Abstract
EBV-encoded microRNAs (miRNAs) have been identified and their functions are being studied. The expression pattern of these miRNAs in clinical samples of EBV-associated non–Hodgkin’s lymphomas is unknown. We analyzed five primary “endemic” pediatric Burkitt’s lymphomas (BL), two acquired immunodeficiency syndrome (AIDS)-related type I latency BL lines, a type III latency line, three EBV+ primary effusion lymphomas (PEL), and three AIDS-related diffuse large B-cell lymphomas (DLBCL) for expression of EBV-encoded miRNAs. A markedly elevated expression of miRNA BHRF1-3 in type III relative to its parental type I BL line was found. Primary unmanipulated type I BLs and EBV+ PELs expressed high levels of BART2 miRNA, whereas DLBCLs expressed both BART2 and BHRF1-3 species. BHRF1-3 miRNA expression inversely correlated with levels of a putative cellular target, the IFN-inducible T-cell attracting chemokine CXCL-11/I-TAC, and suppression of this factor was reversed by transfection of an antisense oligo to the EBV miRNA BHRF1-3. EBV-encoded miRNAs are expressed in primary lymphomas classically linked to the virus and are associated with the viral latency status. Targeted suppression of CXCL-11/I-TAC by a viral-encoded miRNA may serve as an immunomodulatory mechanism in these tumors.
Introduction
MicroRNAs (miRNA) are small noncoding RNAs of ~22 nucleotides processed from primary transcripts (pri-miRNAs) to short stem-loop RNA precursors of ~60 nucleotides (pre-miRNAs) and finally to functional miRNAs. miRNAs regulate gene expression by base pairing with the 3′-untranslated region (UTR) of their target mRNAs and repress target gene translation or induce target mRNA degradation. miRNAs have important regulatory functions as indicated by their gene specificity and modulation (1–3). The absence of miRNAs in all unicellular organisms studied thus far makes a case for their role in highly specialized cell functions (4, 5).
A growing body of data indicates that viruses exploit the miRNA machinery to modulate and/or subvert virus-host cell interactions (6–10). Pfeffer et al. (7) initially identified five EBV-encoded miRNAs from the B95-8 EBV laboratory strain. The EBV-encoded BHRF1 miRNAs cluster map to the 5′-UTR (BHRF1-1) and 3′-UTR (BHRF1-2 and BHRF1-3) of the BHRF1 (BamHI fragment H rightward open reading frame 1) gene, whereas the BART miRNAs (BART1 and BART2) cluster map to intronic regions of the BART (BamHI-A region rightward transcript) gene. Cai et al. (8) identified additional BART miRNAs from an EBV-infected primary effusion lymphoma (PEL) cell line, and Grundhoff et al. (10) identified additional EBV miRNAs in the BART and BHRF1 region, bringing the total to 22 clustered, EBV-encoded miRNAs. The conservation of miRNAs in the lymphocryptoviruses indicates their importance in the viral life cycle, and the elucidation of their function is ongoing albeit incompletely understood.
The typical outcome of EBV infection is viral latency, which may eventually result in transformation (11, 12). Latency patterns have been defined based on EBV gene expression in distinct tumor subtypes. Primary Burkitt’s lymphomas (BL) generally show the type I pattern of latency, whereas posttransplant lymphoproliferative disease, immunoblastic lymphomas [including some acquired immunodeficiency syndrome (AIDS)-related lymphomas], as well as in vitro–transformed lymphoblastoid cell lines (LCL) adopt the latency type III pattern of gene expression (12). Latency variation is consistent with the hypothesis that the more restricted the viral gene expression, the easier it is for the virally infected cell to escape immune surveillance. In the absence of immunologic constraints, such as in vitro, primary BL cell cultures tend to drift from the classic type I latency to a type III lymphoblastoid phenotype (11, 12). Prior studies have focused exclusively on long-term culture propagated cell lines. These can accumulate mutations in viral miRNA loci, such as exemplified by B95-8 (7), or the host genome, which may alter miRNA transcription, processing, or stability. Here, we extend these studies by analyzing viral miRNA levels by two independent methods, RNase protection assay (RPA) and a novel quantitative PCR (QPCR) method, in early-passage primary BL cultures, a lymphoblastoid derivative of BL, and a wide variety of unmanipulated primary EBV-related lymphomas. We show that EBV-encoded BHRF1-3 miRNA levels are markedly elevated in the type III latency cell line and for the first time in primary EBV-associated AIDS-related diffuse large B-cell lymphomas (DLBCL). Primary BLs and PELs expressed high levels of BART2 but not BHRF1-3 miRNA. We further show that BHRF1-3 miRNA represses the expression of the IFN-inducible T-cell attracting chemokine, CXCL-11/I-TAC, a putative cellular target of BHRF1-3 miRNA. This targeted suppression of CXCL-11/I-TAC by BHRF1-3 miRNA may serve as an immunomodulatory mechanism in EBV-related lymphomas.
