Abstract
Gα subunits act to regulate vegetative growth, conidiation, and the mycoparasitic response in Trichoderma atroviride. To extend our knowledge on G protein signalling, we analysed G protein-coupled receptors (GPCRs). As the genome sequence of T. atroviride is not publicly available yet, we carried out an in silico exploration of the genome database of the close relative T. reesei. Twenty genes encoding putative GPCRs distributed over eight classes and additional 35 proteins similar to the Magnaporthe grisea PTH11 receptor were identified. Subsequently, four T. atroviride GPCR-encoding genes were isolated and affliated to the cAMP receptor-like family by phylogenetic and topological analyses. All four genes showed lowest expression on glycerol and highest mRNA levels upon carbon starvation. Transcription of gpr3 and gpr4 responded to exogenously added cAMP and the shift from liquid to solid media. gpr3 mRNA levels also responded to the presence of fungal hyphae or cellulose membranes. Further characterisation of mutants bearing a gpr1-silencing construct revealed that Gpr1 is essential for vegetative growth, conidiation and conidial germination. Four genes encoding the first GPCRs described in Trichoderma were isolated and their expression characterized. At least one of these GPCRs is important for several cellular processes, supporting the fundamental role of G protein signalling in this fungus.
Keywords: Trichoderma, G protein signalling, GPCR, Gene expression
Introduction
Heterotrimeric G protein signalling is basically comprised of three parts: a G protein-coupled receptor (GPCR), a heterotrimeric G protein (α, β, γ subunits), and an effector (Neer 1995). GPCRs constitute the vast majority of receptors, thereby forming one of the largest protein families found in nature with more than 600 members in the human genome (Lander et al. 2001; Venter et al. 2001). GPCRs do not have significant sequence similarity but share a common domain structure containing seven transmembrane helices that are connected by intra- and extracellular loops (Dohlman et al. 1991). Ligand binding to the receptor results in a conformational change leading to release of the G protein and exchange of GDP for GTP on the Gα subunit. GTP-bound α dissociates from its βγ partner, allowing both signalling units to regulate the activities of downstream effectors (Kaziro et al. 1991; Birnbaumer et al. 1992; Neer 1995; Gutkind 1998).
In fungi, two signalling branches defined by cAMP-dependent protein kinase (PKA) or MAPK cascades primarily relay G protein-mediated signals to elicit cellular responses such as growth, mating, cell division, cell–cell fusion, morphogenesis, chemotaxis, and pathogenic development (Bölker 1998; Versele et al. 2001). Fungal Gα subunits are highly conserved and can be divided into three subgroups (Bölker 1998). As heterotrimeric G proteins act as signal transducers that connect cell surface receptors to cytoplasmic effectors, GPCRs are expected to play essential roles in the transduction of extracellular signals. Nevertheless, little information is available on the characteristics and functions of fungal GPCRs other than pheromone receptors which have been examined in fungi such as Saccharomyces cerevisiae (Hagen et al. 1986; Blumer et al. 1988), Schizosaccharomyces pombe (Tanaka et al. 1993), Cryptococcus neoformans (Chang et al. 2003), Aspergillus nidulans (Seo et al. 2004), Ustilago maydis (Bölker et al. 1992), Neurospora crassa (Kim and Borkovich 2004), and Coprinus cinereus (Olesnicky et al. 1999).
The N. crassa genome sequence was the first available database for a filamentous fungus, and during its initial annotation 10 predicted seven-transmembrane helix GPCRs were found and divided into five families (Borkovich et al. 2004). Subsequent to the release of the genome of Magnaporthe grisea, Kulkarni et al. (2005) annotated 76 GPCR-like proteins of which 61 represented a large novel class related to PTH11, a receptor required for development of the appressorium (DeZwaan et al. 1999). Recently, Lafon et al. (2006) reported on in silico exploration of G protein signalling in the genomes of A. nidulans, A. fumigatus, and A. oryzae. The investigations resulted in classification of GPCRs into nine classes: classes I and II: pheromone receptors; classes III and V: putative carbon and cAMP sensors; class IV: nitrogen sensors; class VI: a new class of GPCRs with an RGS (regulator of G protein signalling) domain; classes VII and VIII: members of class VII like MG00532.4 of M. grisea, and GprM and GprN of A. nidulans are related to different mammalian GPCRs including the rat growth hormone releasing factor receptor, and class VIII comprises fungal GPCRs that are homologous to the steroid receptor mPR; and class IX: fungal opsins. In the meantime, a more detailed analysis of the N. crassa proteome resulted in additional 29 putative GPCRs, 25 of them representing homologues of M. grisea PTH11 (Li et al. 2007).
Generally, analyses of fungal genomes confirmed the high diversity of GPCRs compared to the little diversity in G proteins and components of the cAMP-signalling pathway such as PKA and adenylate cyclase. This may reflect the ability of fungi to integrate a broad spectrum of environmental cues into highly conserved cellular processes.
