Abstract
Based on the prediction that histone lysine demethylases may contain the JmjC domain, we examined the methylation patterns of five knock-out strains (ecm5Δ, gis1Δ, rph1Δ, jhd1Δ, and jhd2Δ (yjr119cΔ)) of Saccharomyces cerevisiae. Mass spectrometry (MS) analyses of histone H3 showed increased modifications in all mutants except ecm5Δ. High-resolution MS was used to unequivocally differentiate trimethylation from acetylation in various tryptic fragments. The relative abundance of specific fragments indicated that histones K36me3 and K4me3 accumulate in rph1Δ and jhd2Δ strains, respectively, whereas both histone K36me2 and K36me accumulate in gis1Δ and jhd1Δ strains. Analyses performed with strains overexpressing the JmjC proteins yielded changes in methylation patterns that were the reverse of those obtained in the complementary knock-out strains. In vitro enzymatic assays confirmed that the JmjC domain of Rph1 specifically demethylates K36me3 primarily and K36me2 secondarily. Overexpression of RPH1 generated a growth defect in response to UV irradiation. The demethylase activity of Rph1 is responsible for the phenotype. Collectively, in addition to Jhd1, our results identified three novel JmjC domain-containing histone demethylases and their sites of action in budding yeast S. cerevisiae. Furthermore, the methodology described here will be useful for identifying histone demethylases and their target sites in other organisms.
The covalent modification of histones plays a pivotal role in the epigenetic control of gene expression (1). Histone methylation, which occurs on the side chains of lysine and arginine and is most prominent in histones H3 and H4, is linked to transcriptional activation, differentiation, imprinting, and X inactivation (2–4). In general, histone methylation at H3-K4, H3-K36, and H3-K79 is associated with euchromatin and gene activation, whereas histone methylation at H3-K9, H3-K27, and H4-K20 is associated with heterochromatin and repressed genes. Lysine may be mono-, di-, or trimethylated, where the degree of modification is likely related to different biological events. In the budding yeast Saccharomyces cerevisiae, histone H3 lysines Lys-4, Lys-36, and Lys-79 are methylated to differing degrees by histone methyltransferases Set1, Set2, and Dot1, respectively (5–8). Although methylation of these sites has been suggested to be a marker of active transcription (9), it may also play critical roles in silencing and DNA damage responses (6, 7, 10, 11).
The above site-specific histone methyltransferases have been studied for a number of years. It was believed that histone methylation was stable and irreversible. However, emerging evidence has shown that histone methylation is dynamic (12, 13), which suggests that there are histone demethylases. The 2004 landmark discovery by Shi et al. (14) of human amine oxidase LSD1, the first unequivocal histone demethylase, demonstrates that histone methylation is indeed reversible. LSD1 demethylates mono- and dimethylated H3-K4 or H3-K9 (14–16), and its homologues are found in organisms ranging from Schizosaccharomyces pombe to mammals. However, because of the nature of the amino oxidation demethylation mechanism (which requires a free lone pair of electrons on the lysine ε-nitrogen), LSD1 is unable to demethylate trimethylated lysine. Thus it was clear that an additional class of histone lysine demethylases would be requisite. We posited, as described in the following paragraph, that JmjC domain-containing proteins might be new histone demethylases. This hypothesis, apparently shared by other research groups, has been verified empirically recently with the discovery of novel human JmjC-containing histone demethylases (17–22).
Nonetheless, histone demethylation in budding yeast is still poorly understood. In particular, the trimethylated lysine demethylases remain to be determined. The fact that histone lysine methylation is prevalent in S. cerevisiae and that it does not encode an LSD1 homologue (14) suggests that it is imperative to identify yeast histone demethylases. In our search for new yeast demethylases, we postulated that candidate histone demethylases would satisfy four specific criteria; as described below, proteins containing the JmjC domain satisfy each of these. (i) Candidates must be able to cleave the stable N–CH3 bond. We surmised that an Fe2+/α-ketoglutarate-dependent dioxygenase could potentially catalyze histone demethylation via an oxygen radical mechanism in which an unstable carbinolamine intermediate is generated and followed by spontaneous elimination of formaldehyde; several groups have independently suggested a similar mechanism (12, 23, 24). Importantly, an Fe2+/α-ketoglutarate family protein, AlkB, was recently shown to catalyze the hydroxylation and subsequent demethylation of the DNA lesions 1-methyladenine and 3-methylcytosine (25, 26). The fact that the JmjC domain displays structural similarity to Fe2+/α-ketoglutarate enzymes suggests that it might be capable of catalyzing demethylation (27, 28). (ii) Candidates must be known to influence histone modifications. In satisfaction of this criterion, two S. pombe JmjC proteins, Msc1 and Epe1, have been reported to be associated with histone modifications (29, 30). (iii) Candidates must be involved in transcription regulation, such as gene silencing. Consistent with this, several JmjC-containing proteins interact with either the tumor suppressor Rb or histone deacetylases and regulate transcription. For example, RBP2, RBP1, and Jumonji interact with Rb (31, 32), and JMJD2A forms complexes with histone deacetylases and Rb (33). The JmjC proteins SMCX and SMCY are known to be involved in X inactivation. Moreover, the JmjC domain is found in conjunction with the zinc finger, PHD,5 ARID, or TUDOR domains, all of which are protein-protein interaction or DNA binding domains involved in transcription. Additionally, the PHD and Tudor domains have been shown to be methylated histone binding domains (34–36). (iv) Given the conservation of histone methylation, candidate histone demethylases should be conserved in all eukaryotic organisms, from S. cerevisiae to human, which is true for JmjC domain proteins.
Herein, we increased the repertoire of JmjC demethylases by demonstrating that four of the five JmjC-containing proteins encoded by S. cerevisiae influence histone methylation levels in vivo. The five yeast proteins are: Ecm5 (YMR176W), Gis1 (YDR096W), Rph1 (YER169W), Jhd1 (YER051W), and Jhd2 (YJR119C) (Fig. 1A). YJR119C gene product is named Jhd2 (Jmjc domain-containing histone demethylase 2 following the naming of Jhd1). Here, we utilized proteomic MS techniques that have been used successfully in previous studies of individual yeast histone methyltransferases (8, 37). We systematically analyzed global (overall pattern) and site-specific histone H3 methylation in wild type (WT) and the five yeast histone demethylase candidate knock-out strains. The changes in methylation patterns led us to predict that Rph1, Jhd1, Gis1, and Jhd2 are histone demethylases. Subsequent overexpression data were complementary to the deletion data. Particularly, overexpression of RPH1 and JHD2 significantly decreased histone H3-K36me3 and H3-K4me3 levels, respectively. In vitro enzymatic assays demonstrated that Rph1 demethylates histone H3-K36me3 primarily and H3-K36me2 secondarily. RPH1 overexpression strain is sensitive to UV irradiation damage. In addition, its histone H3-K36me3 demethylase activity is responsible for the phenotype. Rph1 and Jhd1 might be involved in transcription elongation.
