SUMMARY
Recent studies have defined a group of muscular dystrophies, now termed the dystroglycanopathies, as novel disorders of glycosylation. These conditions include Walker–Warburg syndrome, muscle–eye–brain disease, Fukuyama-type congenital muscular dystrophy, congenital muscular dystrophy types 1C and 1D, and limb-girdle muscular dystrophy type 2I. Although clinical findings can be highly variable, dystroglycanopathies are all characterized by cortical malformations and ocular defects at the more severe end of the clinical spectrum, in addition to muscular dystrophy. All of these disorders are defined by the underglycosylation of α-dystroglycan. Defective glycosylation of dystroglycan severs the link between this important cell adhesion molecule and the extracellular matrix, thereby contributing to cellular pathology. Recent experiments indicate that glycosylation might not only define forms of muscular dystrophy but also provide an avenue to the development of therapies for these disorders.
Keywords: dystroglycan, glycosylation, laminin, lissencephaly, neuromuscular junction, skeletal muscle
INTRODUCTION
Mutations in 12 different genes have been shown to cause forms of congenital muscular dystrophy (CMD).1–8 Approximately one-third of all CMDs are caused by mutations in the LAMA2 gene, which encodes the α2 chain of laminin. Mutations in COL6A1, COL6A2 and COL6A3, which encode the three chains of collagen type VI, give rise to Ullrich congenital muscular dystrophy and Bethlem myopathy. Mutations in the selenoprotein N gene (SEPN1) give rise to rigid spine syndrome, multiminicore disease, and a desmin-related myopathy with MALLORY-BODY-like inclusions.
The other genes that are associated with CMD all code for molecules that affect cell surface receptors for the extracellular matrix molecule laminin. These genes include ITGA7—the gene that encodes integrin α7 (the predominant integrin α chain in skeletal muscle)—and six genes (fukutin [FCMD], fukutin-related protein [FKRP], protein O-linked mannose β1,2-N-acetyl-glucosaminyltransferase [POMGnT1], protein-O-mannosyltransferases 1 and 2 [POMT1/2], and like-glycosyltransferase [LARGE]), the products of which affect the glycosylation of α-dystroglycan.1–8 α-Dystroglycan is an important membrane protein that, like integrins, binds to the extracellular matrix. Diseases resulting from mutations that affect α-dystroglycan glycosylation have been grouped together under the heading dystroglycanopathies. Mutations in the dystroglycanopathy-associated genes give rise to characteristic alterations in the glycosylation of the α-dystroglycan protein, which can alter its function, although the precise molecular mechanisms are poorly understood in some cases.9 The ‘dystroglycanopathy genes’ now account for half of the genes that have been implicated in CMD.
This review will summarize the genetic and clinical features of the dystroglycanopathies. In addition, it will describe the relationship between glycosylation and the function of dystroglycan, and the function of dystroglycanopathy-associated genes. The article will also highlight recent work that indicates that altering glycosylation might be a viable therapeutic approach to treating certain forms of muscular dystrophy.
THE DYSTROGLYCANOPATHIES: CLINICAL AND GENETIC FINDINGS
The original studies on dystroglycanopathy-associated genes indicated that defects in each gene yielded a distinct disorder with unique clinical findings. In recent years, however, this idea has given way to the realization that almost all of these genes can be mutated so that they present the same spectrum of clinical findings. The dystroglycanopathies include Walker–Warburg syndrome (WWS),10,11 muscle–eye–brain disease (MEB),12 Fukuyama-type congenital muscular dystrophy (FCMD),13 congenital muscular dystrophy types 1C and 1D (MDC1C14 and MDC1D15), and limb-girdle muscular dystrophy type 2I (LGMD2I;16 Table 1). WWS, MEB and FCMD have common clinical findings, including brain malformations and muscular dystrophy;1–8 brain malformations are far less common in MDC1C, and are not present in LGMD2I. The most common brain finding is type II (‘cobblestone’) LISSENCEPHALY, which can be AGYRIC in WWS. This finding is most probably attributable to aberrant migration of neurons through gaps in the glia limitans–basement membrane complex during cortical development. Work in the Largemyd animal model, in which the Large gene is deleted, also implicates cell-autonomous defects in neuronal migration in aberrant cortical development.17–19
Table 1.
Known dystroglycanopathy genes and the disorders caused by their mutation.
| Disorder | Gene | Reference |
|---|---|---|
| Fukuyama-type congenital muscular dystrophy (FCMD) |
FCMD | Kobayashi et al. (1998)13 |
| Walker–Warburg syndrome (WWS) | POMT1 | Beltran-Valero de Bernabe et al. (2002)10 |
| POMT2 | van Reeuwijk et al. (2005)11 | |
| FCMD | Beltran-Valero de Bernabe et al. (2003)25 | |
| POMGnT1 | Taniguchi et al. (2003)27 | |
| FKRP | Beltran-Valero de Bernabe et al. (2004)22 | |
| Congenital muscular dystrophy type 1C (MDC1C) |
FKRP | Brockington et al. (2001)14 |
| Limb-girdle muscular dystrophy type 2I (LGMD2I) |
FKRP | Brockington et al. (2001)16 |
| Congenital muscular dystrophy type 1D (MDC1D) |
LARGE | Longman et al. (2003)15 |
| Muscle–eye–brain disease (MEB) | POMGnT1 | Yoshida et al. (2001)12 |
| FKRP | Beltran-Valero de Bernabe et al. (2004)22 | |
| Limb-girdle muscular dystrophy with mental retardation (LGMD2K) |
POMT1 | Balci et al. (2005)26 |
| Severe autistic features | POMGnT1 | Haliloglu et al. (2004)28 |
FCMD, fukutin; FKRP, fukutin-related protein; LARGE, like-glycosyltransferase; POMGnT1, protein O-linked mannose β1,2-N-acetylglucosaminyltransferase; POMT1, protein-O-mannosyltransferase 1; POMT2, protein-O-mannosyltransferase 2.
Other brain findings for the dystroglycanopathies include dilation of the cerebral ventricles, flattened brainstem, absent corpus collosum, aberrant myelination, and occasional occipital encephalocele. Ocular findings are common in WWS and MEB, but less so in FCMD and MDC1C. These findings include myopia, cataracts, retinal detachment, microphthalmia, BUPHTHALMOS, PERSISTENT HYPERPLASTIC PRIMARY VITREOUS, PETER’S ANOMALY, and congenital glaucoma. Muscular dystrophy is present in all of the dystroglycanopathies, as signified by raised serum creatine kinase levels, which can be elevated by several orders of magnitude, and by dystrophic muscle pathology—including necrosis, fibrosis and evidence of muscle regeneration—on biopsy. Cardiac involvement is also common in FCMD, MDC1C and LGMD2I after the first decade of life.