Analysis of G protein signalling in T. atroviride revealed that subgroup I and III Gα proteins play crucial roles during vegetative growth and conidiation and the attack of phytopathogenic host fungi (Rocha-Ramirez et al. 2002; Reithner et al. 2005; Zeilinger et al. 2005). Until now, no GPCRs have been isolated or studied from any Trichoderma spp. although their characterisation should provide a deeper insight into the interrelation between the signal recognizing receptor and the triggered cellular response. Clarifying the role of receptors would be a fundamental step toward understanding the role of G protein signalling in Trichoderma.
Recently, the first genome sequence of a member of the genus Trichoderma, T. reesei, became publicly available. Here we present a genomic exploration of the T. reesei database resulting in the identification of 20 GPCRs distributed over classes I to VIII according to Lafon et al. (2006) and additional 35 GPCRs related to the M. grisea PTH11 protein. Based on this analysis, we isolated four GPCR-encoding genes from the mycoparasite T. atroviride and analysed their expression under different growth conditions. Further characterization of the isolated T. atroviride GPCR-encoding genes showed that at least gpr1 plays a major role during vegetative growth and conidiation in T. atroviride. To our knowledge, this is the first report on GPCRs from a biocontrol fungus.
Materials and methods
Genome mining for the identification of genes encoding GPCR-like proteins
The genomic sequence of T. reesei is available at http://genome.jgi-psf.org/Trire2/Trire2.home.html. The following genomic sequences and deduced proteomes were searched as basis for identifying GPCR-like proteins from T. reesei: A. nidulans (http://www.broad.mit.edu/annotation/fungi/aspergillus/index.html), A. fumigatus (http://www.tigr.org/tdb/e2k1/afu), A. oryzae (http://www.bio.nite.go.jp/dogan/MicroTop?GENOME_ID=ao), N. crassa (http://www.broad.mit.edu/annotation/genome/neurospora/Home.html), M. grisea (http://www.broad.mit.edu/annotation/fungi/magnaporthe), Podospora anserina (http://podospora.igmors.u-psud.fr), Chaetomium globosum (http://www.broad.mit.edu/annotation/genome/chaetomium_globosum/Home.html), Fusarium graminearum (http://mips.gsf.de/genre/proj/fusarium), and Nectria haematococca (http://genome.jgi-psf.org/Necha2/Necha2.home.html).
Besides BLAST searches, a more sensitive database searching using hidden Markov models (HMM) was performed to identify putative GPCRs within the T. reesei proteome that lack significant sequence similarity to known GPCR-like proteins. To this end, the program HMMER (http://hmmer.wustl.edu; Eddy 1998) was applied.
For manual gene prediction the FGENESH algorithm (Softberry, Mount Kisco, NY) was applied to the raw sequence data and the predicted proteins were tested with TMHMM (Krogh et al. 2001) for transmembrane helices. ClustalW (http://www.ebi.ac.uk/clustalw) was used for alignment and phylogenetic analysis.
Strains and media
Trichoderma atroviride P1 (ATCC 74058; teleomorph Hypocrea atroviridis) was used throughout this study and maintained on potato dextrose agar (PDA). To monitor the transcription levels of GPCR-encoding genes in liquid medium augmented with different carbon sources the fungus was grown in synthetic medium (SM) containing [g/l]: (NH4)2SO4 1.4, KH2PO4 2, CaCl2·2H2O 0.3, MgSO4·7H2O 0.3, FeSO4·7H2O 0.01, ZnSO4·H2O 0.0028, CoCl2·6H2O 0.0032; with 2% glycerol as carbon source. After 36 h the mycelia were transferred to SM with 1% (w/v) of glucose, glycerol, N-acetyl-glucosamine, or colloidal chitin. For the determination of growth rates, fungi were inoculated at the centre of PDA plates and the colony diameter recorded every 24 h. Rhizoctonia solani strain 1450 (strain collection of the Institute of Plant Pathology, University of Naples Federico II, Italy) was used as a plant pathogenic host and was maintained on PDA. Escherichia coli JM109 served as a host for plasmid amplification and was grown as described by Sambrook et al. (1989).
For plate confrontation assays, 5-mm disks of T. atroviride and T. atroviride, or T. atroviride and R. solani, were placed on cellophane-covered PDA plates at a distance from each other of 4 cm. The plates were incubated at 25°C for 3 days in the absence of light. For dual-plate assays avoiding physical contact the two opposite organisms were separated by a tape of cellophane or a dialysis membrane (cutoff 12 kDa) embedded in the agar in the middle of the Petri dish in a way forming a barrier between the fungi as described previously by Zeilinger et al. (1999).
Transmitting light microscopy
Microscopic studies were mainly performed as previously described by Lu et al. (2004). Glass slides, on which 500 μl of PDA was spread onto, were inoculated and incubated on a moistened filter paper (Gel-blotting paper; Roth, Karlsruhe, Germany) at 28°C in a petri dish sealed with Parafilm. After 48–72 h, the fungal hyphae were viewed under a Leitz Aristoplan (Wetzlar, Germany) microscope, and pictures were taken using an Olympus DP 10 (Olympus America Inc., Melville, NY) camera.