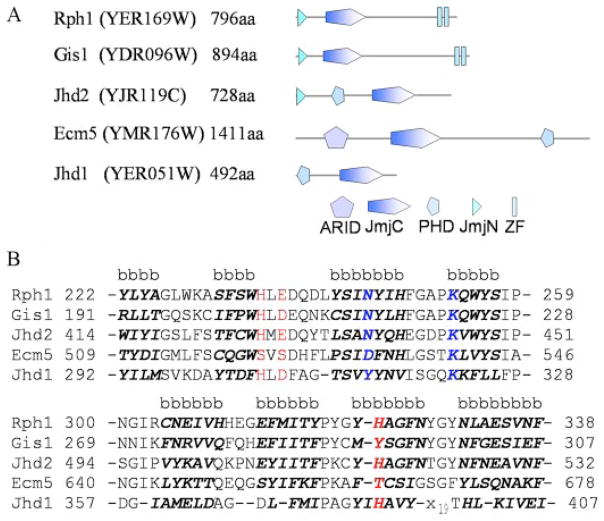
FIGURE 1. The five JmjC domain-containing proteins in S. cerevisiae.
A, domain structures. The diagrams were adapted from the SMART data base. Each protein name is followed by the open reading frame name of the gene. ZF represents a zinc finger domain. B, core β-sheet sequence alignment of the five yeast JmjC proteins. Predicted β-sheet sequences are indicated with a letter b and highlighted by bold italic letters. The predicted co-factor Fe2+ binding sites are shown in red and the predicted α-ketoglutarate binding sites in blue.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids
S. cerevisiae BY4743 was used as the WT yeast strain in this study (38). Homozygous diploid knock-out strains of ECM5, JHD1, GIS1, RPH1, and JHD2 in the BY4743 background were obtained from the Saccharomyces Genome Deletion Project (39). The JHD1, RPH1, and JHD2 overexpression strains were constructed by transforming BY4743 with GAL1 promoter-derived expression vector (BG1805) carrying the coding sequence of each gene (40); for the MS studies a stop codon was inserted into the 3′-end of each gene via site-directed mutagenesis (to enable expression of proteins without a C-terminal tag). BY4743 transformed with a BG1805 vector lacking an inserted gene was used as the WT strain in overexpression studies. For the control spot assays, the rph1 deletion strain (rph1Δ) was transformed with plasmids containing no insert (vector BG1805 only), BG1805 with wild type RPH1 insertion, or BG1805 with the activity-deficient mutant (rph1H235A) insertion, respectively. All S. cerevisiae strains and the unmodified BG1805-based expression vectors were obtained from Open Biosystems.
Culture Growth and Media
In knock-out studies 10 ml of YPD cultures were inoculated with a freshly streaked single colony. Cultures were grown at 30 °C and 200 rpm for 9 h and then diluted into 100 ml of YPD culture. Growth was continued until cultures reached the late logarithmic/stationary phase (A600 of 1.7–1.9). Cells were harvested for histone extraction by centrifugation at 5500 × g for 10 min. Overexpression strains were grown by inoculating 20 ml of SC-uracil medium containing 2% glucose and incubated at 30 °C and 200 rpm for 9 h. Aliquots from the 20-ml starter cultures were then diluted into 200 ml of SC-uracil with 2% sucrose to give an initial A600 of 0.02. Incubation was continued until an A600 of 1.0–1.2 was reached. Cells were pelleted by centrifugation at 4000 × g for 5 min and were resuspended in 200 ml of SC-uracil with 2% galactose. Cells were grown under the inducing condition for 4 h and harvested as described previously.
Histone Extraction and Mass Spectrometry Analyses
Histones were purified as described (41). Histone H3 was separated from other histones on a 16.5% Tris-Tricine SDS-polyacrylamide gel. Trypsin in-gel digestion, liquid chromatography mass spectrometry (LC-MS), nano LC-MS/MS, and MADLI-TOF MS procedures were adapted from previous work (42). High-resolution LC-MS/MS experiments were performed on an LTQ-FTICR mass spectrometer (Thermo Electron). Quantification of Lys-4 isoforms was obtained by calculating the peak areas in MALDI-TOF MS spectra, whereas quantification of Lys-36, and Lys-79 isoforms was performed by analyzing the precursor ions in nano-LC-MS/MS spectra. The reported values represent an average from four trials.
Assays of in Vitro Activities
To obtain a GST fusion protein of Rph1, yeast genomic DNA was amplified and subcloned into the pGEX6p2 vector to give the construct GST-Rph1-(1–373), with the JmjC domain at the C terminus. The plasmid was transformed into BL21-CodonPlus(DE3)-RIL cells for overexpression. Cells were induced at 25 °C and 0.5 mM isopropyl-1-thio-β-D-galactopyranoside for 8 h. The protein was purified with glutathione-agarose beads (Sigma) and then dialyzed into HEPES buffer (30 mM HEPES, pH 7.5, and 1 mM β-mercaptoethanol). Similar procedures were used to generate recombinant Gis1, Jhd2, and Ecm5 JmjC domains in the fusion forms with GST. The GST-Rph1-(1–373) H235A mutant plasmid was obtained using the Stratagene QuikChange system. Full-length His6-tagged recombinant proteins Rph1, Jhd1, and Jhd2 (in pET28a vector) were overexpressed in Escherichia coli and purified with nickel-nitrilotriacetic acid resin. To purify the full-length proteins from yeast, cells from 1 liter of SC-uracil culture were lysed by the liquid nitrogen method and purified with nickel-nitrilotriacetic acid resin.
H3-K4, -K36, and -K79 peptide sequences for the assays are ARTK(me1–3)QTARKST, STGGVK(me1–3)KPHRY, and IAQDFK(me1–3)TDLRFQ. In the following assays, all reagents were freshly made. The assay buffer consisted of 30 mM HEPES (pH 7.5), 5 mM ascorbic acid, 1 mM dithiothreitol, 1 mM α-ketoglutarate, and 100 μM (NH4)2Fe(SO4)2 6H2O. In a typical assay, 20 μg of protein was incubated with 0.5 μl of 5 mM peptide in 60 μl of assay buffer at 37 °C for 30 min to 2 h. The reaction was quenched by the addition of 1 μl of 100 mM EDTA. 1 μl of the reaction mixture was then diluted with 5 μl of saturated matrix solution (α-cyano-4-hydroxycinnamic acid) in 50% acetonitrile/0.1% trifluoroacetic acid/50% H2O, and the samples were spotted for MADLI-TOF MS analysis.