The relationship between gene mutations and clinical findings can be highly variable in the dystroglycanopathies. For example, mutations in FKRP that were originally identified as causing MDC1C indicated that this disease had no brain involvement,14 but FKRP mutations (V405L and A455D) discovered subsequently have been linked with mental retardation, microcephaly and cerebellar cysts.20,21 Further mutations in this gene that present as MEB or WWS are now known.22 By contrast, patients who are homozygous for the L276I mutation have LGMD2I, which is milder in presentation than MDC1C.16 Clinical variability of all of these disorders is probably modulated by secondary genetic factors. For example, a group of LGMD2I patients from a consanguineous Bedouin tribe share the same FKRP mis-sense mutation, but have an age of disease onset that varies from birth to the second decade.23
It is also becoming increasingly clear that mutations in each of the dystroglycanopathy-associated genes give rise to a wider spectrum of clinical findings than was previously thought. For example, FCMD was originally thought not to give rise to ocular findings.13 This observation, however, was later attributed to the fact that most FCMD patients of Japanese descent share a common retrotransposonal insertion in the FCMD gene. This insertion, which occurs in the 3' untranslated region of the gene, causes reduced expression of normal fukutin protein, resulting in a milder phenotype than would occur from a null mutation. Once compound heterozygotes were identified, the clinical spectrum was expanded to include ocular defects and a poorer prognosis,24 and homozygous nonsense mutations were shown to yield WWS.25 Likewise, mutations in POMT1 were originally reported only to give rise to WWS, which presents with the most severe clinical features of all the dystroglycanopathies (the average life expectancy of WWS patients is 0.8 years).2 A recent report, however, identified a POMT1 mutation (A200P) as causing a far milder limb-girdle muscular dystrophy with mental retardation (LGMD2K).26 Similarly, recent findings by Toda and colleagues have expanded the clinical spectrum of POMGnT1 mutations to include FCMD-like and WWS-like phenotypes in addition to MEB.27 Perhaps the most intriguing development of the past year has been the finding of Topaloglu and colleagues of a patient with a defect in the POMGnT1 gene (IVS17-2A→G), in whom severe autistic features were the dominant presenting sign.28 This finding could significantly expand the clinical spectrum for the dystroglycanopathies.
GLYCOSYLATION OF DYSTROGLYCAN
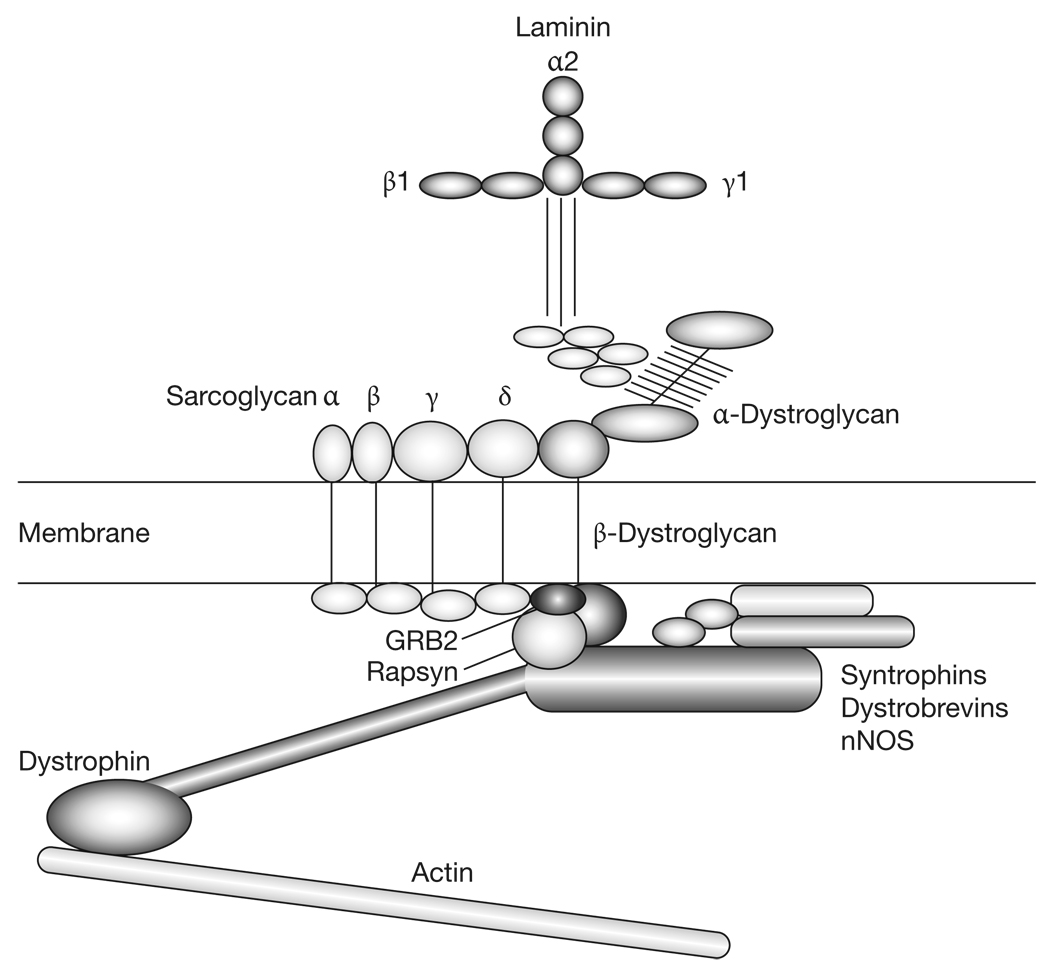
The dystroglycan gene (DAG1) encodes a single polypeptide that is post-translationally cleaved into two protein chains, α-dystroglycan and β-dystroglycan.29,30 α-Dystroglycan is a highly glycosylated peripheral membrane protein that binds tightly but non-covalently to β-dystroglycan, which is a transmembrane protein.29 This complex of α-dystroglycan and β-dystroglycan chains is a vital component of the dystrophin–glycoprotein complex, which links the extracellular matrix that surrounds myofibers (and many other cell types) through the membrane to the actin cytoskeleton (Figure 1).5,31,32 α-Dystroglycan contributes to this complex by binding to proteins in the basal lamina, including laminins, agrin and perlecan, as well as to neurexins, which are transmembrane proteins. α-Dystroglycan also binds to infectious agents (e.g. Mycobacterium leprae, lymphocytic choriomeningitis virus and Lassa virus) and can provide a means of entry for these agents into cells.5,31 β-Dystroglycan binds α-dystroglycan on the extracellular face of the membrane, and binds dystrophin, utrophin, growth factor receptor-bound protein 2 (GRB2) and rapsyn via its intracellular domain. Dystrophin, the protein that is absent in Duchenne type muscular dystrophy, provides a vital link between the muscle cell membrane and the cytoskeleton by virtue of its ability to bind filamentous actin.5,31,32 Dystrophin also binds to or associates with a number of other structural and signaling proteins, including syntrophins, dystrobrevins, and neuronal nitric oxide synthase (nNOS). Also present in this complex are the α to δ sarcoglycans, transmembrane proteins that are defective in limb-girdle muscular dystrophies types 2C to 2F, respectively.33 Utrophin, a homolog of dystrophin that is confined to the neuromuscular junction in skeletal muscle, serves a similar function to dystrophin in synaptic myofiber membranes that are innervated by motor neurons.31,32
Figure 1.