DNA and RNA procedures
Standard molecular techniques were performed according to Sambrook et al. (1989); DNA and RNA isolation was carried out as described by Peterbauer et al. (1996). For standard PCR amplication, recombinant Taq Polymerase (Fermentas, Vilnius, Lithuania) was used according to the manufacturer’s recommendations.
Cloning and sequencing of T. atroviride reference genes
Diagnostic fragments of the following genes were cloned and sequenced for their subsequent use as reference for relative-quantitative real-time PCR: act1 (actin-encoding), gpd1 (glyceraldehyde 3-phosphate-dehydrogenase-encoding), tef1 (translation elongation factor-encoding), sar1 (small GTP binding protein-encoding), and a fragment of the 28S rDNA. Primer pairs for amplification of the respective fragments from genomic DNA are given in Table 1a. The PCR amplicons were cloned into pGEM-T (Promega, Madison, WI) and sequenced (MWG Biotech, Ebersberg, Germany) using the standard sequencing primers SP6 and T7. All nucleotide sequences have been submitted to GenBank (GenBank accession numbers: EF581847, EF581848, EF581849, EF581850, EF591763). Primers for real-time PCR monitoring of these genes were designed with Vector-NTI (Invitrogen, Carlsbad, CA).
Table 1.
Oligonucleotides used in this study
| Gene | Forward primera (5′ to 3′) | Reverse primer (5′ to 3′) | Product (bp) |
|---|---|---|---|
| (a) Degenerate primer pairs used to amplify fragments of reference genes | |||
| act1 | GTIGCICCIGARGARCAYCCI | ACRTCIACRTCRCAYTTCAT | 548 |
| gpd1 | GTIAARATHGTIAAYGAYAAYTT | GCIACIACISWITRYTCIGGYTT | 873 |
| tef1 | GTCCTCAGTCTTGTCATTTTTTTTCCT | GGAGGGGCCTGTCTGTGGGAC | 906 |
| sar1 | CTCGACAAYGCCGGAAAGAC | TTGCCAAGGATGACAAAGGGG | 833 |
| 28S | GGAACCTTTCCCCACTTC | AGTACCCGCTGAACTTAA | 1,461 |
| (b) Primer pairs used for quantification of transcripts by real-time PCR | |||
| gpr1 | TTGATCCAGACCTTCATGCCAGC | CATAAAAGGCCGCGACACGAA | |
| gpr2 | CTGCCCTCGTGTACATCTTCC | GCCTTCAGATGAGTGAAAGTCG | |
| gpr3 | GCCAGCATTGGAACCATCATCG | CCCAACACCAGATCGTAGCTCCA | |
| gpr4 | ATTCTAACTGGAGCCTGGTC | GTACTTTGTGGCTCTGGTTG | |
| act1 | GCACGGAATCGCTCGTTG | TTCTCCACCCCGCCAAGC | |
| gpd1 | CGTCGTAGTCCTTGTGGTTGACACC | TCCTCCCACGGTCTCTTCAAGG | |
| sar1 | CTCGACAATGCCGGAAAGACCA | TTGCCAAGGATGACAAAGGGG | |
| tef1 | TACTTCCCAGGCTGACTGCGCTAT | GGAGGGGCCTGTCTGTGGGAC | |
| 28S | TTTGAGTAAGAGCATACGGGGCC | GTTGATACATTCGAATGCCCACGT | |
R = A or G, Y = C or T, I = inosine (panel A)
Cloning of four T. atroviride genes coding for putative GPCRs
Two pairs of degenerate primers were used for PCR on genomic T. atroviride DNA: Gpr-I-f: 5′-AAYMGRCTGGTCTTTTATGCKTCGTTYGGC-3′, Gpr-I-r: 5′-TCGGTTGGCGCTCGASGGMATCC-3′, and Gpr-II-f: 5′-ATGTTYATGCARWIGAYCCITGGTGG-3′, Gpr-II-r: 5′-CKIARRTAIGCIMDYTTDATIGG-3′. For amplification, the following PCR program was applied: 3 min initial denaturation at 95°C followed by 35 cycles with 30 s at 95°C, 30 s at 56.5°C for Gpr-I or 30 s at 53°C for Gpr-II, and 1 min elongation at 72°C.
A LambdaBlueStar genomic library (Novagen/Merck, Darmstadt, Germany) of T. atroviride P1 was screened with the respective PCR fragments to obtain full length clones of the putative GPCR-encoding genes. The screening was performed according to the manufacturer’s manual. The nucleotide and putative protein sequences were deposited at GenBank (accession numbers: DQ284755, DQ384865, EF534712, EU391628).
