 1.24 pg/mg to 3.90 pg/mg; for HETEs ranged from 2.16 pg/mg to 12.14 pg/mg; for PGE2 ranged from 1.51 pg/mg to 3.60 pg/mg; and for AA ranged from 12,512 pg/mg to 47,727 pg/mg wet weight. The quantitative results of independent samples from the ipsi-lateral (to the injured site) 72 h post-injured brains are given in Table 7. The representative chromatograms are shown in Fig. 5.
Table 7.
Levels of PGs, HETEs, EETs, and AA in ipsi-lateral 72 h post-injured brains (n = 4)
| Concentration (pg/mg wet weight) | Concentration (pg/µg protein) | |||
|---|---|---|---|---|
| Mean | Standard deviation | Mean | Standard deviation | |
| PGE2 | 3.97 | 0.92 | 0.93 | 0.31 |
| PGD2 | 1.76 | 0.58 | 0.40 | 0.13 |
| 20-HETE | 2.05 | 0.29 | 0.47 | 0.09 |
| 12-HETE and/or 9-HETE | 9.73 | 2.99 | 2.25 | 0.88 |
| 8-HETE | 3.19 | 0.20 | 0.73. | 0.13 |
| 5-HETE | 3.10 | 0.52 | 0.71 | 0.17 |
| 14,15-EET | 2.57 | 0.82 | 0.57 | 0.10 |
| 11,12-EET | 2.99 | 0.47 | 0.61 | 0.08 |
| 5,6-EET | 3.51 | 0.42 | 0.72 | 0.23 |
| AA | 42373.21 | 6285.97 | 9632.31 | 1549.41 |
Fig. 5.
Representative extracted ion chromatograms of sample from ipsi-lateral 72 h post-injured brain.
4. Conclusion
This study is first to report a sensitive, specific, robust and validated LC/MS method that allows simultaneous analysis of parent compound AA and its COX, CYP450 and LOX pathway metabolites PGs, DiHETrEs, HETEs and EETs, including PGF2α, PGE2, PGD2,PGJ2, 14,15-DiHETrE, 11,12-DiHETrE, 8,9-DiHETrE, 5,6-DiHETrE, 20-HETE, 15-HETE, 12-HETE, 9-HETE, 8-HETE, 5-HETE, 14,15-EET, 11,12-EET, 8,9-EET, and 5,6-EET in rat brain tissues. LC/MS conditions were optimized for the best sensitivity and separation of 19 eicosanoids and 7 deuterated internal standards within 35 min. The sample preparation procedure was fine-tuned for the purpose of robust and ease of operation. The method validation, including LC/MS instrument qualification, specificity, calibration model, accuracy, precision (without brain matrix and with brain matrix), and extraction efficiency were performed. Furthermore, this method can be adapted easily to a more specific LC/MS/MS method and a high-throughput robotic liquid handling system using 96-well plates SPE. Overall, the methodology described in this paper provides a quantitative way to study eicosanoids derived from AA in various biological fluids and tissues, and has important potential use in elucidating the mechanism of eicosanoid metabolites in TBI as well as other disease.
Acknowledgement
The authors thank Mamta Amin for expert technical assistance.