Dystroglycan glycosylation and its place in the dystrophin–glycoprotein complex. Laminin, which is present in the extracellular matrix surrounding the myofiber membrane, is composed of three different chains, α, β and γ. In skeletal muscle, the primary laminin expressed in the basal lamina surrounding muscle fibers contains α2 (merosin), β1 and γ1 chains. The C-terminal region of the laminin α2 protein binds to α-dystroglycan. This interaction requires the O-mannose-linked carbohydrates that are present in the middle third of the α-dystroglycan molecule. α-Dystroglycan binds tightly, but non-covalently, to β-dystroglycan, which spans the muscle membrane. The intracellular domain of β-dystroglycan binds to signaling and structural molecules, including growth factor receptor-bound protein 2 (GRB2), rapsyn, and dystrophin. Dystrophin, in turn, links the complex both to filamentous actin and to other structural and signaling proteins, including syntrophins, dystrobrevins, and neuronal nitric oxide synthase (nNOS). The sarcoglycans (α–δ) are additional transmembrane components within the dystrophin-associated glycoprotein complex. Absence or deficiency of most of the proteins in this complex give rise to different forms of muscular dystrophy. Loss of dystrophin gives rise to Duchenne type muscular dystrophy, loss of laminin α2 gives rise to merosin-dependent congenital muscular dystrophy (MDC1A), loss of α to δ sarcoglycans gives rise to limb-girdle muscular dystrophies 2C to 2F, respectively, and loss of genes that modify α-dystroglycan glycosylation gives rise to Walker–Warburg syndrome, muscle–eye–brain disease, Fukuyama-type congenital muscular dystrophy, congenital muscular dystrophy types 1C and 1D, and limb-girdle muscular dystrophy type 2I. GRB2, growth factor receptor-bound protein 2; nNOS, neuronal nitric oxide synthase.
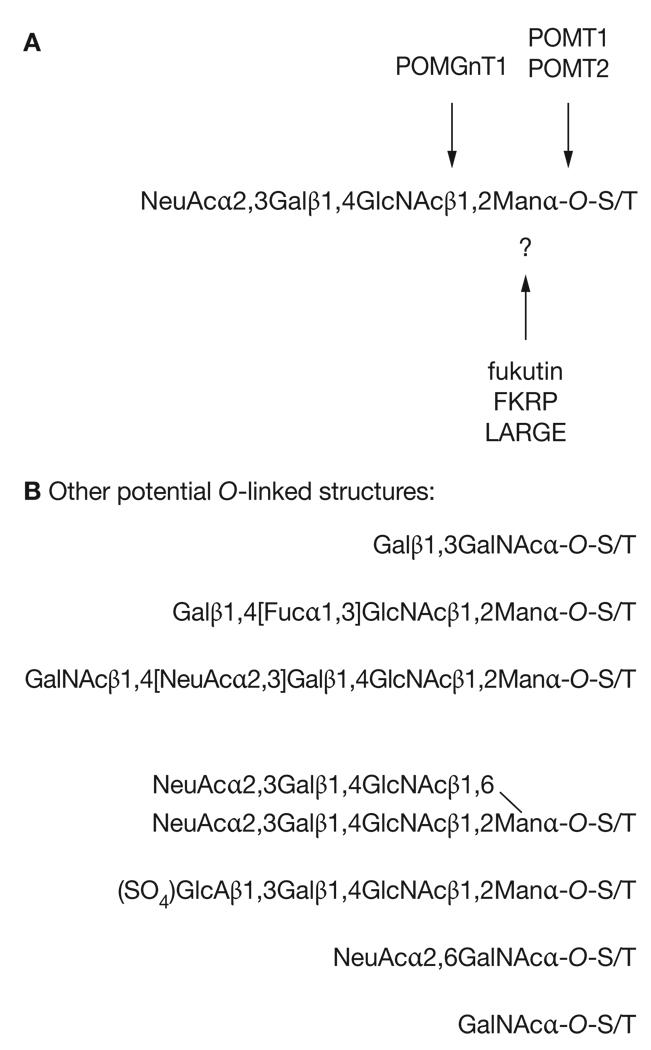
The α-dystroglycan protein is predicted to be 72 kDa in size, yet native α-dystroglycan protein migrates as a diffuse band centered on 160 kDa in skeletal muscle tissue.29,30 Therefore, α-dystroglycan is roughly half carbohydrate by molecular weight. In large part, this characteristic can be attributed to the presence of a serine–threonine (S–T)-rich mucin domain in the middle of the protein that contains up to 55 sites for O-linked glycosylation. O-linked carbohydrates are attached to proteins via S or T residues. Several groups have sequenced the carbohydrates present in this region of α-dystroglycan using tissues from several sources, including skeletal muscle.34–36 All of these studies have identified the presence of two types of O-linked carbohydrate. The first sequence, Galβ1,3GalNAcα-O-S/T (Gal, galactose; GalNAc, N-acetylgalactosamine), is a common carbohydrate structure found on many proteins containing O-linked glycosylation. By contrast, the second sequence, NeuAcα2,3Galβ1,4GlcNAcβ1,2Manα-O-S/T (NeuAc, N-acetylneuraminic acid [or sialic acid]; GlcNAc, N-acetylglucosamine; Man, mannose), is a structure that has only been described on α-dystroglycan in mammalian tissues. O-linked mannose is found on cell membranes of many lower organisms, including yeast,37 but appears to be highly specific to α-dystroglycan in mammals.