Materials and Methods
Cell culture, collection, and histologic confirmation of primary tumors
This study was approved by the University of Miami and the Federal University of Bahia Institutional Review Boards. Informed consent was obtained from patients or their guardians before obtaining clinical samples, and only tissue deemed unnecessary for pathologic diagnosis was used. BL-5 and BL-8 cells are primary cultures derived from patients with AIDS-related EBV+ BL. The tumors carry the typical t8:14 c-MYC translocation (13). A lymphoblastoid derivative (BL-5R) was obtained after serially passaging BL-5 cells over a 4- to 5-week period. BC-1 and JSC-1 are PEL lines that are dually infected with human herpes virus type 8 (HHV-8) and EBV (14). Cells were cultured in RPMI 1640 (Life Technologies) supplemented with 10% fetal bovine serum, 1% penicillin and streptomycin, 1% glutamine, and 1 mmol/L sodium pyruvate at 37°C in a 5% CO2-humidified atmosphere. The PEL cell lines were maintained in the same medium with the addition of 1 unit/mL human interleukin-6.
Primary EBV+ lymphomas were obtained from an area where an extremely high (~90%) association between EBV and pediatric Burkitt’s type histology has been reported (15, 16). Pediatric BLs (BL-1, BL-2, BL-3, BL-4, and BL-6) and EBV+ DLBCLs from AIDS patients (DLBCL-1, DLBCL-2, and DLBCL-3) were obtained from the residual tumor of patient biopsies. A primary PEL specimen (PELmo1) coinfected with EBV and HHV-8 was obtained from residual material from a diagnostic procedure. The EBV− primary DLBCL (DLBCL-4), the BJAB line, and the EBV− PEL lines BCBL-1 and BC-3 served as controls. Classification of lymphomas was made by an experienced hematopathologist (I.A.) and association with EBV was determined by eber staining. RNA was prepared as described below from tumors before any culture step or other manipulation.
Total RNA and miRNA extraction
RNA was extracted with the RNeasy kit (Qiagen, Inc.) for analyses of cellular and EBV RNAs by reverse transcription-PCR (RT-PCR). For RNase protection analysis of miRNAs, total RNA and miRNA-enriched fractions were prepared with the mirVana miRNA extraction kit (Ambion, Inc.). For real-time QPCR analysis, total RNA was isolated by RNAzol (Sigma, Inc.) and residual DNA was removed using Turbo DNA-free (Ambion). RNA was quantitated by UV absorption at 260 nm/280 nm using an Ultrospec 1000 machine (Pharmacia Biotech, Inc.).
PCR analyses
cDNA was synthesized using Moloney murine leukemia virus reverse transcriptase (Promega, Inc.) according to the manufacturer’s instructions. cDNA (2 μL) was PCR amplified with gene-specific primers as previously described (17). Real-time QPCR primers for BART1, BART2, and BHRF1-3 are shown in Supplementary Table S1. Reverse transcription, real-time QPCR, and data analysis were conducted following our established procedures (17, 18). The final PCR contained 7.5 μL primer mix (final concentration, 166 nmol/L), 15 μL of 2× SYBR PCR mix (Applied Biosystems), and a 7.5 μL sample. To guard against contamination and handling errors, all real-time QPCRs were assembled in a segregated clean room using a CAS-1200 robot (Corbett Research, Inc.) with 0.1 μL accuracy, liquid level sensing, and filtered pipette tips. Real-time PCR was performed using an Opticon 2 unit (MJ Research, Inc.) and universal cycle conditions.
RNase protection assay
RPAs were performed using the mirVana miRNA detection kit in accordance with the manufacturer’s instructions (Ambion). 32P-labeled RNA probes were generated from DNA templates corresponding to the miRNA target sequence using the mirVana miRNA probe construction kit (Ambion) according to the manufacturer’s instructions.
BHRF1-3 miRNA or anti-miRNA transfection
Chemically modified oligonucleotides corresponding to the antisense to the BHRF1-3 miRNA target sequence (BHRF1-3 anti-miR), the BHRF1-3 pre-miR miRNA precursor, scrambled sequence, and a negative control anti-miR inhibitor were obtained from Ambion. Oligonucleotides were delivered using the Amaxa nucleofection technology (Amaxa) according to the manufacturer’s protocols.
ELISA
CXCL-11/I-TAC protein levels in cell culture conditioned medium were determined with the Quantikine human CXCL-11/I-TAC ELISA kit (R&D Systems, Inc.) as per the manufacturer’s instructions.
Results
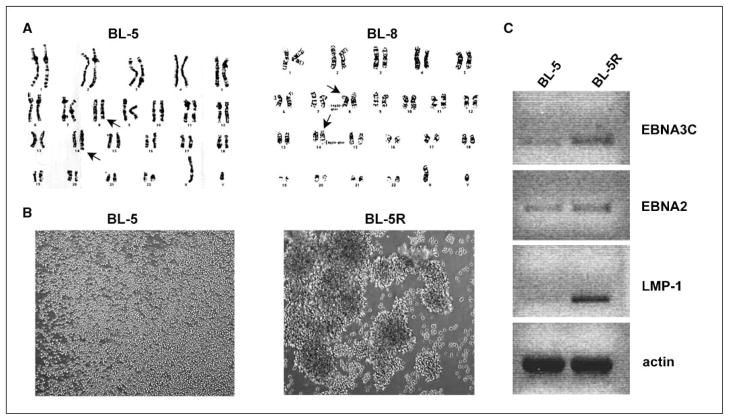
To determine the expression pattern of EBV miRNAs in type I and III EBV-related lymphomas, and to investigate their role in EBV pathogenesis, we initially used two BL early-passage cell lines (BL-5 and BL-8) derived from primary tumors that carry the typical t8:14 chromosomal translocation (Fig. 1A; ref. 13). Serially passaged primary cultures evolved from single-cell suspension (BL-5) to macroscopic clumps (BL-5R), indicating a drift to lymphoblastoid phenotype (Fig. 1B). The expression of viral-encoding proteins, EBNA3C, EBNA2, and LMP-1, is associated with the lymphoblastoid phenotype (11, 12). We therefore assessed the transcription of these mRNAs in BL-5 and its lymphoblastoid derivative BL-5R by RT-PCR. Although cultured BL-5 cells contained some detectable EBNA2 and EBNA3C transcripts, the BL-5R cells, by comparison, expressed higher levels of EBNA2 and EBNA3C (Fig. 1C). Consistent with its role in the lymphoblastoid phenotype, LMP-1 was exclusively expressed in BL-5R cells (Fig. 1C) and LMP-1 protein expression was confirmed by Western blot (data not shown).
