Abstract
Histidyl-transfer ribonucleic acid (tRNA) synthetase (HRS), coded by the hisS gene, appears to play two roles in regulation of the histidine operon of Salmonella typhimurium: (i) in synthesis of a critical effector molecule, histidyl-tRNA, and (ii) a more direct effect elicited by the presence of the enzyme protein itself. The specific activity of HRS was elevated either by mutations in the strB locus or in hisS+ merodiploids of Escherichia coli/S. typhimurium and S. abony/S. typhimurium. In each case, an increase in HRS was accompanied by an increase in histidine operon expression, indicating that HRS may be involved in positive control of the histidine operon. It is unlikely that HRS leads to increased histidine operon expression merely by acting as a “sponge” for charged tRNA. Rather, HRS appears to influence operon expression by interaction with some effector molecule other than charged tRNA or by a direct interaction with the histidine operator-promoter region. The functional level of histidine operon expression has no effect on HRS specific activity.
Full text
PDF


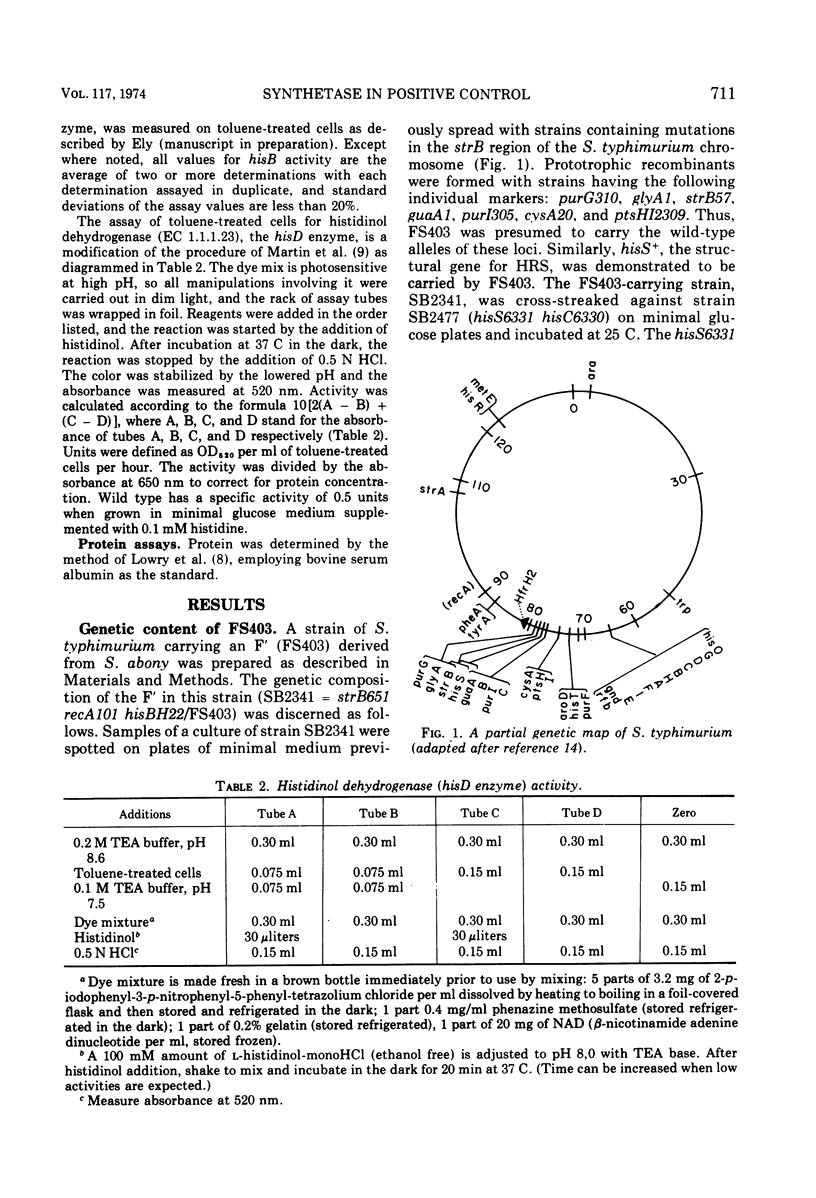

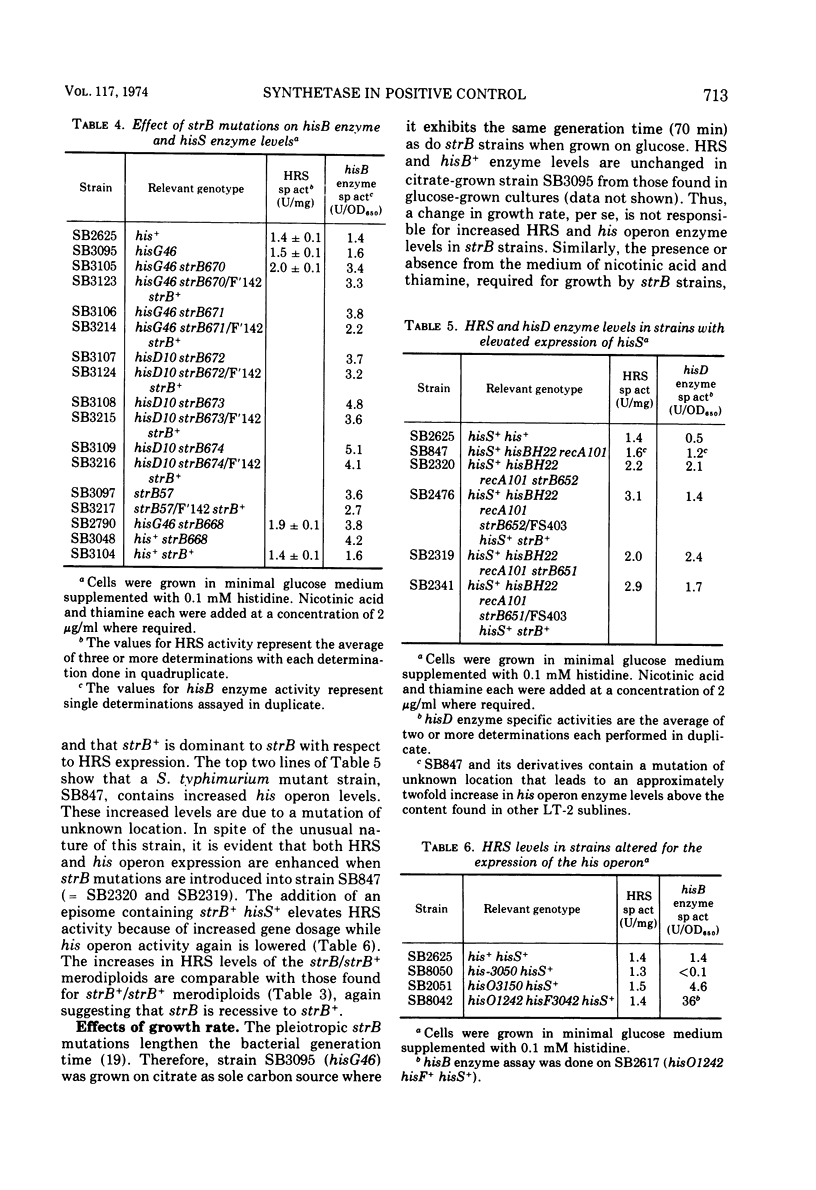



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- AMES B. N., HARTMAN P. E., JACOB F. Chromosomal alterations affecting the regulation of histidine biosynthetic enzymes in Salmonella. J Mol Biol. 1963 Jul;7:23–42. doi: 10.1016/s0022-2836(63)80016-9. [DOI] [PubMed] [Google Scholar]
- Cordaro J. C., Roseman S. Deletion mapping of the genes coding for HPr and enzyme I of the phosphoenolpyruvate: sugar phosphotransferase system in Salmonella typhimurium. J Bacteriol. 1972 Oct;112(1):17–29. doi: 10.1128/jb.112.1.17-29.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lorenzo F., Ames B. N. Histidine regulation in Salmonella typhimurium. VII. Purification and general properties of the histidyl transfer ribonucleic acid synthetase. J Biol Chem. 1970 Apr 10;245(7):1710–1716. [PubMed] [Google Scholar]