Loss of dystroglycan protein from brain, peripheral nerves or skeletal muscle recapitulates most of the cellular pathology found in the dystroglycanopathies.38–40 Admittedly, loss of the entire dystroglycan protein might have effects beyond those caused by defects in its glycosylation, but these data nevertheless support the possibility that all glycosylation defects in the dystroglycanopathies could be centered on dystroglycan. This characterization represents the current state of the literature. It is important, however, to acknowledge that little is known about whether additional mammalian proteins bear the O-mannose linkage, and whether defects in the glycosylation of these proteins would contribute to pathogenesis in the dystroglycanopathies.
Other O-linked glycan structures might also be present on α-dystroglycan (Figure 2), and their absence might affect the function of this protein.6,31 These glycan sequences include additional modifications of the O-linked mannose structure NeuAcα2,3Galβ1,4GlcNAcβ1,2Manα-O-S/T, such as addition of terminal β1,4GalNAc to create the CT carbohydrate antigen, which is found at the neuromuscular junction,41 addition of β1,6-linked sialyl-N-acetyllactosamine to create branched O-mannose structures, addition of the sulfated glucuronic acid to create the HNK-1 carbohydrate, or fucosylation to create the Lewis X antigen. Additional structures that are O-linked to GalNAc might also be present (Figure 2). Many of these other glycan stuctures have been shown to be present on α-dystroglycan only using methods such as antibody binding, which are far less sophisticated than the monosaccharide linkage analysis used to identify NeuAcα2,3Galβ1,4GlcNAcβ1,2Manα-O-S/T and Galβ1,3GalNAcα-O-S/T. Lewis X is the one exception here, but this antigen has been identified on α-dystroglycan only in brain tissue and not in skeletal muscle or peripheral nerve.34–36 Loss of these less well characterized glycan modifications on α-dystroglycan might contribute to the loss of dystroglycan function and to pathology in the dystroglycanopathies.
Figure 2.
Glycosylation of α-dystroglycan. (A) Muscular-dystrophy-associated gene products that are involved in dystroglycan glycosylation. The O-mannose-linked glycans on α-dystroglycan are composed of a linear chain of four carbohydrates. Six genes implicated in muscular dystrophy encode products that alter the glycosylation of α-dystroglycan. Protein-O-mannosyltransferases 1 and 2 (POMT1/2) are both required for the synthesis of O-linked mannose to serines and threonines on the α-dystroglycan protein, whereas protein O-linked mannose β1,2-N-acetylglucosaminyltransferase (POMGnT1) synthesizes the second linkage, β1,2GlcNAc, on the O-mannose chain. Three other proteins, fukutin (FCMD), fukutin-related protein (FKRP) and like-glycosyltransferase (LARGE), are also involved in this pathway, though their function is not yet clear. (B) Additional structures that might be attached to α-dystroglycan by O-linkages. FKRP, fukutin-related protein; Fuc, fucose; Gal, galactose; GalNAc, N-acetylgalactosamine; GlcA, glucuronic acid; GlcNAc, N-acetylglucosamine; LARGE, like-glycosyltransferase; Man, mannose; NeuAc, N-acetylneuraminic acid (or sialic acid); POMGnT1, protein O-linked mannose β1,2-N-acetylglucosaminyltransferase; POMT1/2, protein-O-mannosyltransferases 1/2; S, serine; SO4, sulfate; T, threonine.
All dystroglycanopathies have the common molecular finding that α-dystroglycan is underglycosylated, as defined by a combination of immunostaining and immunoblotting. Only two commercially available monoclonal antibodies to α-dystroglycan are commonly used for this purpose: IIH6, a laminin-blocking monoclonal antibody, and VIA4-1.29 Both of these antibodies require proper glycosylation of α-dystroglycan to bind to the protein at all. Consequently, some original reports in which these reagents were used unwittingly reported a loss of α-dystroglycan protein expression. Antibodies raised against the α-dystroglycan polypeptide, however, have been used to demonstrate that α-dystroglycan protein is indeed present in the myofiber membrane of biopsy material in the dystroglycanopathies, and that the real defect in these patients is a loss of dystroglycan glycosylation.9 This characteristic is commonly shown by demonstrating a lowered molecular weight on sodium-dodecyl-sulfate polyacrylamide-gel electrophoresis for α-dystroglycan isolated from affected tissues. Campbell and colleagues have shown that underglycosylated α-dystroglycan protein that is present in the muscles of patients with FCMD, MEB and WWS binds poorly, if at all, to laminin.9 Defective glycosylation of α-dystroglycan, therefore, divorces the extracellular matrix from the cell membrane, thereby contributing to cellular pathology.
DYSTROGLYCANOPATHY-GENE FUNCTION: SOME MYSTERIES REMAIN
There is good reason to believe that most dystroglycanopathy-associated gene products are involved in the synthesis of the unique O-linked mannose chains on α-dystroglycan (Figure 2); this link is still far from proven, however, for about half of the dystroglycanopathy genes that have so far been identified. As regards the known entities, two protein-O-mannosyltransferase-encoding genes—POMT1 and POMT2—have been identified in humans. Mutations in both genes have now been linked to WWS,10,11 and are associated with hypoglycosylation of α-dystroglycan.11,42 The idea that mutations in both of these genes cause disease is consistent with the demonstration by Endo and colleagues that in flies RNA INTERFERENCE knockdown of POMT1 and POMT2 yields the same phenotype (rotated abdomen),43 and that coexpression of POMT1 and POMT2 is required for O-mannosyltransferase activity in mammalian cell lines.44
Pomt1 has also been eliminated in mice.45 The phenotype of these animals—embryonic lethality due to disruption of REICHERT’S MEMBRANE—is the same phenotype that is found in mice lacking Dag1.46 Reichert’s membrane is a structure that is specific to rodent embryos, so it is unclear whether embryonic lethality in Dag1-deficient mice would be reflective of a similarly lethal phenotype in humans. If this were shown to be the case, it might explain why DAG1 mutations have not yet been identified in the dystroglycanopathies. At least one mutation in Dag1 does cause muscular dystrophy in mice,47 however, so mutations in the dystroglycan gene might ultimately be found in these disorders in humans. Mutations in POMT1 that cause WWS have been shown to be defective in protein-O-mannosyltransferase enzyme activity.42 WWS is therefore very probably caused by a dearth of O-mannose-linked carbohydrates on α-dystroglycan, although such a relationship has never been shown using actual patient tissue.