Figure 1.
Cytogenetic and morphologic analysis of BL lines and differential expression of EBV genes in latency type I and III BL lines. A, cytogenetic analysis of BL lines BL-5 and BL-8 reveals the characteristic t8:14 c-MYC translocation. Arrows point to translocated chromosomes. B, bright-field photomicrograph of BL-5 and BL-5R cells in culture. Magnification, ×100. BL-5 cells grow as a single-cell suspension, whereas BL-5R cells grow as macroscopic clumps, characteristic of LCLs. C, RT-PCR analysis of EBV-encoded EBNA3C, EBNA2, and LMP-1 mRNA levels in BL-5 and BL-5R cells. β-Actin mRNA level was analyzed as loading control. Agarose gels were exposed under UV light in a Bio-Rad Gel Doc 2000 imager and inverted images of DNA bands captured.
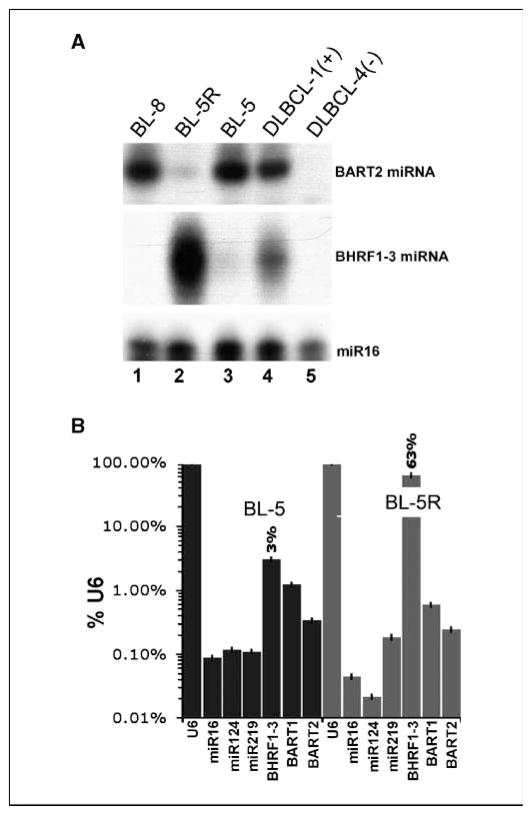
Small RNAs (<200 nucleotides) were purified from BL-5, BL-8, and BL-5R cells and EBV miRNA levels were determined by RPA. We observed elevated levels of BART2 miRNA in the parental type I BL-5 cells and a marked expression of BHRF1-3 miRNA (and concomitantly low levels of BART2 miRNA) in BL-5R cells (Fig. 2A, lanes 2 and 3). The early-passage type I BL-8 cells also expressed high levels of BART2 miRNA, but BHRF1-3 miRNA was not detectable in these cells (Fig. 2A, lane 1). This finding was in agreement with recently published data on long-term established EBV+ cell lines that showed an increased level of BHRF1 miRNAs in type III latency cell lines (8).
Figure 2.
Differential expression of BHRF1-3 and BART2 miRNAs in BL cell lines. A, RPA analysis of BHRF1-3 and BART2 miRNA expression in BL lines BL-8 and BL-5, a lymphoblastoid derivative BL-5R, an EBV+ DLBCL-1(+), and an EBV− DLBCL-4(−). Expression of a cellular miRNA, miR16, was analyzed as loading control. B, real-time QPCR analysis of EBV miRNA expression in BL-5 and BL-5R cells. Bars, relative level of miRNAs in percentage to U6 RNA on the vertical axis on a logarithmic scale. Expression of EBV miRNAs (BHRF1-3, BART1, and BART2) and expression of cellular miRNAs (miR16, miR124, and miR219) were analyzed.
We also analyzed viral miRNAs from an EBV+ primary DLBCL-1 biopsy. RPA analysis revealed high levels of both BART2 and BHRF1-3 miRNAs in this tumor (Fig. 2A, lane 4), but neither miRNA was detectable in its EBV− DLBCL counterpart (DLBCL-4; Fig. 2A, lane 5).