Growth Phenotype Assays
Cells were grown to mid-log phase (A600 0.5–1.0). Five μl of 5- or 10-fold serial dilutions of each culture were plated. For UV irradiation assays, YP plates were subjected to UV irradiation (Stratalinker UV Crosslinker, Stratagene). For 6-azauracil (6-AU) sensitivity assays, SC-uracil plates were supplemented with 30 μg/ml 6-AU. Plates were photographed after cells were grown for 2–5 days at 30 °C.
RESULTS
Global Changes of Histone H3 Modification in JmjC Knock-out Strains
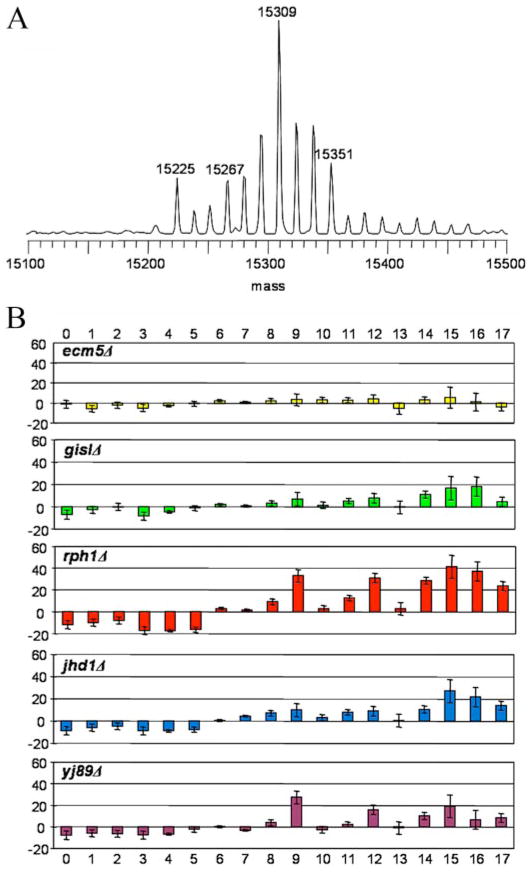
We first studied the overall modifications of histone H3 in WT and the deletion strains by LC-MS. Fig. 2A shows a typical MS spectrum of histone H3 from WT strain. The peak at 15,225 Da corresponds to the unmodified histone H3; the subsequent 17 modified species are each separated by an interval of 14 Da. Although this series of peaks could represent incremental methylation, a shift of 42 Da (3 × 14) can also arise via acetylation. Before the ambiguity between acetylation and methylation is clarified, “methylation equivalents” will be used to describe the mass differences between these peaks.
FIGURE 2. Global changes of histone H3 methylation in JmjC knock-out strains.
A, typical LC-MS spectrum of WT H3. The highest peak has six methylation equivalents. B, bar plots showing “relative percentage change” for each peak in knock-out strains relative to WT. For a given peak (i), the following equation was used to calculate the relative percentage change: 100 × [percentage (mutant, i) − percentage (WT, i)]/percentage (WT, i).
To address changes of histone H3 modification in the five mutants, the relative percentage change for each peak was analyzed. First, the percentage of each peak in a given spectrum was calculated to reflect the abundance of this species in the sample. Then this percentage was compared with its counterpart in the WT strain to generate the relative percentage change. As shown in Fig. 2B, a consistent shift in histone H3 modification pattern was observed. Relative to WT, except for ecm5Δ, each knock-out strain displayed a discernable and reproducible decrease in histone H3 species containing less than 6 methylation equivalents. In contrast, each knock-out strain showed an increase in histone H3 species with 6–17 methylation equivalents. These results suggest that highly modified histones accumulate in the knock-out strains.
When analyzed carefully, the pattern of changes shown in Fig. 2B provided clues about the substrate specificities of the JmjC proteins. The magnitude of changes followed the order rph1Δ > jhd1Δ ≥ jhd2Δ > gis1Δ. Furthermore, the shift in histone H3 modification patterns of the rph1Δ and jhd2Δ strains appeared to be similar; those of jhd1Δ and gis1Δ were also similar. These differences were interesting and potentially significant. Focusing on the data of 7–15 methylation equivalents, the rph1Δ and jhd2Δ strains showed significant accumulation of 9, 12, and 15 methylation equivalents, whereas the jhd1Δ and gis1Δ strains did not show such a pattern. If trimethylation is the predominant state of methylation (which is in fact the case, as described below), then a reasonable interpretation is that Rph1 and Jhd2 are trimethyllysine demethylases, whereas Gis1 and Jhd1 are likely mono- or dimethyllysine demethylases. This was our working hypothesis from this point forward.
To test the above hypothesis, the following experiments were performed: (i) tryptic digestion of histone H3 from WT and the five knock-out mutants; (ii) analysis of the digested samples using high-resolution LTQ-FTICR to identify fragments containing methylated lysine and differentiate the trimethylated fragments from the corresponding acetylated fragments; and (iii) analysis of the relative abundance of unmodified, mono-, di-, and trimethylated species of each fragment.
High-resolution MS Analysis of S. cerevisiae Histone Methylation Patterns
To decipher the chemical identity of the H3 modifications that we previously had described as “methylation equivalents,” high-resolution mass spectrometry of the digested peptides was used. Based on the fact that acetylated and trimethylated peptides (on the same lysine) have different masses (differing by 0.0364 Da), trimethylated peptides (H3-K4me3, H3-K36me3, and H3-K79me3) and acetylated peptides (H3-K9ac, H3-K14ac, H3-K18ac, H3-K23ac, H3-K27ac, and H3-K56ac) were assigned unambiguously. The data demonstrate that H3-K4, -K36, and -K79 are solely methylated. The results are consistent with yeast histone modification patterns (43).
Changes of Methylation Levels at Specific Sites in the Knock-out Strains
After unambiguous methylation assignments, quantitative analyses of histone H3 methylation were conducted to compare the abundance of each methylated fragment between WT and the five mutant strains by using both LC-MS/MS or MALDI-TOF MS. In all of the samples, H3-K4, -K36, and -K79 predominantly exist as trimethylated forms, with the simultaneous presence of various degrees of di-, mono-, and unmethylated forms.