Along with POMT1 and POMT2, the third known entity in the dystroglycanopathies is POMGnT1. POMGnT1 synthesizes the second carbohydrate on the O-mannose chains of α-dystroglycan, transferring N-acetylglucosamine in a β1,2 linkage to O-mannose.12 Because this enzyme requires O-linked mannose for activity and does not have activity on typical N-linked structures, its ability to glycosylate proteins might be relatively specific for α-dystroglycan. Mutations in POMGnT1 that cause MEB disease have been shown to result in reduced or absent enzyme activity.48 Therefore, it is highly probable that MEB is also caused by a deficit in O-mannosyl-linked glycans on α-dystroglycan. Schachter and colleagues have developed an assay to identify MEB on the basis of loss of POMGnT1 enzyme function, which has enabled MEB to be definitively diagnosed.49,50
Three ‘mystery’ genes exist, the function of which remains to be identified in the dystroglycanopathies: FCMD, FKRP and LARGE. FCMD mutations give rise to FCMD, which is a common form of CMD in Japan13 and was the first CMD in which dystroglycan underglycosylation was demonstrated.51 Fukutin protein is localized to the cis compartment of the Golgi apparatus, where it would be well positioned to affect the glycosylation of α-dystroglycan.52 Fukutin is partially homologous to yeast proteins that are involved in mannosyl phosphorylation of oligosaccharides and, on the basis of this evidence, it has been suggested that this protein is a glycosyltransferase. To date, however, there has been no demonstration of enzymatic activity for this protein. Global loss of Fcmd in mice is lethal at an early embryonic stage,53 and chimeric mice lacking this gene in a subset of cells demonstrate most facets of the cellular pathology found in FCMD.53 FKRP was named on the basis of its sequence homology with fukutin. Like fukutin, FKRP has a domain structure and sequence that is indicative of a glycosyltransferase, but again the enzymatic activity is unknown. FKRP has been localized to the Golgi apparatus in neuroblastoma, oligodendroglial and cardiac muscle cell lines, and shows the same intracellular distribution in skeletal muscle tissue taken from control, MDC1C and LGMD2I patients.54 Muntoni and colleagues have shown a correlation between the extent of dystroglycan underglycosylation in MDC1C and LGMD2I and the clinical phenotype, the L276I mutation being the least severe on both fronts.55 Therefore, underglycosylation of α-dystroglycan appears to be the causative factor in all of these disorders.
Perhaps the most important dystroglycanopathy-related gene in which the function has yet to be described is LARGE. LARGE was originally cloned by Hewitt and colleagues56 on the basis of its deletion in the myodystrophy (myd; Largemyd) mouse. The phenotype of the Largemyd mouse shares many features of WWS, MEB and FCMD. The importance of this gene derives not only from its deletion in an animal model, but also from the fact that its overexpression might be therapeutic for some of these disorders.57 Overexpression of LARGE can stimulate the glycosylation of α-dystroglycan and stimulate laminin binding to α-dystroglycan in cells cultured from patients with WWS, MEB or FCMD.57 Because underglycosylation of α-dystroglycan in the cells used for these experiments was caused by defects in genes other than LARGE, this experiment demonstrates that overexpression of LARGE can have a dominant effect on both α-dystroglycan glycosylation and function. LARGE overexpression might, therefore, be therapeutic in many of the dystroglycanopathies.
So, what is the function of LARGE? Unlike fukutin and FKRP, the primary sequence of LARGE is highly indicative of a Golgi-localized tandem glycosyltransferase. LARGE has two independent domains that might possess the ability to synthesize a repeating disaccharide, much as is known to occur with enzymes that synthesize glycosaminoglycans (GAGs) such as heparan sulfate or chondroitin sulfate.58 There is no evidence, however, that LARGE synthesizes GAGs.
What has become clear in the past year is what LARGE does not do. Using CHO CELL mutants, Patnaik and Stanley have shown that LARGE does not require sialic acid, galactose or fucose to glycosylate α-dystroglycan.59 Similarly, Combs and Ervasti have shown that enzymatic removal of sialic acid, galactose and N-acetylglucosamine from α-dystroglycan actually stimulates its binding to laminin,60 much as LARGE overexpression does.57 Again, this evidence indicates that these carbohydrates are not synthesized by LARGE. Enyzmes that remove GAGs also have no effects that are consistent with LARGE being a GAG synthase.61 Therefore, the most logical conclusion seems to be that LARGE creates a novel carbohydrate structure, which might be linked to structures other than, or in addition to, O-mannose.
Recent papers have shown that there is a second gene in the LARGE family, LARGE2.62,63 Like LARGE, LARGE2 can stimulate the glycosylation of α-dystroglycan such that laminin binding is increased.61–63 Unlike LARGE, however, LARGE2 is not appreciably expressed in brain or muscle.62 It appears, therefore, that LARGE2 might not compensate for the absence of LARGE in these tissues and is therefore unable to compensate for the loss of LARGE in the dystroglycanopathies. By contrast, Hewitt and colleagues showed that there is no defect in α-dystroglycan glycosylation in the Largemyd kidney, where Large2 is highly expressed.62
One final mystery, which could be a whole slew of mysteries, involves the genes yet to be identified for the dystroglycanopathies. The combination of mutations in POMT1, FKRP and fukutin has been estimated to account for between 20% and 33% of WWS cases.1,2 POMT2 mutations appear to be similarly rare.10,11 It is anticipated, therefore, that other genes will be found to be associated with WWS, and that they too will be involved in pathways that regulate the glycosylation of α-dystroglycan.