Real-time RT-PCR
The sequences for the respective primer pairs for cDNA amplification of the reference genes and the four receptors are given in Table 1b. All quantifications were performed with the same PCR program: initial denaturation for 90 s at 95°C, 50 cycles with 95°C for 20 s, 60°C for 20 s and 72°C for 30 s. For cDNA synthesis the Revert Aid H Minus First Strand cDNA Synthesis Kit (Fermentas, Vilnius, Lithuania) was used according to the manufacturer’s instructions with a combination of the provided oligo-dT and random primers. All real-time PCR experiments were conducted on a Bio-Rad (Hercules, CA) iCycler IQ. For iCycler reaction the IQ SYBR Green Supermix (Bio-Rad, Hercules, CA) was prepared for 25 μl assays with standard MgCl2 concentration (3 mM) and with final primer concentrations of 100 nM each. All assays were carried out in 96-well plates which were covered with optical tape. PCR efficiency was determined from a single tube reaction set-up as described by Tichopad et al. (2003) and expression ratio was calculated according to the equation published by Pfaffl (2001). All samples were analyzed in three independent experiments with three replicates in each run.
Gene silencing procedure and construction of pSilent2
The method used to silence gpr1 and gpr3 gene expression in T. atroviride P1 was based on the construction of self-complementary RNAs (inverted repeats). These should activate the short interfering (si)RNA pathway mediated by the action of the Dicer complex resulting at the end in the endonucleolytic cleavage of the target mRNAs (Dykxhoorn et al. 2003).
An 850-bp fragment of the A. nidulans gpdA promoter was cloned between the XhoI and the ClaI sites of the vector pBluescriptSK+ (Stratagene, Cedar Creek, TX) and a 720-bp fragment of the A. nidulans trpC terminator was inserted between BamHI and XbaI. The hygromycin resistance-conferring cassette of pRLMex30 (Mach et al. 1994) was inserted at the NotI site of pBluescriptSK+, resulting in vector pSilent2.
Fragments of the gpr1 and gpr3 target genes were amplified with the following modified primers (a HindIII site was inserted into the forward primer and a XbaI site into the reverse primer): gpr1-fw, 5′-AAGCTTCCCATATCAATCGACGAGGTCACC-3′ and gpr1-rev, 5′-TCTAGATTGTCTCGCTCGTGATTTCGC-3′ (amplicon length 359-bp); gpr3-fw, 5′-AAGCTTGGATCTACTGCCTAATTTGTTACG-3′ and gpr3-rev, 5′-TCTAGAGAACTTCGGTAGTGACGGTGC-3′ (amplicon length 464-bp). All PCR reactions were performed at an annealing temperature of 56°C. Subsequently, the amplicons were digested with XbaI and self-ligated to obtain inverted repeats flanked by HindIII sites. 5 μl of the ligation mixture were used for a 35-cycle single-primer PCR with the forward primers previously used to amplify the respective fragments. The amplified inverted repeats were digested with HindIII and inserted into pSilent2 resulting in pSilent2-gpr1 and pSilent2-gpr3, respectively.
Transformation of T. atroviride
Trichoderma atroviride protoplasts were transformed with 10 μg of each silencing vector as described by Peterbauer et al. (2002). HygromycinB-resistant strains were transferred to PDA for sporulation and colonies obtained from three subsequent single spore isolations were used for further analysis. The integration of the respective dsRNA expressing constructs was tested by PCR with primers binding to the gpdA and trpC regulatory sequences: SilTest-f: 5′-AATGTGAAGCCAGGGGTGTATAGC-3′ and SilTest-r: 5′-GTCTCCCGAAAATGAAAATAGCTC-3′.
Biolog phenotype array analysis
The Biolog FF MicroPlate assay (Biolog Inc., Hayward, CA) which comprises 95 wells with different carbon-containing compounds and one well with water was used to investigate growth rates on selected carbon sources. Nutrients and test reagents are prefilled and dried into the 96 wells of the microplate. Conidia were collected from the Trichoderma strains and used as inoculums as described (Druzhinina et al. 2006). Inoculated microplates were incubated in darkness at 25°C, and OD750 readings determined after 18, 24, 42, 48, 66, 72, 96 and 168 h using a microplate reader (Biolog), which measures the turbidity and reflects mycelial production on the tested substrate. Analyses were performed in triplicate.
Results
Identification of genes predicted to encode GPCRs in the genome of T. reesei
At the beginning of our study, seven proteins were annotated as GPCRs in the T. reesei genome database. To get a more complete picture, we searched the T. reesei proteome database for additional GPCR-like proteins based on their similarity to known fungal receptors. To this end, GPCR sequences GprA—GprQ and NopA from A. nidulans, A. fumigatus, an A. oryzae (Lafon et al. 2006) were used as query in a BLASTP search against the predicted proteomes of species of the Sordariomycetes (N. crassa, M. grisea, P. anserina, C. globosum, F. graminearum, N. haematococca, and T. reesei), a subgroup within the Pezizomycotina/Ascomycota, to find putative orthologues in these species. The retrieved proteins from each species were subsequently used as query in similar BLAST searches of the proteomes from the other species to end up with each possible combination.