To verify the differential expression of the BHRF1-3 and BART2 miRNAs by another method, we used real-time QPCR. Primers specific for the BHRF1-3, BART1, and BART2 pre-miRNAs were designed. As control for RNA isolation, we used published primers against pre-miRNAs for cellular miRNAs, miR16, miR124, and miR219 (19, 20). All quantitative analyses were normalized to account for differences in cell number, RNA quality, and reverse transcription efficiency (17, 18). We used U6 as the common denominator for these studies because it was the most abundant RNA, normally distributed and the least variable RNA in our target set (data not shown). Control experiments were done to discount mRNA or DNA contamination of miRNA QPCR amplification (data not shown). The miRNA QPCR confirmed the marked induction (~21-fold) of BHRF1-3 pre-miRNA in BL-5R cells relative to BL-5 cells. BART pre-miRNAs did not show marked differences between BL-5 and BL-5R (<2-fold) compared with BHRF1-3 pre-miRNA in the QPCR assay (Fig. 2B). In contrast to the viral pre-miRNAs, none of the three cellular pre-miRNAs showed a pattern that correlated with EBV status, indicating that specific viral miRNA regulation rather than global defects in miRNA processing accounts for the pattern of viral miRNA levels. We also analyzed several well-characterized PEL cell lines, wherein EBV assumes a tight latency I phenotype based on latent EBV mRNA splicing patterns (18). BART1, BART2, and, to a lesser degree, BHRF1-3 pre-miRNAs were detectable in the EBV+ PEL cell lines BC-1 and JSC-1, but not in the EBV− BCBL-1 and BC-3 cell lines, and the expression levels were comparable with the latency I BL-5 cells (data not shown).
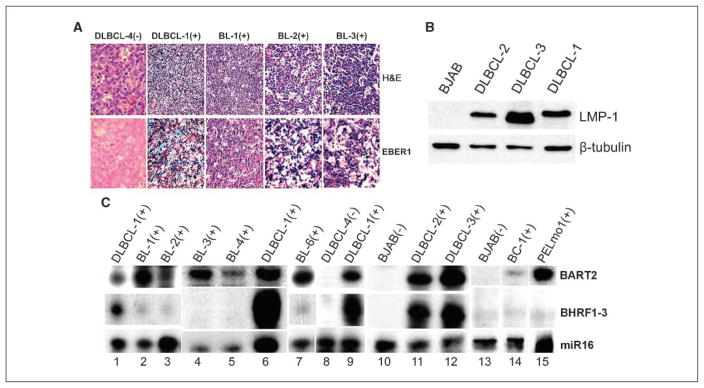
As all prior studies were conducted entirely in long-term culture adopted cell lines, we assessed the expression of EBV miRNAs in primary B-cell lymphomas classically associated with EBV. Endemic-type pediatric BLs from an area of high incidence and DLBCLs from HIV+ patients were confirmed to be EBV+ by in situ analysis of constitutively expressed EBV-encoded small RNAs (EBER; Fig. 3A) and by RT-PCR of EBV-encoded mRNAs (data not shown). As a control, an EBV (EBER)-negative DLBCL (DLBCL-4) was used in these analyses (Fig. 3A). Each of the EBV+ DLBCLs expressed high levels of LMP-1 (Fig. 3B), confirming that they are associated with type III EBV latency pattern. We also analyzed a primary EBV+ PEL specimen (PELmo1) as well as the established EBV+ PEL line (BC-1). RPA analysis showed that, as was observed in the EBV-associated BL lines (BL-5 and BL-8), endemic-type BLs (BL-1, BL-2, BL-3, BL-4, and BL-6) also expressed predominantly BART2 miRNA (Fig. 3C, lanes 2, 3, 4, 5, and 7), whereas the EBV+ DLBCLs (DLBCL-1, DLBCL-2, and DLBCL-3) expressed both BART2 and BHRF1-3 miRNAs at high levels (Fig. 3C, lanes 1, 6, 9, 11, and 12). The established PEL line and the primary PEL specimen both expressed predominantly BART2 miRNAs (Fig. 3C, lanes 14 and 15), consistent with the reported type I EBV latency transcription pattern in PEL. In EBV− controls (DLBCL-4 and BJAB), expression of both viral miRNAs was not detected (Fig. 3C, lanes 8, 10, and 13). Therefore, as in the type I latency cell line (e.g., BL-5), both EBV-associated primary BLs and PELs express high levels of BART2 miRNA, whereas the expression of BHRF1-3 miRNA is markedly elevated in the type III latency cell line (BL-5R) as well as in primary EBV+ AIDS-related DLBCLs. The types of primary lymphomas and cell lines analyzed and their expression pattern of EBV miRNA are summarized in Table 1.
Figure 3.
Differential expression of BHRF1-3 and BART2 miRNAs in primary unmanipulated clinical specimens. A, representative immunohistochemical analysis in primary unmanipulated clinical specimens. Endemic BLs (BL-1, BL-2, and BL-3) and primary DLBCLs (DLBCL-1 and DLBCL-4) were stained with H&E (top row) or stained for EBER1 (bottom row). Positive staining to EBV/EBER is visualized as dark blue spots. (−), EBV negative; (+), EBV positive. B, Western blot of LMP-1 protein in primary DLBCLs. EBV+ DLBCL-1, DLBCL-2, and DLBCL-3 were probed with specific antibody to LMP-1. An EBV− BL line, BJAB, was used as a negative control, and the same membrane was also probed with β-tubulin for a loading control. C, RPA analysis of BHRF1-3 and BART2 miRNA expression in primary EBV+ BLs (BL-1, BL-2, BL-3, BL-4, and BL-6), EBV+ primary DLBCLs (DLBCL-1, DLBCL-2, and DLBCL-3), a primary EBV+ PEL specimen (PELmo1), and an established EBV+ PEL line (BC-1). EBV− DLBCL-4 and BJAB were also analyzed as negative controls, and miR16 as a loading control.