CONCLUSIONS
Work over the past half decade has demonstrated that mutations in genes involved in the glycosylation of α-dystroglycan cause forms of muscular dystrophy that are now termed dystroglycanopathies. The carbohydrates created by these genes are required for the binding of extracellular matrix proteins, including laminin, to dystroglycan, and this is the probable cause of most muscle, brain and eye pathology in these disorders. In the coming years, new dystroglycanopathy-associated genes will undoubtedly be identified in this pathway, and much work remains to be done in the effort to understand the function of genes that have already been implicated in these disorders. Defects in glycosylation are often highly PLEIOTROPIC, owing to the common nature of this type of post-translational modification. Therefore, the relatively specific nature of the pathology in the dystroglycanopathies, coupled with the fact that this pathology is linked to a defect in a single glycoprotein, makes this group of disorders one of the best models in which to understand the relationship between protein glycosylation and disease. The demonstration that overexpression of dystroglycanopathy gene products that stimulate dystroglycan glycosylation can have therapeutic benefit in multiple forms of these disorders makes it all the more important to understand these relationships.
KEY POINTS
Mutations in 12 different genes have been shown to cause forms of congenital muscular dystrophy
Six of the genes associated with congenital muscular dystrophy code for molecules that are involved in the glycosylation of α-dystroglycan, a membrane protein that binds to the extracellular matrix
Dystroglycanopathies—diseases resulting from mutations that affect α-dystroglycan glycosylation—include Walker–Warburg syndrome, muscle–eye–brain disease, Fukuyama-type congenital muscular dystrophy, congenital muscular dystrophy types 1C and 1D, and limb-girdle muscular dystrophy type 2I
α-Dystroglycan is roughly half carbohydrate by molecular weight, which is largely attributable to the presence of a serine–threonine-rich domain that contains up to 55 sites for O-linked glycosylation
All dystroglycanopathies have the common molecular finding that α-dystroglycan is underglycosylated, and most dystroglycanopathy-associated gene products are thought to be involved in the synthesis of O-linked mannose chains on α-dystroglycan
GLOSSARY
- MALLORY BODY
Hyaline (glassy) cytoplasmic inclusion
- LISSENCEPHALY
A brain disorder that is characterized by absence or reduction of the cerebral convolutions
- AGYRIC
Absence of developed folds or convolutions in the cerebral cortex
- BUPHTHALMOS
Abnormal enlargement of the eyes
- PERSISTENT HYPERPLASTIC PRIMARY VITREOUS
A congenital anomaly due to persistence of the embryonic remnants of the fibromuscular tunic of the eye and part of the hyaloid vascular system
- PETER’S ANOMALY
A congenital disorder characterized by a central leukoma (dense white corneal opacity) with adhesions at the periphery
- RNA INTERFERENCE
Post-transcriptional gene silencing in which double-stranded RNA mediates the destruction of messenger RNAs in a sequence-specific fashion
- REICHERT’S MEMBRANE
An extraembryonic membrane that surrounds the rodent embryo
- CHO CELL
Chinese hamster ovary cell
- PLEIOTROPIC
Producing multiple effects; that is, a single agent acting on several different processes
Footnotes
Competing interests
The author declared he has no competing interests.
References
- 1.Jimenez-Mallebrera C, et al. Congenital muscular dystrophy: molecular and cellular aspects. Cell Mol Life Sci. 2005;62:809–823. doi: 10.1007/s00018-004-4510-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Reeuwijk J, et al. Glyc-O-genetics of Walker–Warburg syndrome. Clin Genet. 2005;67:281–289. doi: 10.1111/j.1399-0004.2004.00368.x. [DOI] [PubMed] [Google Scholar]
- 3.Martin PT, Freeze HH. Glycobiology of neuromuscular disorders. Glycobiology. 2003;13:67R–75R. doi: 10.1093/glycob/cwg077. [DOI] [PubMed] [Google Scholar]
- 4.Muntoni F, et al. Defective glycosylation in muscular dystrophy. Lancet. 2002;360:1419–1421. doi: 10.1016/S0140-6736(02)11397-3. [DOI] [PubMed] [Google Scholar]
- 5.Michele DE, Campbell KP. Dystrophin–glycoprotein complex: post-translational processing and dystroglycan function. J Biol Chem. 2003;278:15457–15460. doi: 10.1074/jbc.R200031200. [DOI] [PubMed] [Google Scholar]
- 6.Endo T. Structure, function and pathology of O-mannosyl glycans. Glycoconjugate J. 2004;21:3–7. doi: 10.1023/B:GLYC.0000043740.26062.2c. [DOI] [PubMed] [Google Scholar]
- 7.Schachter H, et al. The role of defective glycosylation in congenital muscular dystrophy. Glycoconjugate J. 2004;20:291–300. doi: 10.1023/B:GLYC.0000033626.65127.e4. [DOI] [PubMed] [Google Scholar]
- 8.Endo T, Toda T. Glycosylation and congenital muscular dystrophies. Biol Pharm Bull. 2003;26:1641–1647. doi: 10.1248/bpb.26.1641. [DOI] [PubMed] [Google Scholar]
- 9.Michele DE, et al. Post-translational disruption of dystroglycan–ligand interactions in congenital muscular dystrophies. Nature. 2002;418:417–422. doi: 10.1038/nature00837. [DOI] [PubMed] [Google Scholar]
- 10.Beltran-Valero de Bernabe D, et al. Mutations in the O-mannosyltransferase gene POMT1 give rise to the severe neuronal migration disorder Walker–Warburg syndrome. Am J Hum Genet. 2002;71:1033–1043. doi: 10.1086/342975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Reeuwijk J, et al. POMT2 mutations cause alpha-dystroglycan hypoglycosylation and Walker–Warburg syndrome. J Med Genet. 2005;42:907–912. doi: 10.1136/jmg.2005.031963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshida A, et al. Muscular dystrophy and neuronal migration disorder caused by mutations in a glycosyltransferase, POMGnT1. Dev Cell. 2001;1:717–724. doi: 10.1016/s1534-5807(01)00070-3. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi K, et al. An ancient retrotransposonal insertion causes Fukuyama-type congenital muscular dystrophy. Nature. 1998;394:388–392. doi: 10.1038/28653. [DOI] [PubMed] [Google Scholar]