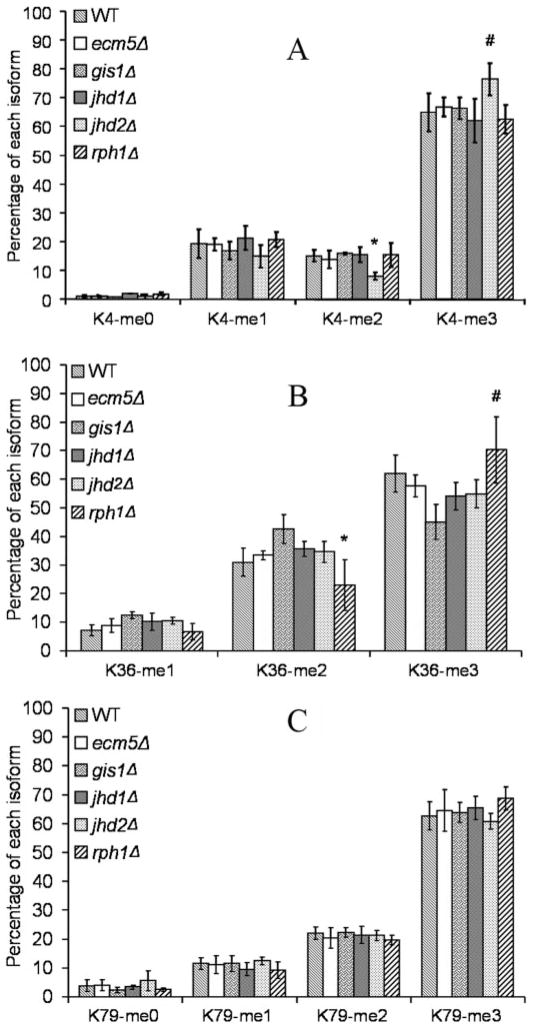
Quantitative results from the above analyses are summarized in Fig. 3. Statistical analysis was performed by using t tests. Although overall the changes were small, the major changes were reproducible and consistent. Detailed analyses of the five knock-out strains are presented as follows below.
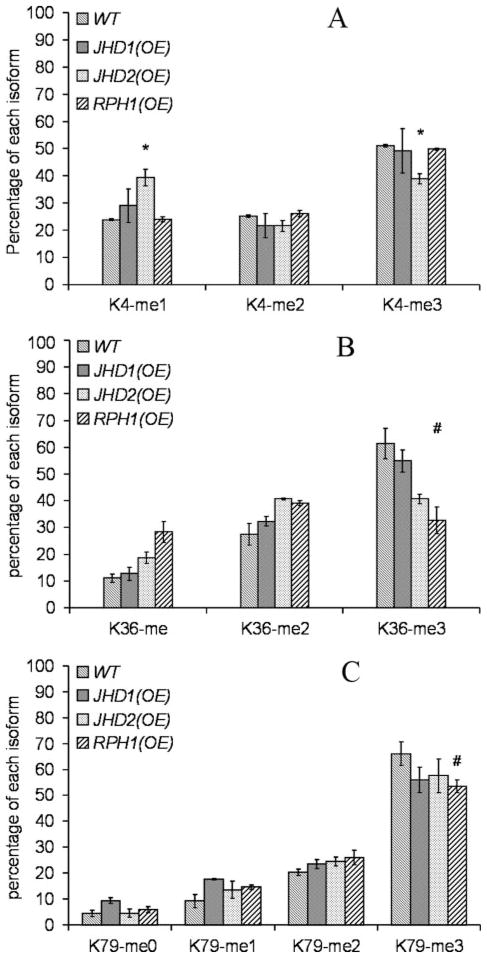
FIGURE 3. Site-specific quantification of histone H3 methylation in knock-out strains.
The relative levels of unmodified, mono-, di-, and trimethylated peptides are shown as percentages, with panels A, B, and C representing H3-K4, -K36, and -K79, respectively. *, p < 0.05; #, p < 0.1.
rph1Δ and jhd2Δ
The jhd2Δ strain showed an increased level of K4me3 and decreased levels of K4me2 and K4me. Similarly, rph1Δ strain showed a higher level of K36me3 and reduced levels of K36me2 and K36me. The data support the prediction from the global analysis that Rph1 and Jhd2 are histone trimethyllysine demethylases, further suggesting that they target histone H3-K36 and -K4, respectively.
gis1Δ and jhd1Δ
These strains showed modest changes in methylation levels at Lys-36 only (Fig. 3B). In contrast to the rph1Δ strain, the gis1Δ and jhd1Δ strains displayed decreased levels of K36me3 but increased levels of di- and monomethylated species. Therefore, we predict that Gis1 and Jhd1 are H3-K36me or H3-K36me2 demethylases.
ecm5Δ
In agreement with the results of the global analysis, the methylation patterns in the ecm5Δ strain did not display significant differences relative to the WT strain. We did not have enough experimental evidence to conclude whether Ecm5 is a histone demethylase. In summary, we predicted that four of the five yeast JmjC proteins are histone H3-K4 or H3-K36 demethylases.
Changes of Methylation in RPH1, JHD1, and JHD2 Overexpression Strains
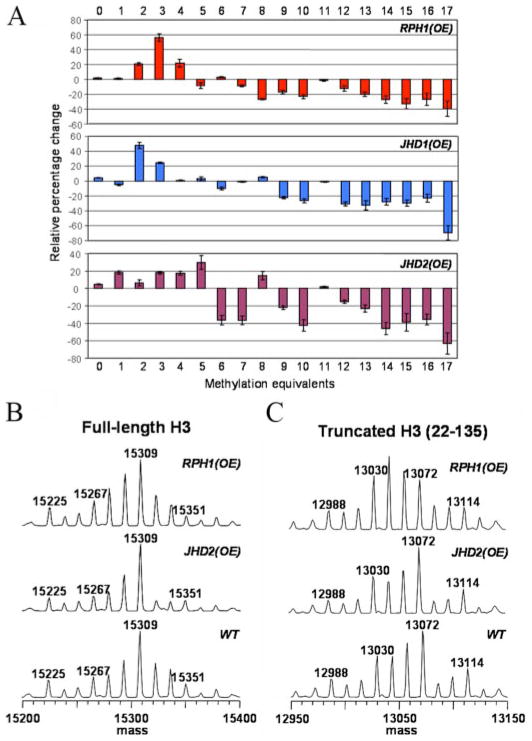
A potential argument with the knock-out strains is that the full impact on methylation levels could be masked because of redundancy or dominating effects of histone methyltransferases. If an effect that is complementary to that observed for a knock-out strain could be observed in the corresponding overexpression strain, this would establish a stronger link between a given enzyme and its proposed site of demethylation. We therefore examined the consequences of individually overexpressing RPH1, JHD1, and JHD2. JmjC protein overexpression was verified by Western blot (supplemental Fig. S1). As shown in Fig. 4A, global analysis of histone H3 from the overexpression strains demonstrated that the relative percentages of the lower methylated species were increased, whereas those of the higher methylated species were decreased. Compared with the patterns observed in the knock-out strains in Fig. 2B, the patterns in Fig. 4A were in the opposite direction and of higher magnitude, strongly suggesting that these JmjC proteins are directly involved in histone H3 methylation. Specifically, as Fig. 5A indicates, in the JHD2 overexpression strain, but not in the JHD1 or RPH1 overexpression strains, the trimethylated H3-K4 level was reduced, but monomethylated H3-K4 level was increased significantly. This effect was complementary to that observed in the jhd2Δ strain shown in Fig. 3A, which supports our previous assertion that Jhd2 catalyzes demethylation of H3-K4me3. Similarly, in Fig. 5B, histone H3-K36me3 level in RPH1 overexpression strain was reduced by half, which was consistent with our predication based on the knock-out data that Rph1 is an H3-K36me3 demethylase. We noticed that the histone H3-K36me3 level was also reduced modestly in the JHD2 overexpression strain in Fig. 5B. It is possible that overexpression of JHD2 mildly affects histone H3 methylation patterns by a mechanism yet to be established. In Fig. 5C, similar to Fig. 3C, the changes in histone H3-K79 methylation levels were very mild. Thus, no histone H3-K79 demethylases were predicted here.