Table 1.
EBV miRNA expression in primary lymphomas and cell lines
| BART2 | BHRF1-3 | ||
|---|---|---|---|
| EBV+ | |||
| Early-passage BL lines | BL-5 | High | Low |
| BL-8 | High | Low | |
| Primary pediatric BLs | BL-1 | High | Low |
| BL-2 | High | Low | |
| BL-3 | High | Low | |
| BL-4 | High | Low | |
| BL-6 | High | Low | |
| Type III derivative line | BL-5R | Low | High |
| Primary AIDS-related DLBCLs | DLBCL-1 | High | High |
| DLBCL-2 | High | High | |
| DLBCL-3 | High | High | |
| PELs | BC-1 | High | Low |
| JSC-1 | High | Low | |
| PELmo1 (primary) | High | Low | |
| EBV− | |||
| BL line | BJAB | None | None |
| Primary AIDS-related DLBCL | DLBCL-4 | None | None |
| PELs | BCBL-1 | None | None |
| BC-3 | None | None | |
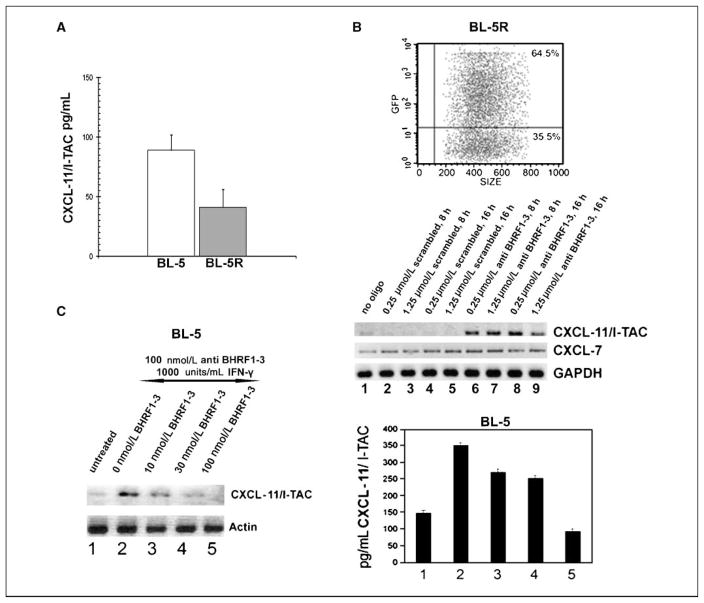
The IFN-inducible T-cell attracting chemokine CXCL-11/I-TAC is one of three CC chemokines, including CXCL-9/Mig and CXCL-10/IP-10, that bind to and activate the chemokine receptor CXCR3, which is predominantly expressed on T cells (21, 22). Although these cytokines are functionally redundant, CXCL-11/I-TAC is by several orders of magnitude the most potent T-cell chemo-attractant (21). Based on published in silico analysis, the 3′-UTR of CXCL-11/I-TAC contains a sequence that is 100% complimentary to the BHRF1-3 miRNA sequence and therefore constitutes a putative BHRF1-3 miRNA target (7). Because of significantly differing BHRF1-3 miRNA levels, we hypothesized that CXCL-11/I-TAC might also be differentially expressed in BL-5 and BL-5R. The basal level of CXCL-11/I-TAC proteins secreted from culture supernatants of BL-5 and BL-5R was analyzed using ELISA. BL-5 cells expressed higher levels of CXCL-11/I-TAC than BL-5R cells (Fig. 4A), suggesting that the expression of CXCL-11/I-TAC in these cells inversely correlates with the expression of EBV-encoded BHRF1-3 miRNA.
Figure 4.
Differential expression and modulation of putative BHRF1-3 miRNA target CXCL-11/I-TAC. A, ELISA analysis of basal level expression of CXCL-11/I-TAC proteins secreted from culture supernatants of BL-5 and BL-5R. B, RT-PCR analysis of CXCL-11/I-TAC mRNA levels in BL-5R cells following BHRF1-3 miRNA antisense transfection. Top, green fluorescent protein (GFP) control to determine transfection efficiency; bottom, mRNA level of CXCL-11/I-TAC following BHRF1-3 antisense transfection in BL-5R cells. BL-5R cells were transfected with RNA oligonucleotides at the following dosages: control transfections with no oligo (lane 1); transfection with a scrambled RNA oligo at 0.25 μmol/L for 8 h (lane 2), 1.25 μmol/L for 8 h (lane 3), 0.25 μmol/L for 16 h (lane 4), and 1.25 μmol/L for 16 h (lane 5); and transfection with anti-BHRF1-3 miRNA oligo at 0.25 μmol/L for 8 h (lane 6), 1.25 μmol/L for 8 h (lane 7), 0.25 μmol/L for 16 h (lane 8), and 1.25 μmol/L for 16 h (lane 9). CXCL-11/I-TAC, CXCL-7, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels in transfected cells were also analyzed by RT-PCR. PCR products were visualized under UV in a Bio-Rad Gel Doc 2000 imager and inverted images of DNA bands were captured. C, effect of BHRF1-3 miRNA on induced CXCL-11/I-TAC. BL-5 cells were transfected with 100 nmol/L BHRF1-3 anti-miR and treated with 1,000 units/mL IFN-γ (lanes 2–5) and then transfected with BHRF1-3 miRNA sense oligos (lane 2, 0 nmol/L; lane 3, 10 nmol/L; lane 4, 30 nmol/L; lane 5, 100 nmol/L). CXCL-11/I-TAC and β-actin mRNA levels in transfected cells were then analyzed by RT-PCR (left) and CXCL-11/I-TAC protein level was analyzed by ELISA (right). Agarose gels were exposed under UV in a Bio-Rad Gel Doc 2000 imager and inverted images of DNA bands captured.