All obtained putative GPCR sequences were evaluated for seven transmembrane regions with the TMHMM algorithm (Krogh et al. 2001). For nearly all retrieved proteins the algorithm predicted the seven-span helix topology. For those not predicted to contain seven transmembrane regions, the respective encoding gene and flanking sequences were retrieved from the genome and inspected. In individual cases, mistakes at the intron-exon boundaries were found and manual correction resulted in the detection of the missing membrane-spanning domain(s). Only those proteins fulfilling the topological characteristics of GPCRs (seven transmembrane-spanning domains, amino-terminal domain predicted to face outside the cell, and the C-terminus predicted to face the cytoplasm) were included in further analyses.
BLASTP searches using the M. grisea PTH11 receptor as query against the predicted proteomes of the above listed species of the Sordariomycetes resulted in a number of proteins with seven transmembrane regions. Again, the retrieved proteins from each species were subsequently used to BLAST in a second round the proteomes from the other species to end up with each possible combination.
In total, these combinatorial BLAST searches resulted in the identification of 52 T. reesei GPCR-like proteins including 32 PTH11-like receptors. To also identify putative GPCRs that lack significant sequence similarity to known GPCR-like proteins and therefore may escape detection by homology search, the T. reesei genome database was again queried using a HMM method. Applying the program HMMER, three additional PTH11-like receptors (Tr_69904, Tr_105224, Tr_122795) could be identified (Table 2). Phylogenetic analysis based on an alignment of all identified putative T. reesei GPCRs other than PTH11-like receptors with those identified in Aspergillus spp. was performed. The resulting tree (Fig. 1) exhibits the characteristic nine classes previously described for Aspergillus spp. (Lafon et al. 2006), grouping the 20 T. reesei GPCRs into classes I to VIII: (I and II) Tr_57526 and Tr_64018 (numbers indicate protein ID in the genome database) similar to pheromone receptors GprA (Ste2-like) and GprB (Ste3-like) from Aspergillus sp., respectively; (III) Tr_59778 similar to the putative carbon sensors GprC, GprD and GprE from Aspergillus sp.; (IV) Tr_4508, Tr_80125 and Tr_111600 similar to the putative Aspergillus nitrogen sensors GprF, GprG and GprJ; (V) Tr_28731, Tr_38672, Tr_72605, and Tr_123806 similar to the putative Aspergillus cAMP sensors GprH, GprI and GprL; (VI) Tr_23297, Tr_37525, Tr_63981, and Tr_81383 similar to the RGS domain-containing receptor GprK from Aspergillus sp.; (VII) Tr_53238 similar to Aspergillus GprM and GprN; (VIII) Tr_56426, Tr_68212, Tr_70139, Tr_82246, and Tr_119819 similar to the mPR_dom domain-containing proteins GprO, GprP, and GprQ from Aspergillus sp. Interestingly, we could not identify any member of class IX which contains receptors similar to bacterial opsins.
Table 2.
GPCRs identified in the T. reesei genome database
| Receptor class | T. reesei | Class | T. reesei | |
|---|---|---|---|---|
| I Pheromone | Tr_57526 | PTH11-like | Tr_5647 | Tr_82041 |
| II Pheromone | Tr_64018 | Tr_27983 | Tr_103694 | |
| III Carbon sensor | Tr_59778 | Tr_27992 | Tr_105224 | |
| IV Nitrogen sensor | Tr_4508 | Tr_39587 | Tr_106082 | |
| Tr_80125 | Tr_40156 | Tr_107042 | ||
| Tr_111600 | Tr_41260 | Tr_109146 | ||
| V cAMP receptor-like | Tr_72605 | Tr_41425 | Tr_110339 | |
| Tr_38672 | Tr_45573 | Tr_110744 | ||
| Tr_28731 | Tr_53452 | Tr_111861 | ||
| Tr_123806 | Tr_55561 | Tr_121990 | ||
| VI RGS domain-containing | Tr_63981 | Tr_57101 | Tr_122795 | |
| Tr_81383 | Tr_58767 | Tr_122824 | ||
| Tr_37525 | Tr_61354 | Tr_124113 | ||
| Tr_23297 | Tr_62462 | |||
| VII | Tr_53238 | Tr_66673 | ||
| VIII Similar to mPR | Tr_119819 | Tr_66786 | ||
| Tr_68212 | Tr_67334 | |||
| Tr_70139 | Tr_69500 | |||
| Tr_82246 | Tr_69904 | |||
| Tr_56426 | Tr_70967 | |||
| IX Fungal opsins | nd | Tr_76763 | ||
| Tr_78499 |
Fig. 1.