FIGURE 4. MS analyses of overexpression strains.
A, bar plots showing “relative percentage change” of each peak in overexpression strains relative to WT. B, full-length histone H3 MS spectra for WT and the overexpression strains of RPH1 and JHD2. C, corresponding spectra from truncated H3 (residues 22–135).
FIGURE 5. Site-specific quantification of histone H3 methylation in overexpression strains.
Quantification of H3-K4, -K36, and -K79 peptides is shown in A, B, and C, respectively, *, p < 0.05; #, p < 0.1.
As reported previously, loss of the first 21 amino acids was observed in some histone H3 purification trials (10). Within the first 21 amino acids of S. cerevisiae histone H3, the only possible methylation site is Lys-4. In Fig. 4C, if we look at the peaks between 13,072 and 13,114 (heavily modified populations), truncated H3 (residues 22–135) showed similar methylation patterns and intensities in both the WT and JHD2 overexpression strains, indicating that: (i) JHD2 does not affect Lys-36 or Lys-79 methylation levels; and that (ii) the JHD2-induced difference in the region of 15,309–15,351 in Fig. 4B is solely the consequence of H3-K4 demethylation, which is consistent with the prediction from the jhd2 knock-out and overexpression results shown in Figs. 3A and 5A. Taken together, the data indicate that Jhd2 is an H3-K4me3 demethylase.
In Vitro Enzymatic Assays of Rph1
To confirm that it is the JmjC domain specifically that catalyzes histone demethylations, in vitro assays were performed with recombinant JmjC proteins. To obtain active enzymes, the full-length proteins (from E. coli and yeast) and truncated JmjC proteins were purified and tested. When full-length recombinant Rph1, Jhd2, and Jhd1 were overexpressed in E. coli, most of them had solubility problems, so no further assays were performed. When the full-length proteins were overexpressed and purified from yeast, only Jhd2 showed slight enzyme activities toward Lys-4 peptides. When the truncated recombinant Rph1, Gis1, Jhd2, and Ecm5 purified from E. coli were tested, only Rph1 showed robust enzyme activities. Here we report the results of Rph1 enzyme assays.
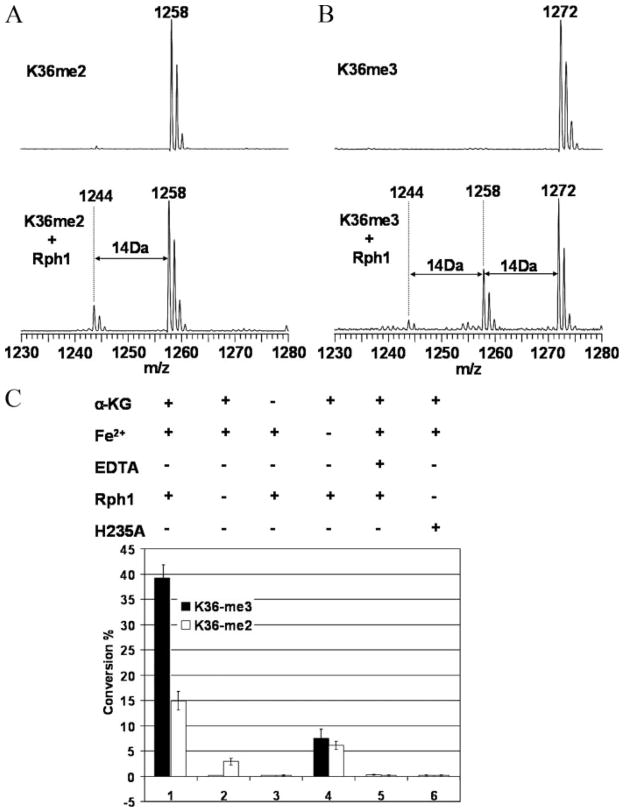
A truncated Rph1 construct, Rph1-(1–373), which contains the JmjC domain, was overexpressed as a GST fusion protein, purified, and then assayed for demethylase activity. Nine synthesized peptides containing mono-, di-, and trimethylated lysine at Lys-4, Lys-36, and Lys-79 were tested. As predicted, the K36me3 peptide was readily demethylated to K36me2, which was further converted to K36me but with lower efficiency (Fig. 6, A and B). No activity toward methylated Lys-4 or Lys-79 peptides was observed. Importantly, the activity of Rph1 toward K36me3 and K36me2 was ablated by omission of Fe2+, omission of α-ketoglutarate, addition of excess EDTA, or replacement of the WT enzyme with the H235A mutant (Fig. 6C). Although the dependence on α-ketoglutarate confirms this as a co-substrate, the residual activity observed in the absence of added Fe2+ could be explained by the presence of trace Fe2+ in the purified protein sample. That a metal cofactor is indeed essential for catalysis is suggested by the complete loss of activity in the presence of excess EDTA. The fact that mutation of residue 235, which based on sequence alignment (Fig. 1B) is predicted to bind Fe2+, inactivated Rph1 reinforced the conclusion that it is the JmjC domain of Rph1 that is responsible for catalysis. Collectively, the above data unequivocally demonstrate that Rph1 is an Fe2+/α-ketoglutarate-dependent histone lysine demethylase with specificity for H3-K36me3.
FIGURE 6. In vitro enzyme assays of GST-Rph1-(1–373).