References
- 1.Beare-rogers J, Dieffenbacher A, Holm JV. Pure Appl. Chem. 2001;73:685–744. [Google Scholar]
- 2.Roman RJ. Physiol. Rev. 2002;82:131–185. doi: 10.1152/physrev.00021.2001. [DOI] [PubMed] [Google Scholar]
- 3.Vink R, Heuvel CVD. Expert Opin. Investig. Drugs. 2004;13:1263–1274. doi: 10.1517/13543784.13.10.1263. [DOI] [PubMed] [Google Scholar]
- 4.Mcintosh TK, Smith DH, Garde E. Eur. J. Anaesthesiol. 1996;13:291–309. doi: 10.1046/j.1365-2346.1996.00981.x. [DOI] [PubMed] [Google Scholar]
- 5.Bradl M, Hohlfeld R. Neurosci. Neurologists. 2003;74:1364–1370. doi: 10.1136/jnnp.74.10.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enriquez P, Bullock R. Curr. Pharm. Des. 2004;10:2131–2143. doi: 10.2174/1381612043384060. [DOI] [PubMed] [Google Scholar]
- 7.Strauss KI, Barbe MF, Marshall RM, Raghupathi R, Mehta S, Narayan RK. J. Neurotrauma. 2000;17:695–711. doi: 10.1089/089771500415436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilroy DW, Colville-Nash PR. J. Mol. Med. 2000;78:121–129. doi: 10.1007/s001090000094. [DOI] [PubMed] [Google Scholar]
- 9.Vane J. Pharmacology. 1994;367:215–216. [Google Scholar]
- 10.Hurley SD, Olschowka JA, O’Banion MK. J. Neurotrauma. 2002;19:1–15. doi: 10.1089/089771502753460196. [DOI] [PubMed] [Google Scholar]
- 11.Willoughby DA, Moore AR, Colville-Nash PR. Lancet. 2000;355:646–648. doi: 10.1016/S0140-6736(99)12031-2. [DOI] [PubMed] [Google Scholar]
- 12.Consilvio C, Vincent AM, Feldman EL. Exp. Neurol. 2004;187:1–10. doi: 10.1016/j.expneurol.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 13.Node K, Huo Y, Ruan B, Yang M, Spiecker K, Ley DC, Zeldin JK. Liao, Science. 1999;285:1276–1279. doi: 10.1126/science.285.5431.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hampson AJ, Grimaldi M. J. Neurosci. 2002;22:257–264. doi: 10.1523/JNEUROSCI.22-01-00257.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis BB, Thompson DA. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2222–2227. doi: 10.1073/pnas.261710799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colville-Nash P, Gilroy D. Drug News Perspect. 2000;13:587–597. doi: 10.1358/dnp.2000.13.10.661378. [DOI] [PubMed] [Google Scholar]
- 17.Cernak I, O’connor C, Vink R. Exp. Brain Res. 2002;147:193–199. doi: 10.1007/s00221-002-1245-z. [DOI] [PubMed] [Google Scholar]
- 18.Strauss KI, Marini AM. J. Neurotrauma. 2002;19:627–638. doi: 10.1089/089771502753754091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gopez JJ, Yue H, Vasudevan R, Malik AS, Fogelsanger LN, Lewis S, Panikashvili D, Shohami E, Jansen SA, Narayan RK, Strauss KI. Neurosurgery. 2005;56:590–597. doi: 10.1227/01.NEU.0000154060.14900.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsikas D. J. Chromatogr. B. 1998;717:201–245. doi: 10.1016/s0378-4347(98)00210-2. [DOI] [PubMed] [Google Scholar]
- 21.Tsukamoto H, Hishinuma T, Mikkaichi T, Nakamura H, Yamazaki T, Tomioka Y, Mizugaki M. J. Chromatogr. B. 2002;774:205–214. doi: 10.1016/s1570-0232(02)00220-9. [DOI] [PubMed] [Google Scholar]