- De Lorenzo F., Straus D. S., Ames B. N. Histidine regulation in Salmonella typhimurium. X. Kinetic studies of mutant histidyl transfer ribonucleic acid synthetases. J Biol Chem. 1972 Apr 25;247(8):2302–2307. [PubMed] [Google Scholar]
- Ito K. Regulatory mechanism of the tryptophan operon in Escherichia coli: possible interaction between trpR and trpS gene products. Mol Gen Genet. 1972;115(4):349–363. doi: 10.1007/BF00333173. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lewis J. A., Ames B. N. Histidine regulation in Salmonella typhimurium. XI. The percentage of transfer RNA His charged in vivo and its relation to the repression of the histidine operon. J Mol Biol. 1972 Apr 28;66(1):131–142. doi: 10.1016/s0022-2836(72)80011-1. [DOI] [PubMed] [Google Scholar]
- McGinnis E., Williams L. S. Regulation of histidyl-transfer ribonucleic acid synthetase formation in a histidyl-transfer ribonucleic acid synthetase mutant of Salmonella typhimurium. J Bacteriol. 1972 Sep;111(3):739–744. doi: 10.1128/jb.111.3.739-744.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis E., Williams L. S. Role of histidine transfer ribonucleic acid in regulation of synthesis of histidyl-transfer ribonucleic acid synthetase of Salmonella typhimurium. J Bacteriol. 1972 Feb;109(2):505–511. doi: 10.1128/jb.109.2.505-511.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J. R., Ames B. N. Histidine regulatory mutants in Salmonella typhimurium II. Histidine regulatory mutants having altered histidyl-tRNA synthetase. J Mol Biol. 1966 Dec 28;22(2):325–333. doi: 10.1016/0022-2836(66)90135-5. [DOI] [PubMed] [Google Scholar]