A, MALDI-TOF MS data on demethylation of the K36me2 peptide. B, MS data on demethylation of the K36me3 peptide. C, control experiments. Solid and open columns represent the percentages of conversion from K36me3 and K36me2 peptides, respectively. Columns: 1, normal assays; 2, control assays without the enzyme; 3, control assays without α-ketoglutarate (α-KG); 4, control assays without Fe2+; 5, control assays with excess EDTA; 6, assay with Rph1 H235A mutant.
Overexpression of RPH1 Causes a Growth Defect upon UV Irradiation
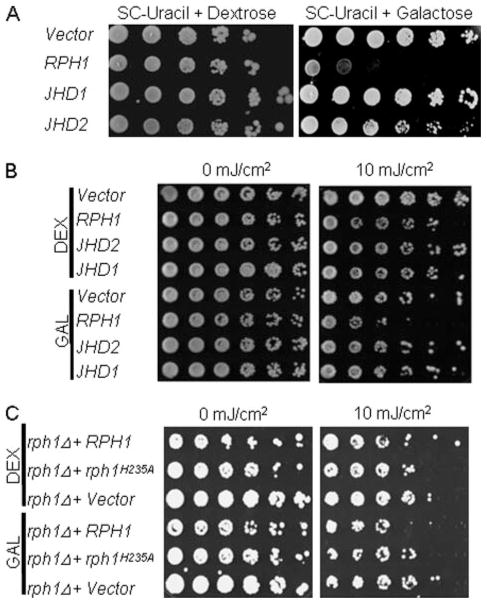
As a first step to elucidate the specific biological contexts in which the five S. cerevisiae JmjC proteins are requisite, phenotype screening was performed. Under normal conditions, no discernible growth phenotype was observed in any of the individual knock-out strain. However, when galactose was used as the carbon source in synthetic medium (SC-uracil), the growth rate of the RPH1 overexpression strain dropped significantly (Fig. 7A, right panel). By contrast, there was no difference among these strains in SC-uracil medium with dextrose (glucose) (Fig. 7A, left panel). Interestingly, in rich medium (YP-glucose or YP-galactose), the growth defect was not observed. The results suggest that overexpression of RPH1 results in a growth defect in synthetic media but not in rich media.
FIGURE 7. Growth phenotypes of yeast strains with overexpression of histone demethylase genes.
A, overexpression of RPH1 in synthetic medium SC-uracil has a growth defect. Cells grown in raffinose until A600 reached 1.0 were diluted 10-fold and spotted on SC-uracil plates with 2% dextrose (left panel) or galactose (right panel). B, hypersensitivity of the RPH1 overexpression strain to UV irradiation. Cells were grown in rich medium (YP) with 2% glucose (DEX) or galactose (GAL). Cell viability was assayed by plating serial dilutions of cells on YP-galactose plates followed by exposure to UV irradiation (10 mJ/cm2). C, histone demethylase activity of Rph1 was associated with cell survival to DNA damage. The rph1 deletion strain (rph1Δ) was transformed with plasmids containing no insert (Vector) or GAL1 promoter-driven wild type (RPH1) and activity-deficient mutant (rph1H235A), respectively. UV sensitivity assays were performed as described in B.
To avoid the growth defect observed in synthetic media, the following assays were performed on YP plates. As reported previously, Rph1 is a repressor of PHR1, which encodes for a photolyase in response to UV damage (44). To determine whether the histone demethylase activities are required in the UV damage responses, an UV sensitivity assay was performed. Deletion of individual JmjC genes had no discernible effect on the viability of cells in response to UV irradiation (supplemental Fig. S2A). However, overexpression of RPH1, but not JHD1 or JHD2, resulted in a growth defect in response to UV irradiation (Fig. 7B). The phenotype was observed in the range of 5–20 mJ/cm2 UV irradiation. To test whether the trimethylated H3-K36 demethylase activity is responsible for the phenotype, a demethylase activity-defective mutant, rph1H235A, was tested in the UV sensitivity assay. Under the same UV irradiation treatment, rph1H235A mutant abolished the phenotype (Fig. 7C). The results indicated that the demethylase activity is required for the RPH1-dependent phenotype. Furthermore, MS data (Fig. 5B) demonstrated that overexpression of RPH1, not JHD1, reduced H3-K36me3 level by 50%. Collectively, the data indicate that Rph1 is involved in cell survival in response to DNA damage, likely by changing the level of histone H3-K36me3.
RPH1 and JHD1 Overexpression Strains Are Slightly Resistant to 6-AU
The methylation status of H3-K36 is associated with transcriptional elongation. Previous studies have shown that deletion of Set2, the histone H3-K36 methyltransferase, results in a modest resistance to 6-AU treatment (45, 46). 6-AU, a commonly used drug for testing transcription elongation defects, inhibits nucleotide metabolism and leads to depletion of cellular GTP and UTP. Nucleotide depletion affects elongation efficiency (47). To test whether the JmjC proteins are involved in transcription elongation, we performed 6-AU sensitivity assays. Preliminary studies showed that overexpression of RPH1 or JHD1 resulted in slight resistance to 6-AU (supplemental Fig. S2B). The result was observed only in the strains overexpressing H3-K36 demethylase (Rph1 and Jhd1) but not in that overexpressing H3-K4 demethylase (Jhd2). The data are consistent with previous observations that deletion of the H3-K36 methyltransferase gene SET2 results in a modest resistance to 6-AU (45, 46). Our data support the idea that demethylation of H3-K36 by Rph1 and Jhd1 acts to decrease the methylation level of H3-K36 and to interfere with transcription.
DISCUSSION
JmjC Domain Encodes Histone Demethylase Activities
While our work was in progress, YER051W, newly named JHD1, was shown to encode a yeast H3-K36 demethylase (17); as far as we know, this is the only yeast JmjC-containing histone demethylase reported to date. It is not clear whether Jhd1 can demethylate H3-K36me3. Our in vivo MS data predict that Jhd1 is an H3-K36me2 or H3-K36me demethylase. Our data also suggest that Rph1, Jhd2, and Gis1 are demethylases. Among them, Jhd2 and Rph1 are trimethyllysine demethylases with specificity on H3-K4 and H3-K36, respectively. Gis1 is predicted to be similar to Jhd1, acting as an H3-K36me or H3-K36me2 demethylase. The specific demethylase activities of Rph1 toward histone H3-K36 have been verified by in vitro enzyme assays. It is the JmjC domain that is responsible for Rph1 demethylase activity. We also observed very weak but reproducible demethylase activities of Jhd2 and Gis1 with specificity on K4 and K36 peptides, respectively.6 In summary, four of the five yeast JmjC proteins (Jhd1, Rph1, Jhd2, and Gis1) are histone demethylases.