We investigated the effect of BHRF1-3 miRNA on its putative target, CXCL-11/I-TAC. First, we studied the high BHRF1-3 miRNA BL-5R cell line. BL-5R cells were transfected with increasing amounts of BHRF1-3 miRNA antisense oligonucleotide (BHRF1-3 anti-miR) at ~65% efficiency (Fig. 4B, top) and the CXCL-11/I-TAC level was assessed by RT-PCR. By competing the BHRF1-3 miRNA targeting sequence with its antisense RNA oligonucleotide, we repeatedly elicited basal induction of CXCL-11/I-TAC expression in BL-5R (Fig. 4B, bottom, lanes 6–9). Control transfections with a scrambled RNA oligonucleotide (Fig. 4B, bottom, lanes 2–5), as well as mock transfection (Fig. 4B, bottom, lane 1), showed no modulation of CXCL-11/I-TAC expression. This result indicates that BHRF1-3 miRNA antisense competes out BHRF1-3 miRNA and reverses its repressive effect on CXCL-11/I-TAC expression. We also analyzed the expression of another chemokine, CXCL-7, in the same set of experiments. RT-PCR showed that CXCL-7 mRNA level was not modulated by transfection of either BHRF1-3 anti-miR or scrambled oligonucleotides (Fig. 4B, bottom), which indicates that the effect of BHRF1-3 miRNA on CXCL-11/I-TAC expression is specific. Similarly, CXCL-11/I-TAC expression was markedly induced in the low BHRF1-3 miRNA BL-5 cells transfected with BHRF1-3 anti-miR, and we also observed a much greater induction of CXCL-11/I-TAC expression in BL-5 cells dually treated with both the antisense oligo of BHRF1-3 miRNA and IFN-γ (data not shown). This showed that expression of CXCL-11/I-TAC is suppressed by the EBV-encoded BHRF1-3 miRNA.
We further examined the direct effect of BHRF1-3 miRNA on CXCL-11/I-TAC expression. CXCL-11/I-TAC was stimulated to a higher level in BL-5 cells when transfected with 100 nmol/L BHRF1-3 anti-miR and induced with 1,000 units/mL IFN-γ (Fig. 4C, left and right, lanes 2–5). Cells were then transfected with increasing amounts of BHRF1-3 miRNA sense oligos (Fig. 4C, left and right, lanes 3–5) and analyzed for CXCL-11/I-TAC by RT-PCR (Fig. 4C, left) and ELISA (Fig. 4C, right). We observed a significant reduction of CXCL-11/I-TAC at both mRNA and protein levels when cells were transfected with BHRF1-3 miRNA. This result further shows that CXCL-11/I-TAC expression is suppressed by EBV miRNA BHRF1-3, suggesting that cellular chemokines can be targeted by the viral miRNA.
Discussion
Following primary EBV infection, B cells undergo blast transformation during which they express the full spectrum of EBV genes (latency III program; refs. 11, 12). Presumably, to evade host viral immune defenses, the cells later adopt a latency I program, in which EBV gene expression is restricted to the relatively nonimmunogenic EBNA1 protein, the BamHI transcripts including the BART miRNAs, and the noncoding EBER transcripts. It is thought that BL arises from a B-cell subset in which EBV naturally adopts a tight latency I program or that a clone with limited antigen expression is selected from an EBV-transformed latency III progenitor pool (23). EBV latency III lymphomas typically arise in immunodeficient patients, such as those infected with HIV, or following pharmacologic immunosuppression after organ transplantation or they arise during in vitro culture of latency type I cells, which underscores the importance of a robust immunosurveillance system to counter the expression of EBV genes (23, 24).
Although there are distinct forms of EBV latency types in established cell lines, the clinical scenario is often quite different. Clinical specimens from patients with the diagnosis of BL have been found to express a heterogeneous latency program (25–27). The heterogeneity of EBV latency patterns in clinical specimens may be due to variant EBV clones, the establishment of immunoprivileged sites in the tumor microenvironment, and the selection of a clone from a type III progenitor pool (23, 28). It may also be part of an immune evasion strategy that includes EBV-mediated regulation of antigen processing and presentation and the down-modulation of the cytotoxic T-cell (CTL) cytokine networks (29). It will be important to study the biology of these primary EBV+ lymphomas to advance diagnosis and treatment.
The identification of virus-encoded miRNAs has generated new insights on virus transformation and immune evasion. Sullivan et al. (9) showed that SV40-encoded miRNAs target early viral mRNAs for cleavage, thus reducing the expression of viral tumor antigens, CTL recognition, and cytolysis. Recently, Samols et al. (30) identified thrombospondin 1, a strong tumor suppressor and anti-angiogenic factor, as a potential target for HHV-8–encoded miRNAs, and Lo et al. (31) reported modulation of LMP-1 protein expression by EBV-encoded BART miRNAs, highlighting the role of viral miRNAs in regulating viral pathogenesis.