- 22.Catella F, Lawson JA, Fitzgerald DJ, Fitzgerald GA. Proc. Natl. Acad. Sci. U.S.A. 1990;87:5893–5897. doi: 10.1073/pnas.87.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capdevila J, Chacos N, Wehrincloer J, Prough RA, Estabrook RW. Proc. Natl. Acad. Sci. U.S.A. 1981;78:5362–5366. doi: 10.1073/pnas.78.9.5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capdevila JH, Wei J, Yan A, Karara A, Jacobson HR, Falck JR, Guengerich FP, Dubois RN. J. Biol. Chem. 1992;267:21720–21726. [PubMed] [Google Scholar]
- 25.VanderNoot VA, VanRollins M. Anal. Chem. 2002;74:5859–5865. doi: 10.1021/ac025909+. [DOI] [PubMed] [Google Scholar]
- 26.Nithipatikom K, Pratt PF, Campbell WB. Am. J. Physiol. Heart Circ. Physiol. 2000;279:H857–H862. doi: 10.1152/ajpheart.2000.279.2.H857. [DOI] [PubMed] [Google Scholar]
- 27.Maier KG, Henderson L, Narayanan J, Alonso-Galicia M, Falck JR, Roman RL. Am. J. Physiol. Heart Circ. Physiol. 2000;279:H863–H871. doi: 10.1152/ajpheart.2000.279.2.H863. [DOI] [PubMed] [Google Scholar]
- 28.Yue H, Strauss KI, Borenstein MR, Barbe MF, Rossi LJ, Jansen SA. J. Chromatogr. B. 2004;803:267–277. doi: 10.1016/j.jchromb.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 29.Bolcato CA, Frye RF, Zemaitis MA, Poloyac SM. J. Chromatogr. B. 2003;794:363–372. doi: 10.1016/s1570-0232(03)00496-3. [DOI] [PubMed] [Google Scholar]
- 30.Takabatake M, Hishinuma T, Suzuki N, Chiba S, Tsukamoto H, Nakamura H, Saga T, Tomioka Y, Kurose A, Sawai T, Mizugaki M. Leuko. Essent. Fatty Acids. 2002;67:51–56. doi: 10.1054/plef.2002.0381. [DOI] [PubMed] [Google Scholar]
- 31.Margalit A, Duffin KL, Isakson PC. Anal. Biochem. 1996;235:73–81. doi: 10.1006/abio.1996.0093. [DOI] [PubMed] [Google Scholar]
- 32.Kempen KC, Yang P, Felix E, Madden T, Newman RA. Anal. Biochem. 2001;297:183–190. doi: 10.1006/abio.2001.5325. [DOI] [PubMed] [Google Scholar]
- 33.Nithipatikom K, Laabs ND, Isbell MA, Campbell WB. J. Chromatogr. 2003;B 785:135–145. doi: 10.1016/s1570-0232(02)00906-6. [DOI] [PubMed] [Google Scholar]
- 34.MacPherson JC, Pavlovich JG, Jacobs RS. Biochim. Biophy. Acta. 1996;1303:127–136. doi: 10.1016/0005-2760(96)00097-5. [DOI] [PubMed] [Google Scholar]
- 35.Schwedhelm E, Tsikas D, Durand T, Gutzki F-M, Guy A, Rossi J-C, Frolich JC. J. Chromatogr. B. 2000;744:99–112. doi: 10.1016/s0378-4347(00)00236-x. [DOI] [PubMed] [Google Scholar]
- 36.Newby CS, Mallet AI. Rapid Commun. Mass Spectrom. 1997;11:1723–1727. doi: 10.1002/(SICI)1097-0231(19971015)11:15<1723::AID-RCM71>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 37.Wheelan P, Zirrolli JA, Murphy RC. J. Am. Soc. Mass Spectrom. 1996;7:140–149. doi: 10.1016/1044-0305(95)00628-1. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura T, Baratton DL, Murphy RC. J. Mass Spectrom. 1997;32:888–896. doi: 10.1002/(SICI)1096-9888(199708)32:8<888::AID-JMS548>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 39.Murphy RC, Khaselev N, Nakamura T, Hall LM. J. Chromatogr. B. 1999;731:59–71. doi: 10.1016/s0378-4347(99)00207-8. [DOI] [PubMed] [Google Scholar]