As for human enzymes, several groups have independently reported six JmjC proteins to be H3 demethylases. Although JHDM1 and JHDM2A demethylate mono- and dimethyl H3-K36 and -K9, respectively (17, 19), JMJD2 family histone demethylases (JMJD2A, JMJD2B, JMJD2C, and JMJD2D) demethylate trimethyl H3-K9/K36 (18, 20–22). It is interesting to note that each of these six JmjC proteins has a zinc finger, PHD, or Tudor domain. This matches one of our search criteria and implies a role for each of these proteins in transcription.
In summary, by using different approaches, we and other groups have reached the same conclusion that JmjC domain encodes histone demethylase activity. Our study is the first systematic MS proteomic report on all JmjC proteins in one single organism. Our in vivo MS analysis results matched in vitro enzyme assay results. In our knock-out MS data (Fig. 3), the changes were relatively small in comparison with those in the overexpression data (Fig. 5). Phenotype screening assays demonstrated that overexpression of RPH1 resulted in phenotypes that were not observed in the deletion strains. The differences suggest that deletion strains have a lesser impact on histone H3 methylation. Considering that all three sites (H3-K4, H3-K36, and H3-K36) are heavily methylated (more than 60%), we believe that in yeast, histone methyltransferases are dominant. Furthermore, localized effects of the demethylases may not contribute to changes in global MS data. Despite these effects, small but consistent changes were observed. The good agreement between in vitro activity assays and in vivo MS data for Rph1 provides solid support for the reliability of our approach. This approach could be applied in other organisms to identify histone demethylases.
Rph1 Is an H3-K36me3 Demethylase Involved in DNA Damage Response and Transcription Elongation
In the current study, in vivo MS data indicated that Rph1 is an H3-K36me3 demethylase, which was verified by in vitro assays. Its JmjC domain specifically catalyzes demethylation on H3-K36me3 and H3-K36me2, without any detectable activity toward H3-K4 and H3-K79 peptides. To the best of our knowledge, this would be the first reported yeast H3-K36me3 demethylase. Our phenotype screening experiments demonstrated that the RPH1 overexpression strain is sensitive to UV irradiation and the catalytic activity of Rph1 JmjC domain is responsible for the defect. The phenotype indicated that Rph1, an H3-K36me3 demethylase, is involved in DNA damage responses. Consistently, it has been reported that H3-K36 methyltransferase Set2 is involved in the DNA damage response (11). Furthermore, the UV irradiation defect caused by RPH1 overexpression strain was not observed in JHD1 over-expression strain. The difference can be explained by our prediction that Jhd1 is not an H3-K36me3 demethylase, whereas Rph1 is an H3-K36me3 demethylase. These results indicate that UV damage responses are correlated to H3-K36 methylation levels, particularly on the level of H3-K36me3.
As shown in Fig. 5B, the MS data of RPH1 overexpression strain showed a sharp decrease in H3-K36me3 level. The decrease in the global level suggests that overexpression of RPH1 has global effects. Because the link between H3-K36 methylation and transcription in yeast is well established and H3-K36 methyltransferase Set2 has been shown to interact with RNA polymerase II and play important roles in transcription elongation (45, 46, 48–50), H3-K36 demethylase Rph1 might affect transcription elongation. Indeed, when RPH1 was overexpressed, there was a slight 6-AU phenotype, suggesting its participation in transcription elongation. Furthermore, another H3-K36 demethylase, Jhd1, not the H3-K4 demethylase Jhd2, showed the same phenotype as Rph1. The H3-K36 demethylases Rph1 and Jhd1 might shift H3-K36 to less methylated species and result in lower transcription activity. This could be the reason that they are mildly resistant to 6-AU. The biochemical details need to be studied further.
Jhd2 Is an H3-K4 Demethylase
In vivo MS data suggested that Jhd2 is an H3-K4 demethylase. A weak H3-K4 demethylase activity was observed in in vitro assays. To the best of our knowledge, no JmjC domain containing H3-K4 demethylase has been reported previously. Jhd2 is a nuclear protein with unknown function. A recent large scale yeast protein-protein interaction study identified three Jhd2-interacting proteins, Sas10, Fun30, and YIL091C (51). Sas10 overexpression results in derepression of mating type genes at HML and HMR (52). Fun30 is a putative ATP-helicase and homologue of Snf2. Over-expression of Fun30 affects chromosome stability and integrity (53). Interestingly, the Snf2 family yeast protein Isw1, an ATP-dependent chromatin remodeling factor, has been shown to be recruited to active genes by H3-K4 methylation (54). These studies suggest that Jhd2 could be a component of a histone modification and chromatin remodeling complex related to silencing. Our current data have demonstrated that Jhd2 is a novel histone demethylase acting on H3-K4.
Methylation on H3-K4 has many interesting consequences. On the one hand, Lys-4 trimethylation occurs in the promoter region of genes, and it is involved in transcription activation (49). On the other hand, Lys-4 methylation also mediates rDNA and telomere silencing. The H3-K4 methyltransferase Set1 is required for silencing at rDNA region, telomeres, and HML mating type loci (5, 55). Therefore, Jhd2 might negatively regulate transcription, or it could repress silencing on the rDNA region, the telomeres, or the HML mating type loci.
Possible H3-K79 Demethylases
Of the three known lysine methylation sites in histone H3 (Lys-4, Lys-36, and Lys-79), only Lys-4 and Lys-36 thus far have been shown to be acted upon by demethylases. Because H3-K79 is highly (90%) methylated in yeast (8), small changes in the demethylase knock-out mutants could be masked by the relatively high histone methyltransferase activity. Therefore, it cannot be ruled out that Ecm5 or a non-JmjC protein may be an H3-K79 demethylase. Further investigation is needed to resolve the question.
Yeast JmjC Proteins in Transcription
Of the five JmjC proteins in S. cerevisiae, Jhd1, Jhd2, and Ecm5 each has a PHD domain, whereas Rph1 and Gis1 have zinc finger domains. These domains might be associated with transcription regulations related to histone demethylation. Rph1 and Gis1 have been reported to act as repressors (44). Rph1 binds to a promoter element, URSPHR1, likely via the zinc finger domains. We speculate that the PHD domains in Jhd1 and Jhd2 may bind to chromatin via methylated histone. Genome-wide location analysis by using chromatin immunoprecipitation and DNA-microarray (CHIP-chip) of the JmjC proteins will provide details regarding their roles in transcription.