Cladogram of the phylogenetic relationship between putative GPCRs identified in the T. reesei predicted proteome (Table 2) and those previously identified in A. nidulans, A. fumigatus and A. oryzae (Lafon et al. 2006). The tree was generated using the CLUSTALX alignment
Cloning and in silico analysis of putative GPCR-encoding genes from T. atroviride
A PCR-based approach was followed for the isolation of GPCR-encoding genes from T. atroviride. Until now, only GPCRs from groups I, II (Kim and Borkovich 2004; Seo et al. 2004), III (Han et al. 2004; Li and Borkovich 2006), V (Krystofova and Borkovich 2006) and IX (Bieszke et al.2007) and the M. grisea PTH11 receptor (DeZwaan et al.1999; Kulkarni et al. 2005) have been analysed in more detail in filamentous fungi. Members of the other groups have only been characterised in silico and it is still unknown if they are actually expressed. Because of these considerations, we focused on class V GPCRs from T. atroviride. Members of this class exhibit similarity to cAMP receptor-like (CRL) proteins from Dictyostelium discoideum and are absent from the genomes of the ascomycete yeasts S. cerevisiae and S. pombe.
To amplify fragments of putative T. atroviride CRL-encoding genes, two degenerate primer pairs were designed based on protein alignments of putative class V receptors obtained from the genome databases of the close relatives T. reesei (four CRL proteins) and F. graminearum (five CRL proteins). Using genomic T. atroviride DNA in a PCR approach, primer pair Gpr-I yielded two fragments of ~800 and ~1,000-bp, respectively; the Gpr-II primers produced amplicons of ~900 and ~1,000-bp, respectively. Screening of a genomic library of T. atroviride with these fragments as probes resulted in the isolation of four different clones, each corresponding to one probe. The inserts were sequenced and the derived nucleotide sequences were scanned with FGENESH (Softberry, Mount Kisco, NY) resulting in detection of seven transmembrane helices in all four predicted proteins with a distribution typical for the CRL class: five domains at the N-terminal end followed by a long intracellular loop and by two helices close to the C-terminus (Fig. 2).
Fig. 2.
Alignment of T. atroviride Gpr1–4 with predicted fungal CRL proteins from other filamentous fungi. CLUSTALX was used to align GPCR sequences from T. atroviride (Gpr1, Gpr2, Gpr3, Gpr4) with predicted proteins from T. reesei (Tr_72605, Tr_38672, Tr_28731, Tr_123806), F. graminearum (fg01861, fg07716, fg03023, fg05239, fg09693), and the N. crassa GPR–1 (Nc_GPR1) receptor. Conserved residues are shaded in black (identical) or in grey (similar). The predicted seven transmembrane domains are marked by the bars above the sequence and are numbered (TM1–TM7)
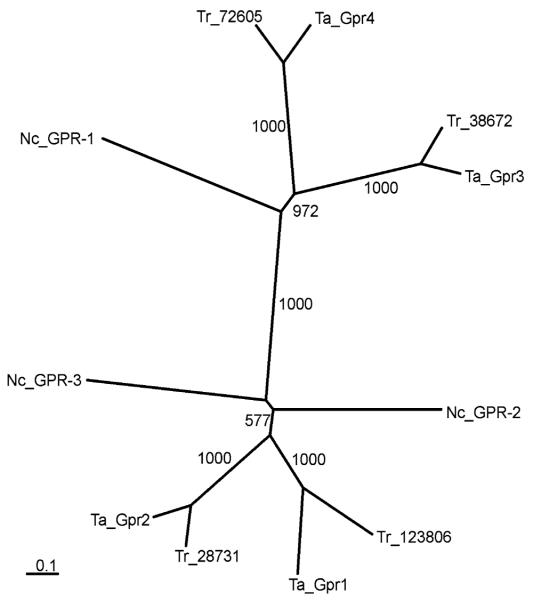
Alignment of the isolated putative T. atroviride GPCRs Gpr1, Gpr2, Gpr3, and Gpr4 with N. crassa GPR-1, the only CRL protein characterized in ascomycete fungi until now (Krystofova and Borkovich 2006), and predicted CRL proteins from T. reesei and F. graminearum (Fig. 2) and subsequent phylogenetic analysis resulted in two branches with Gpr1 and Gpr2 in one branch and Gpr3 and Gpr4 in the other branch (Fig. 3). As shown in Fig. 3, the CRL proteins from T. atroviride and T. reesei are closely related, which confirms and validates the approach used.
Fig. 3.
Phylogeny of class V GPCRs isolated from T. atroviride (Ta_Gpr1, Ta_Gpr2, Ta_Gpr3, Ta_Gpr4) and those identified in the T. reesei (Tr_72605, Tr_38672, Tr_28731, Tr_123806) and N. crassa (Nc_GPR-1 = NCU00786.3, Nc_GPR-2 = NCU04626.3, Nc_GPR-3 = NCU9427.3) genome sequences. The sequences were aligned and a tree was generated using CLUSTALX and neighbour-joining algorithm with 1,000 bootstraps
To prove that the isolated T. atroviride GPCR-encoding genes are actually transcribed and do not represent pseudogenes, transcriptional analyses were performed. As gpr1, gpr2, gpr3 and gpr4 mRNA levels were too low to be detected by Northern analysis, we performed quantitative real-time RT-PCR to determine their expression.