- 14.Brockington M, et al. Mutations in the fukutin-related protein gene (FKRP) cause a form of congenital muscular dystrophy with secondary laminin α2 deficiency and abnormal glycosylation of α-dystroglycan. Am J Hum Genet. 2001;69:1198–1209. doi: 10.1086/324412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Longman C, et al. Mutations in the human LARGE gene cause MDC1D, a novel form of congenital muscular dystrophy with severe mental retardation and abnormal glycosylation of α-dystroglycan. Hum Mol Genet. 2003;12:2853–2861. doi: 10.1093/hmg/ddg307. [DOI] [PubMed] [Google Scholar]
- 16.Brockington M, et al. Mutations in the fukutin-related protein gene (FKRP) identify limb girdle muscular dystrophy 2I as a milder allelic variant of congenital muscular dystrophy MDC1C. Hum Mol Genet. 2001;10:2851–2859. doi: 10.1093/hmg/10.25.2851. [DOI] [PubMed] [Google Scholar]
- 17.Qu Q, Smith FI. Alpha-dystroglycan interactions affect cerebellar granule neuron migration. J Neurosci Res. 2004;76:771–782. doi: 10.1002/jnr.20129. [DOI] [PubMed] [Google Scholar]
- 18.Qu Q, Smith FI. Neuronal migration defects in cerebellum of the Largemyd mouse are associated with disruptions in Bergman glia organization and delayed migration of granule neurons. Cerebellum. 2005;4:261–270. doi: 10.1080/14734220500358351. [DOI] [PubMed] [Google Scholar]
- 19.Montanaro F, Carbonetto S. Targeting dystroglycan in the brain. Neuron. 2003;37:193–196. doi: 10.1016/s0896-6273(03)00032-1. [DOI] [PubMed] [Google Scholar]
- 20.Topaloglu H, et al. FKRP gene mutations cause congenital muscular dystrophy, mental retardation, and cerebellar cysts. Neurology. 2003;60:988–992. doi: 10.1212/01.wnl.0000052996.14099.dc. [DOI] [PubMed] [Google Scholar]
- 21.Louhichi N, et al. New FKRP mutations causing congenital muscular dystrophy associated with mental retardation and central nervous system abnormalities: identification of a founder mutation in Tunisian families. Neurogenetics. 2004;5:27–34. doi: 10.1007/s10048-003-0165-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beltran-Valero de Bernabe D, et al. Mutations in the FKRP gene can cause muscle–eye–brain disease and Walker–Warburg syndrome. J Med Gen. 2004;41:e61. doi: 10.1136/jmg.2003.013870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Paula F, et al. Asymptomatic carriers for homozygous novel mutations in the FKRP gene: the other end of the spectrum. Eur J Hum Genet. 2003;11:923–930. doi: 10.1038/sj.ejhg.5201066. [DOI] [PubMed] [Google Scholar]
- 24.Kondo-Iida E, et al. Novel mutations and genotype–phenotype relationships in 107 families with Fukuyama-type congenital muscular dystrophy (FCMD) Hum Mol Genet. 1999;8:2303–2309. doi: 10.1093/hmg/8.12.2303. [DOI] [PubMed] [Google Scholar]
- 25.Beltran–Valero de Bernabe D, et al. A homozygous nonsense mutation in the Fukutin gene causes a Walker–Warburg syndrome phenotype. J Med Genet. 2003;40:835–838. doi: 10.1136/jmg.40.11.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balci B, et al. An autosomal recessive limb girdle muscular dystrophy (LGMD2) with mild mental retardation is allelic to Walker–Warburg syndrome (WWS) caused by a mutation in the POMT1 gene. Neuromuscul Disord. 2005;15:271–275. doi: 10.1016/j.nmd.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 27.Taniguchi K, et al. Worldwide distribution and broader clinical spectrum of muscle–eye–brain disease. Hum Mol Genet. 2003;12:527–534. doi: 10.1093/hmg/ddg043. [DOI] [PubMed] [Google Scholar]
- 28.Haliloglu G, et al. Clinical spectrum of muscle–eye–brain disease: from the typical presentation to severe autistic features. Acta Myol. 2004;23:137–139. [PubMed] [Google Scholar]
- 29.Ervasti JM, Campbell KP. Membrane organization of the dystrophin–glycoprotein complex. Cell. 1991;66:1121–1131. doi: 10.1016/0092-8674(91)90035-w. [DOI] [PubMed] [Google Scholar]
- 30.Ibraghimov-Beskrovnaya O, et al. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature. 1992;355:696–702. doi: 10.1038/355696a0. [DOI] [PubMed] [Google Scholar]
- 31.Martin PT. Dystroglycan glycosylation and its role in matrix binding in skeletal muscle. Glycobiology. 2003;13:55R–66R. doi: 10.1093/glycob/cwg076. [DOI] [PubMed] [Google Scholar]
- 32.Blake DJ, et al. Function and genetics of dystrophin and dystrophin-associated proteins in muscle. Physiol Rev. 2002;82:291–329. doi: 10.1152/physrev.00028.2001. [DOI] [PubMed] [Google Scholar]
- 33.Mathews KD, Moore SA. Limb-girdle muscular dystrophy. Curr Neurol Neurosci. 2003;3:78–85. doi: 10.1007/s11910-003-0042-9. [DOI] [PubMed] [Google Scholar]
- 34.Chiba A, et al. Structures of sialylated O-linked oligosaccharides of bovine peripheral nerve α-dystroglycan. J Biol Chem. 1997;272:2156–2162. doi: 10.1074/jbc.272.4.2156. [DOI] [PubMed] [Google Scholar]
- 35.Sasaki T, et al. Detection of O-mannosyl glycans in rabbit skeletal muscle α-dystroglycan. Biochem Biophys Acta. 1998;1425:599–606. doi: 10.1016/s0304-4165(98)00114-7. [DOI] [PubMed] [Google Scholar]
- 36.Smalheiser NR, et al. Structural analysis of sequences O-linked to mannose reveals a novel Lewis X structure in cranin (dystroglycan) purified from sheep brain. J Biol Chem. 1998;273:23689–23703. doi: 10.1074/jbc.273.37.23698. [DOI] [PubMed] [Google Scholar]
- 37.Willer T, et al. O-mannosyl glycans: from yeast to novel associations with human disease. Curr Opin Stru Biol. 2003;13:621–630. doi: 10.1016/j.sbi.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Moore SA, et al. Deletion of brain dystroglycan recapitulates aspects of congenital muscular dystrophy. Nature. 2002;418:422–425. doi: 10.1038/nature00838. [DOI] [PubMed] [Google Scholar]