Supplementary Material
Acknowledgments
We are grateful to L. J. Juan of the Genomics Research Center and to Brandon Lamarche, Chunhua Yuan, and Mark Parthun of Ohio State University (OSU) for helpful discussion, to Nan Kleinholz at the OSU Campus Chemical Instrument Center for assistance with LC-MS, and to Yu Wang and Xianwen Chen for help in protein purification. The vector BG1805 (without a gene inserted) was obtained from the laboratory of Michael Synder (Yale University).
Footnotes
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs S1 and S2.
The abbreviations used are: PHD, plant homeodomain; ARID, AT-rich interaction domain; 6-AU, 6-azauracil; LC, liquid chromatography; MS, mass spectrometry; MS/MS, tandem mass spectrometry; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; LTQ-FTICR, linear ion trap-Fourier transform ion cyclotron resonance; WT, wild type; GST, glutathione S-transferase; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine; YP, yeast-peptone; YPD, yeast-peptone-dextrose.
S. Tu, L. Yang, M. A. Freitas, and M.-D. Tsai, unpublished data.
This work was supported by funding from the Genomics Research Center of Academia Sinica (to M.-D. T.) and by National Institutes of Health Grants CA69472 (to M.-D. T.) and CA110496 (to M. A. F.).
References
- 1.Jenuwein T, Allis CD. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Lachner M, O’Sullivan RJ, Jenuwein T. J Cell Sci. 2003;116:2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 3.Margueron R, Trojer P, Reinberg D. Curr Opin Genet Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Martin C, Zhang Y. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 5.Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, Allis CD. Mol Cell Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Leeuwen F, Gafken PR, Gottschling DE. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 9.Sims RJ, III, Belotserkovskaya R, Reinberg D. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 10.Lacoste N, Utley RT, Hunter JM, Poirier GG, Cote J. J Biol Chem. 2002;277:30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- 11.Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ. Mol Cell Biol. 2005;25:8430–8443. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bannister AJ, Schneider R, Kouzarides T. Cell. 2002;109:801–806. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- 13.Morillon A, Karabetsou N, Nair A, Mellor J. Mol Cell. 2005;18:723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Lee MG, Wynder C, Cooch N, Shiekhattar R. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 16.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 17.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 18.Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 19.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 20.Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- 21.Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 22.Fodor BD, Kubicek S, Yonezawa M, O’Sullivan RJ, Sengupta R, Perez-Burgos L, Opravil S, Mechtler K, Schotta G, Jenuwein T. Genes Dev. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubicek S, Jenuwein T. Cell. 2004;119:903–906. doi: 10.1016/j.cell.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Trewick SC, McLaughlin PJ, Allshire RC. EMBO Rep. 2005;6:315–320. doi: 10.1038/sj.embor.7400379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 26.Falnes PO, Johansen RF, Seeberg E. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 27.Balciunas D, Ronne H. Trends Biochem Sci. 2000;25:274–276. doi: 10.1016/s0968-0004(00)01593-0. [DOI] [PubMed] [Google Scholar]
- 28.Clissold PM, Ponting CP. Trends Biochem Sci. 2001;26:7–9. doi: 10.1016/s0968-0004(00)01700-x. [DOI] [PubMed] [Google Scholar]
- 29.Ahmed S, Palermo C, Wan S, Walworth NC. Mol Cell Biol. 2004;24:3660–3669. doi: 10.1128/MCB.24.9.3660-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ayoub N, Noma K, Isaac S, Kahan T, Grewal SI, Cohen A. Mol Cell Biol. 2003;23:4356–4370. doi: 10.1128/MCB.23.12.4356-4370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fattaey AR, Helin K, Dembski MS, Dyson N, Harlow E, Vuocolo GA, Hanobik MG, Haskell KM, Oliff A, Defeo-Jones D, Jones RE. Oncogene. 1993;8:3149–3156. [PubMed] [Google Scholar]
- 32.Jung J, Kim TG, Lyons GE, Kim HR, Lee Y. J Biol Chem. 2005;280:30916–30923. doi: 10.1074/jbc.M414482200. [DOI] [PubMed] [Google Scholar]
- 33.Gray SG, Iglesias AH, Lizcano F, Villanueva R, Camelo S, Jingu H, Teh BT, Koibuchi N, Chin WW, Kokkotou E, Dangond F. J Biol Chem. 2005;280:28507–28518. doi: 10.1074/jbc.M413687200. [DOI] [PubMed] [Google Scholar]
- 34.Huyen Y, Zgheib O, DiTullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shahbazian MD, Zhang K, Grunstein M. Mol Cell. 2005;19:271–277. doi: 10.1016/j.molcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 38.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 39.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M’Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 40.Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, Gerstein M, Dumont ME, Phizicky EM, Snyder M, Grayhack EJ. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lo WS, Henry KW, Schwartz MF, Berger SL. Methods Enzymol. 2004;377:130–153. doi: 10.1016/S0076-6879(03)77007-4. [DOI] [PubMed] [Google Scholar]
- 42.Ren C, Zhang L, Freitas MA, Ghoshal K, Parthun MR, Jacob ST. J Am Soc Mass Spectrom. 2005;16:1641–1653. doi: 10.1016/j.jasms.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Millar CB, Grunstein M. Nat Rev Mol Cell Biol. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- 44.Jang YK, Wang L, Sancar GB. Mol Cell Biol. 1999;19:7630–7638. doi: 10.1128/mcb.19.11.7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li B, Howe L, Anderson S, Yates JR, III, Workman JL. J Biol Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- 46.Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. Mol Cell Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Archambault J, Lacroute F, Ruet A, Friesen JD. Mol Cell Biol. 1992;12:4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Moazed D, Gygi SP. J Biol Chem. 2002;277:49383–49388. doi: 10.1074/jbc.M209294200. [DOI] [PubMed] [Google Scholar]
- 49.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 50.Hampsey M, Reinberg D. Cell. 2003;113:429–432. doi: 10.1016/s0092-8674(03)00360-x. [DOI] [PubMed] [Google Scholar]
- 51.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MH, Butland G, Altaf-Ul AM, Kanaya S, Shilatifard A, O’Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 52.Kamakaka RT, Rine J. Genetics. 1998;149:903–914. doi: 10.1093/genetics/149.2.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ouspenski II, Elledge SJ, Brinkley BR. Nucleic Acids Res. 1999;27:3001–3008. doi: 10.1093/nar/27.15.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santos-Rosa H, Schneider R, Bernstein BE, Karabetsou N, Morillon A, Weise C, Schreiber SL, Mellor J, Kouzarides T. Mol Cell. 2003;12:1325–1332. doi: 10.1016/s1097-2765(03)00438-6. [DOI] [PubMed] [Google Scholar]
- 55.Fingerman IM, Wu CL, Wilson BD, Briggs SD. J Biol Chem. 2005;280:28761–28765. doi: 10.1074/jbc.C500097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.