Cloning and evaluation of putative reference genes for real-time PCR
To allow accurate real-time PCR analysis of transcription rates, suitable reference genes with only minimal regulation of gene expression are indispensable. As only few sequences of putative reference genes were available from T. atroviride P1, fragments from the following five genes were cloned by PCR with degenerate primers (Table 1a) and sequenced: the actin-encoding gene act1, the glyceral-dehyde-3-phosphate-dehydrogenase-encoding gene gpd1, the translation elongation factor-encoding gene tef1, the GTP-binding protein-encoding gene sar1 and the gene for 28S rDNA. Primers for real-time PCR monitoring of the transcription of the respective gene were all tested for annealing at a temperature of 60°C to allow a combined use of these references for any run. All reference genes were evaluated for their constant expression under the following cultivation conditions of the fungus: (1) in liquid medium [potato dextrose broth (PDB)] and on solid agar (PDA) to allow monitoring of surface/cultivation-dependent gene expression differences, (2) after replacement of Trichoderma to liquid medium with different carbon sources (glucose, glycerol, N-acetylglucosamine, colloidal chitin, or carbon-starvation), and (3) in dual plate assays when directly interacting with different confrontation partners or when the two fungi were separated by membrane barriers. Three independent real-time PCR runs with three replicates per sample were performed for each experiment and the data were evaluated with genNorm (Vandesompele et al. 2002) to identify the most stable internal control genes for each condition. Although this algorithm leads to two winner genes, the gene that performed best during the exclusion procedure was taken for normalization during the further experiments. To compare gene transcription upon growth on PDB and PDA, tef1 turned out to be the best reference gene. To investigate mRNA levels in liquid media with different carbon sources act1 was chosen for data normalization, and mRNA levels of in vitro biocontrol conditions are best to be referenced to sar1 (Table 3).
Table 3.
Stability of reference genes according to the output of geNorm (Vandesompele et al. 2002)
| Ranking | Cultivation condition |
||
|---|---|---|---|
| PDB/PDA | Various C-sources |
Plate confrontation assays |
|
| 1/2 | act1/tef1 | act1/sar1 | sar1/gpd1 |
| 3 | 28S rRNA | tef1 | 28S rRNA |
| 4 | sar1 | gpd1 | act1 |
| 5 | gpd1 | 28S rRNA | tef1 |
Real-time PCR quantification of receptor gene expression under different cultivation conditions
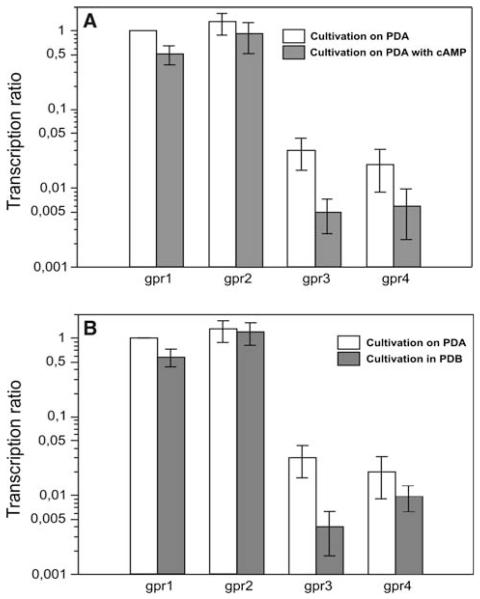
As all four putative T. atroviride receptors, designated Gpr1, Gpr2, Gpr3, and Gpr4, grouped to the CRL class V which was suggested to be involved in cAMP sensing in N. crassa (Galagan et al. 2003), we investigated the influence of extracellular cAMP added to PDA plates in a concentration of 5 mM on their gene transcription using real-time RT-PCR with sar1 as reference gene. gpr1 and gpr2 did not show significantly altered transcription in the presence of cAMP compared to growth on un-supplemented PDA, whereas expression of gpr3 was reduced by 84% and that of gpr4 was reduced by 70% on plates augmented with cAMP (Fig. 4a).
Fig. 4.
Expression of gpr1, gpr2, gpr3, and gpr4 determined by real-time RT-PCR using tef1 as a reference gene upon cultivation of T. atroviride on PDA compared to growth on PDA augmented with 5 mM cAMP (a) and on PDA compared to growth in PDB (b). To allow comparison of mRNA levels of the four GPCR-encoding genes, mRNA levels of gpr1 on PDA were arbitrarily assigned the factor 1. The transcription ratio is presented in a logarithmic scale
To test for a surface-dependent expression (i.e. differential expression between growth in liquid and on solid media) of the isolated receptor-encoding genes, T. atroviride was cultivated in potato dextrose broth (PDB) or on PDA. The compared growth conditions had no influence on gpr2 transcription, whereas gpr1 and gpr4 mRNA levels were reduced to approx. 50% on PDB compared to growth on PDA. The most striking expression decrease was observed for gpr3: its transcription was reduced by 88% when the fungus was cultivated in PDB compared to growth on PDA (Fig. 4b).