- Roth J. R., Antón D. N., Hartman P. E. Histidine regulatory mutants in Salmonella typhimurium. I. Isolation and general properties. J Mol Biol. 1966 Dec 28;22(2):305–323. doi: 10.1016/0022-2836(66)90134-3. [DOI] [PubMed] [Google Scholar]
- Sanderson K. E. Linkage map of Salmonella typhimurium, edition IV. Bacteriol Rev. 1972 Dec;36(4):558–586. doi: 10.1128/br.36.4.558-586.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E., Ross H., Ziegler L., Mäkelä P. H. F + , Hfr, and F' strains of Salmonella typhimurium and Salmonella abony. Bacteriol Rev. 1972 Dec;36(4):608–637. doi: 10.1128/br.36.4.608-637.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Levine M. A phage P22 gene controlling integration of prophage. Virology. 1967 Feb;31(2):207–216. doi: 10.1016/0042-6822(67)90164-x. [DOI] [PubMed] [Google Scholar]
- Straus D. S., Ames B. N. Histidyl-transfer ribonucleic acid synthetase mutants requiring a high internal pool of histidine for growth. J Bacteriol. 1973 Jul;115(1):188–197. doi: 10.1128/jb.115.1.188-197.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L. S., Neidhardt F. C. Synthesis and inactivation of aminoacyl-transfer RNA synthetases during growth of Escherichia coli. J Mol Biol. 1969 Aug 14;43(3):529–550. doi: 10.1016/0022-2836(69)90357-x. [DOI] [PubMed] [Google Scholar]
- Yamada T., Davies J. A genetic and biochemical study of streptomycin- and spectinomycin-resistance in Salmonella typhimurium. Mol Gen Genet. 1971;110(3):197–210. doi: 10.1007/BF00337833. [DOI] [PubMed] [Google Scholar]
- Yamamoto N., Fukuda S., Takebe H. Effect of a potent carcinogen, 4-nitroquinoline 1-oxide, and its reduced form, 4-hydroxylaminoquinoline 1-oxide, on bacterial and bacteriophage genomes. Cancer Res. 1970 Oct;30(10):2532–2537. [PubMed] [Google Scholar]