We analyzed for the first time a variety of primary unmanipulated EBV-associated lymphomas (rather than established cell lines) that are representative of the major forms that occur in immunocompromised patients. Primary BLs expressed high levels of BART2 miRNA, which was expected based on the ubiquitous, high-level transcription across the BamHI locus in all EBV+ tumors (32–34). In contrast, a lymphoblastoid derivative of a BL line derived from an AIDS patient as well as AIDS-related DLBCLs expressed high levels of BHRF1-3 miRNA. The expression pattern of these two EBV miRNAs correlated with that previously seen in long-term cultured cell lines (8). An induced expression of BHRF1 miRNAs and a putative cleavage residue have recently been reported in EBV+ AKATA cells induced from type I latency to type III latency, indicating that the high level of BHRF1 miRNAs is a characteristic of EBV latency III infection (35). It is interesting to note that BHRF1-3 miRNA was low or absent in AIDS-related BL. These tumors typically occur in relatively immunocompetent patients compared with EBV+ DLBCL, which arise in patients with more advanced immunodeficiency (36). In contrast, EBV-associated large cell variants may exert a previously unknown immunosuppressive effect via modulation of cellular chemokines. Therefore, we hypothesize that one of the activities of EBV BHRF1-3 miRNA is the regulation of host cell immune function. This article establishes that EBV miRNAs are readily detectable in primary unmanipulated lymphomas associated with the virus. It is also likely that BHRF1-3 and other EBV miRNAs have additional complementary cellular targets.
Two recent reports have shown a potent antitumor activity for the IFN-inducible T-cell attracting chemokine (CXCL-11/I-TAC) in laboratory animals. However, there is a paucity of studies on the expression of CXCL-11/I-TAC expression in clinical specimens (37, 38). We have identified EBV BHRF1-3 miRNA, which is 100% identical to its target sequence in the 3′-UTR of CXCL-11/I-TAC mRNA, as a potential effector in the modulation of CTL cytokine networks (7). We show that CXCL-11/I-TAC mRNA and protein levels inversely correlate with BHRF1-3 miRNA expression levels. Changes in BHRF1-3 miRNA levels in a pair of isogenic type I/III BL cell lines were inversely correlated with CXCL-11/I-TAC levels. Furthermore, antisense delivery of the BHRF1-3 miRNA could relieve BHRF1-3 miRNA repression of CXCL-11/I-TAC. Therefore, the activity of BHRF1-3 miRNA and other virally encoded miRNAs may have far-reaching implications in cancer immunosurveillance. Alternatively, antisense delivery of BHRF1-3 miRNA may represent a targeted immunotherapeutic tool that can be directed toward enhancing immunosurveillance of EBV-related tumors.
Supplementary Material
Acknowledgments
Grant support: NIH/National Cancer Institute grants CA082274, CA70058, and CA121935 (W.J. Harrington, Jr.); Leukemia Society Translational Award 6021 (D.P. Dittmer and W.J. Harrington, Jr.); the Kulick Trust at the University of Miami Miller School of Medicine (W.J. Harrington, Jr.); and NIH grants CA109232 and DE018304 (D.P. Dittmer).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–52. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 2.He Z, Sontheimer EJ. “siRNAs and miRNAs”: a meeting report on RNA silencing. RNA. 2004;10:1165–73. doi: 10.1261/rna.7900204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sevignani C, Calin GA, Siracusa LD, Croce CM. Mammalian microRNAs: a small world for fine-tuning gene expression. Mamm Genome. 2006;17:189–202. doi: 10.1007/s00335-005-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–24. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 5.Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16:203–8. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Lecellier CH, Dunoyer P, Arar K, et al. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–60. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 7.Pfeffer S, Zavolan M, Grasser FA, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–36. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 8.Cai X, Schafer A, Lu S, et al. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan CS, Grundhoff AT, Tevethia S, et al. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–86. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 10.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human γ-herpes-viruses. RNA. 2006;12:733–50. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F, Gregory C, Sample C, et al. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J Virol. 1990;64:2309–18. doi: 10.1128/jvi.64.5.2309-2318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L, Hong K, Zhang J, et al. Multiple signal transducers and activators of transcription are induced by EBV LMP-1. Virology. 2004;323:141–52. doi: 10.1016/j.virol.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Kurokawa M, Ghosh SK, Ramos JC, et al. Azidothymidine inhibits NF-κB and induces Epstein-Barr virus gene expression in Burkitt’s lymphoma. Blood. 2005;106:235–40. doi: 10.1182/blood-2004-09-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cesarman E, Moore PS, Rao PH, et al. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi’s sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–14. [PubMed] [Google Scholar]