- 40.Nithipatikom K, Grall AJ, Holmes BB, Harder DR, Falck JR, Campbell WB. Anal. Biochem. 2001;298:327–336. doi: 10.1006/abio.2001.5395. [DOI] [PubMed] [Google Scholar]
- 41.Shah VP, Midha KK, Findlay JWA, Hill HM, Hulse JD, McGilveray IJ, McKay G, Miller KJ, Patnaik ML, Powell A, Tonelli A, Viswanathan CT, Yacobi A. Pharm. Res. 2000;17:1551–1557. doi: 10.1023/a:1007669411738. [DOI] [PubMed] [Google Scholar]
- 42.Editorial, J. Chromatogr. B. 1998;707:1. [Google Scholar]
- 43.Editorial, J. Chromatogr. B. 1996;683:133. [Google Scholar]
- 44.Causon R. J. Chromatogr. B. 1997;689:175–180. doi: 10.1016/s0378-4347(96)00297-6. [DOI] [PubMed] [Google Scholar]
- 45.Bressolle F, Bromet-Petit M, Audran M. J. Chromatogr. B. 1996;686:3–10. doi: 10.1016/s0378-4347(96)00088-6. [DOI] [PubMed] [Google Scholar]
- 46.Wieling J, Hendriks G, Tamminga WJ, Hempenius J, Mensink CK, Oosterhuis B, Jonkman JHC. J. Chromatogr. B. 1996;730:381–394. doi: 10.1016/0021-9673(96)00006-4. [DOI] [PubMed] [Google Scholar]
- 47.Hartmann C, Smeyers-Verbeke J, Massart DL, McDowall RD. J. Pharm. Biomed. Anal. 1998;17:193–218. doi: 10.1016/s0731-7085(97)00198-2. [DOI] [PubMed] [Google Scholar]
- 48.Braggio S, Barnaby RJ, Grossi P, Cugola M. J. Pharm. Biomed. Anal. 1996;14:375–388. doi: 10.1016/0731-7085(95)01644-9. [DOI] [PubMed] [Google Scholar]
- 49.Dadgar D, Burnett PE. J. Pharm. Biomed. Anal. 1995;14:23–31. doi: 10.1016/0731-7085(95)01634-1. [DOI] [PubMed] [Google Scholar]
- 50.Dadgar D, Burnett PE, Choc MG, Gallicano K, Hooper JW. J. Pharm. Biomed. Anal. 1995;13:89–97. doi: 10.1016/0731-7085(94)00106-c. [DOI] [PubMed] [Google Scholar]
- 51.Karnes HT, March C. J. Pharm. Biomed. Anal. 1991;9:911–918. doi: 10.1016/0731-7085(91)80022-2. [DOI] [PubMed] [Google Scholar]
- 52.Buick AR, Doig MV, Jeal SC, Land GS, McDowall RD. J. Pharm. Biomed. Anal. 1990;8:629–637. doi: 10.1016/0731-7085(90)80093-5. [DOI] [PubMed] [Google Scholar]
- 53.Fajgelj A, Ambrus A. Principles and Practices of Method Validation. Cambridge: The Royal Society of Chemistry; 2000. [Google Scholar]
- 54.Hubert P, Chiap P, Crommen J, Boulanger B, Chapuzet E, Bervoas-Martin MNS, Chevalier P, Grandjean D, Lagorce P, Lallier M, Laparra MC, Laurentie M, Nivert JC. Anal. Chim. Acta. 1999;391:135–148. [Google Scholar]
- 55.FDA. Guidance for Industry Bioanalytical Method Validation. 2001. pp. 1–98. [Google Scholar]
- 56.ICH. Harmonised Tripartite Guideline—Validation of Analytical Procedures: Methodology. 1996. pp. 1–33. [Google Scholar]
- 57.Powell ML, Unger SE. Bioanalytical Method: Challenges and Opportunities in Drug Development, Applications of Pharmacokinetic Principles in Drug Development. New York: Kluwer Academic/Plenum Publishers; 2004. [Google Scholar]
- 58.Law B, Temesi D. J. Chromatogr. B. 2000;748:21–30. doi: 10.1016/s0378-4347(00)00319-4. [DOI] [PubMed] [Google Scholar]
- 59.Almeida AM, Castel-Branco MM, Falcao AC. J. Chromatogr. B. 2002;744:215–222. doi: 10.1016/s1570-0232(02)00244-1. [DOI] [PubMed] [Google Scholar]