- 39.Saito F, et al. Unique role of dystroglycan in peripheral nerve myelination, nodal structure, and sodium channel stabilization. Neuron. 2003;38:747–758. doi: 10.1016/s0896-6273(03)00301-5. [DOI] [PubMed] [Google Scholar]
- 40.Cohn RD, et al. Disruption of dag1 in differentiated skeletal muscle reveals a role for dystroglycan in muscle regeneration. Cell. 2002;110:639–648. doi: 10.1016/s0092-8674(02)00907-8. [DOI] [PubMed] [Google Scholar]
- 41.Martin PT, et al. Distinct structures and functions of related pre- and postsynaptic carbohydrates at the mammalian neuromuscular junction. Mol Cell Neurosci. 1999;13:105–118. doi: 10.1006/mcne.1999.0737. [DOI] [PubMed] [Google Scholar]
- 42.Akasaka-Manya K, et al. Mutations in the POMT1 gene found in patients with Walker–Warburg syndrome lead to a defect in protein O-mannosylation. Biochem Biophys Res Commun. 2003;325:75–79. doi: 10.1016/j.bbrc.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 43.Ichimiya T, et al. The twisted abdomen phenotype of Drosophila POMT1 and POMT2 mutants coincides with their heterophilic protein O-mannosyltransferase activity. J Biol Chem. 2004;279:42638–42647. doi: 10.1074/jbc.M404900200. [DOI] [PubMed] [Google Scholar]
- 44.Manya H, et al. Demonstration of mammalian protein O-mannosyltransferase activity: coexpression of POMT1 and POMT2 required for enzymatic activity. Proc Natl Acad Sci USA. 2004;101:500–505. doi: 10.1073/pnas.0307228101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willer T, et al. Targeted disruption of the Walker–Warburg syndrome gene Pomt1 in mouse results in embryonic lethality. Proc Natl Acad Sci USA. 2004;101:14126–14131. doi: 10.1073/pnas.0405899101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williamson RA, et al. Dystroglycan is essential for early embryonic development: disruption of Reichert’s membrane in Dag-1 null mice. Hum Mol Genet. 1997;6:831–841. doi: 10.1093/hmg/6.6.831. [DOI] [PubMed] [Google Scholar]
- 47.Jayasinha V, et al. Inhibition of dystroglycan cleavage causes muscular dystrophy in transgenic mice. Neuromuscul Disord. 2003;13:365–375. doi: 10.1016/s0960-8966(03)00040-3. [DOI] [PubMed] [Google Scholar]
- 48.Manya H, et al. Loss-of-function of an N-acetylgl ucosaminyltransferase, POMGnT1, in muscle–eye–brain disease. Biochem Biophys Res Commun. 2003;306:93–97. doi: 10.1016/s0006-291x(03)00924-0. [DOI] [PubMed] [Google Scholar]
- 49.Zhang W, et al. Enzymatic diagnostic test for muscle–eye–brain type congenital muscular dystrophy using commercially available reagents. Clin Biochem. 2003;36:339–344. doi: 10.1016/s0009-9120(03)00036-5. [DOI] [PubMed] [Google Scholar]
- 50.Vajsar J, et al. Carriers and patients with muscle–eye–brain disease can be rapidly diagnosed by enzymatic analysis of fibroblasts and lymphoblasts. Neuromuscul Disord. 2006 doi: 10.1016/j.nmd.2005.11.012. [doi:10.1016/j.nmd.2005.11.012] [DOI] [PubMed] [Google Scholar]
- 51.Hayashi YK, et al. Selective deficiency of α-dystroglycan in Fukuyama-type congenital muscular dystrophy. Neurology. 2001;57:115–121. doi: 10.1212/wnl.57.1.115. [DOI] [PubMed] [Google Scholar]
- 52.Matsumoto H, et al. Subcellular localization of fukutin and fukutin-related protein in muscle cells. J Biochem. 2004;135:709–712. doi: 10.1093/jb/mvh086. [DOI] [PubMed] [Google Scholar]
- 53.Takeda S, et al. Fukutin is required for maintenance of muscle integrity, cortical histiogenesis and normal eye development. Hum Mol Genet. 2003;12:1449–1459. doi: 10.1093/hmg/ddg153. [DOI] [PubMed] [Google Scholar]
- 54.Torelli S, et al. Sub-cellular localisation of fukutin related protein in different cell lines and in the muscle of patients with MDC1C and LGMD2I. Neuromuscul Disord. 2005;15:836–843. doi: 10.1016/j.nmd.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 55.Brown SC, et al. Abnormalities in α-dystroglycan expression in MDC1C and LGMD2I muscular dystrophies. Am J Pathol. 2004;164:727–737. doi: 10.1016/s0002-9440(10)63160-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grewal PK, et al. Mutant glycosyltransferase and altered glycosylation of α-dystroglycan in the myodystrophy mouse. Nature Genet. 2001;28:151–154. doi: 10.1038/88865. [DOI] [PubMed] [Google Scholar]
- 57.Barresi R, et al. LARGE can functionally bypass α-dystroglycan glycoyslation defects in distinct congenital muscular dystrophies. Nature Med. 2004;10:696–703. doi: 10.1038/nm1059. [DOI] [PubMed] [Google Scholar]
- 58.Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 59.Patnaik SK, Stanley P. Mouse large can modify complex N- and mucin O-glycans on α-dystroglycan to induce laminin binding. J Biol Chem. 2005;280:20851–20859. doi: 10.1074/jbc.M500069200. [DOI] [PubMed] [Google Scholar]
- 60.Combs AC, Ervasti JM. Enhanced laminin binding by α-dystroglycan after enzymatic digestion. Biochem J. 2005;390:303–309. doi: 10.1042/BJ20050375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fujimura K, et al. LARGE2 facilitates the maturation of α-dystroglycan more effectively than LARGE. Biochem Biophys Res Commun. 2005;329:1162–1171. doi: 10.1016/j.bbrc.2005.02.082. [DOI] [PubMed] [Google Scholar]
- 62.Grewal PK, et al. Characterization of the LARGE family of putative glycosyltransferases associated with dystroglycanopathies. Glycobiology. 2005;15:912–923. doi: 10.1093/glycob/cwi094. [DOI] [PubMed] [Google Scholar]
- 63.Brockington M, et al. Localization and functional analysis of the LARGE family of glycosyltransferases: significance for muscular dystrophy. Hum Mol Genet. 2005;14:657–665. doi: 10.1093/hmg/ddi062. [DOI] [PubMed] [Google Scholar]