- 15.Hayashi K, Chen WG, Chen YY, et al. Deletion of Epstein-Barr virus latent membrane protein 1 gene in United States and Brazilian Hodgkin’s disease and reactive lymphoid tissue: high frequency of a 30-bp deletion. Hum Pathol. 1997;28:1408–14. doi: 10.1016/s0046-8177(97)90231-8. [DOI] [PubMed] [Google Scholar]
- 16.Araujo I, Foss HD, Bittencourt A, et al. Expression of Epstein-Barr virus-gene products in Burkitt’s lymphoma in Northeast Brazil. Blood. 1996;87:5279–86. [PubMed] [Google Scholar]
- 17.Hilscher C, Vahrson W, Dittmer DP. Faster quantitative real-time PCR protocols may lose sensitivity and show increased variability. Nucleic Acids Res. 2005;21:e182. doi: 10.1093/nar/gni181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papin J, Vahrson W, Hines-Boykin R, et al. Real-time quantitative PCR analysis of viral transcription. Methods Mol Biol. 2005;292:449–80. doi: 10.1385/1-59259-848-x:449. [DOI] [PubMed] [Google Scholar]
- 19.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–7. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmittgen TD, Jiang J, Liu Q, et al. A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res. 2004;32:e43. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cole KE, Strick CA, Paradis TJ, et al. Interferon-inducible T cell α chemoattractant (I-TAC): a novel non-ELR CXC chemokine with potent activity on activated T cells through selective high affinity binding to CXCR3. J Exp Med. 1998;187:2009–21. doi: 10.1084/jem.187.12.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colvin RA, Campanella GS, Sun J, et al. Intracellular domains of CXCR3 that mediate CXCL9, CXCL10, and CXCL11 function. J Biol Chem. 2004;279:30219–27. doi: 10.1074/jbc.M403595200. [DOI] [PubMed] [Google Scholar]
- 23.Kelly G, Bell A, Rickinson A. Epstein-Barr virus-associated Burkitt lymphomagenesis selects for down-regulation of the nuclear antigen EBNA2. Nat Med. 2002;8:1098–104. doi: 10.1038/nm758. [DOI] [PubMed] [Google Scholar]
- 24.Khanna R, Burrows SR. Role of cytotoxic T lymphocytes in Epstein-Barr virus-associated diseases. Annu Rev Microbiol. 2000;54:19–48. doi: 10.1146/annurev.micro.54.1.19. [DOI] [PubMed] [Google Scholar]
- 25.Xue SA, Labrecque LG, Lu QL, et al. Promiscuous expression of Epstein-Barr virus genes in Burkitt’s lymphoma from the central African country Malawi. Int J Cancer. 2002;99:635–43. doi: 10.1002/ijc.10372. [DOI] [PubMed] [Google Scholar]
- 26.Touitou R, Arbach H, Cochet C, et al. Heterogeneous Epstein-Barr virus latent gene expression in AIDS associated lymphomas and in type I Burkitt’s lymphoma cell lines. J Gen Virol. 2003;84:949–57. doi: 10.1099/vir.0.18687-0. [DOI] [PubMed] [Google Scholar]
- 27.Niedobitek G, Agathanggelou A, Rowe M, et al. Heterogeneous expression of Epstein-Barr virus latent proteins in endemic Burkitt’s lymphoma. Blood. 1995;86:659–65. [PubMed] [Google Scholar]
- 28.Frisan T, Sjoberg J, Dolcetti R, et al. Local suppression of Epstein-Barr virus (EBV)-specific cytotoxicity in biopsies of EBV-positive Hodgkin’s disease. Blood. 1995;86:1493–501. [PubMed] [Google Scholar]
- 29.Ploegh HL. Viral strategies of immune evasion. Science. 1998;280:248–53. doi: 10.1126/science.280.5361.248. [DOI] [PubMed] [Google Scholar]
- 30.Samols MA, Skalsky RL, Maldonado AM, et al. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 2007;3:e65. doi: 10.1371/journal.ppat.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lo AK, To KF, Lo KW, et al. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci U S A. 2007;104:16164–9. doi: 10.1073/pnas.0702896104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karran L, Gao Y, Smith PR, Griffin BE. Expression of a family of complementary-strand transcripts in Epstein-Barr virus-infected cells. Proc Natl Acad Sci U S A. 1992;89:8058–62. doi: 10.1073/pnas.89.17.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brooks LA, Lear AL, Young LS, Rickinson AB. Transcripts from the Epstein-Barr virus BamHI A fragment are detectable in all three forms of virus latency. J Virol. 1993;67:3182–90. doi: 10.1128/jvi.67.6.3182-3190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sadler RH, Raab-Traub N. Structural analyses of the Epstein-Barr virus BamHI A transcripts. J Virol. 1995;69:1132–41. doi: 10.1128/jvi.69.2.1132-1141.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xing L, Kieff E. Epstein-Barr virus BHRF1 micro- and stable RNAs during latency III and after induction of replication. J Virol. 2007;81:9967–75. doi: 10.1128/JVI.02244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Navarro WH, Kaplan LD. AIDS-related lymphoproliferative disease. Blood. 2006;107:13–20. doi: 10.1182/blood-2004-11-4278. [DOI] [PubMed] [Google Scholar]
- 37.Yang X, Chu Y, Wang Y, et al. Vaccination with IFN-inducible T cell α chemoattractant (I-TAC) gene-modified tumor cell attenuates disseminated metastases of circulating tumor cells. Vaccine. 2006;24:2966–74. doi: 10.1016/j.vaccine.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Hensbergen PJ, Wijnands PG, Schreurs MW, et al. The CXCR3 targeting chemokine CXCL11 has potent antitumor activity in vivo involving attraction of CD8+ T lymphocytes but not inhibition of angiogenesis. J Immunother. 2005;28:343–51. doi: 10.1097/01.cji.0000165355.26795.27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.