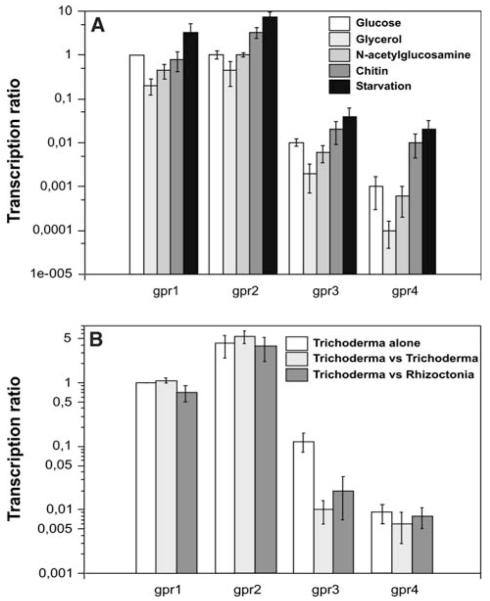
Furthermore, the effect of different carbon sources on the expression of the four receptor genes was investigated using cDNA obtained from replacement cultivations. To this end, pre-grown mycelia of the fungus were transferred to liquid media with glucose, glycerol, colloidal chitin or N-acetyl-glucosamine as sole carbon sources, the latter two representing fungal cell wall components or cell wall-derived degradation products. Additionally, cDNA from carbon-starved mycelium was integrated into this experiment. gpr1, gpr2, gpr3 and gpr4 all followed the same tendency showing lowest expression on glycerol and the highest mRNA levels when T. atroviride was transferred to a medium without carbon source (Fig. 5a) indicating that transcription of these GPCR-encoding genes is induced during carbon starvation.
Fig. 5.
Real-time RT-PCR analyses of gpr1, gpr2, gpr3 and gpr4 mRNA levels upon cultivation of T. atroviride on different carbon sources using act1 as a reference gene (a) and during in vitro biocontrol using sar1 as reference gene (b). The samples from cultures with glucose, glycerol and N-acetyl-glucosamine as sole carbon sources were harvested 12 h after transfer, those from cultures with chitin as sole carbon source or from cultures grown without carbon source were harvested 24 h after transfer. For monitoring gpr expression during in vitro biocontrol, T. atroviride P1 was either grown alone, or confronted with itself, or confronted with R. solani as host fungus. To allow comparison of mRNA levels of the four GPCR-encoding genes, mRNA levels of gpr1 on glucose (a) or PDA (b) were arbitrarily assigned the factor 1. The transcription ratio is presented in a logarithmic scale
To investigate the expression patterns of the receptor-encoding genes under in vitro biocontrol conditions, RNA was isolated from dual plate assays with various confrontation partners. T. atroviride grown alone served as the control condition and was compared with a self-confrontation (T. atroviride–T. atroviride) and with a confrontation against R. solani. gpr1, gpr2 and gpr4 showed no significant transcriptional regulation, whereas gpr3 was significantly down-regulated upon contact of T.atroviride with itself or with R. solani (Fig. 5b). To investigate if this decrease in expression of gpr3 is specific for a direct physical contact of the hyphae, we separated the confrontation partners by either a stripe of cellophane or by a dialysis membrane. As control, Trichoderma was grown against a membrane without another fungus behind this barrier. However, all experimental setups led to the same result namely a significant reduction of gpr3 transcription after contact with the membrane. Therefore, the observed effect seems not to be specific for fungal–fungal interactions but to be more general occurring upon physical contact with any object.
Silencing of gpr1 gene expression results in mutants with alterations in vegetative growth, conidiation, and conidial germination
To get further insights into putative functions of the isolated T. atroviride GPCR-encoding genes, one member of each branch (Fig. 3) was selected for functional studies. Vectors for silencing gpr1 and gpr3 were constructed and transformed into protoplasts of T. atroviride P1. Four strains transformed with the gpr1- silencing construct and five strains transformed with the gpr3- silencing construct were randomly selected for further studies. Integration of the silencing vectors was positively tested by PCR amplification of a fragment containing the gpdA promoter, the trpC terminator and the inverted repeats in-between. The PCR reaction produced diagnostic fragments of 718-bp for gpr1-silenced strains and 928-bp for gpr3-silenced transformants (data not shown).
Transformants bearing the gpr3 silencing constructs exhibited no aberrant phenotype when grown on PDA and real-time RT-PCR analysis using primers gpr3forw and gpr3rev (Table 1b) showed that they, although bearing the gpr3 silencing construct, exhibited unaltered gpr3 mRNA levels.
In contrast, real-time RT-PCR analysis using primers gpr1forw and gpr1rev (Table 1b) revealed levels of gpr1 silencing of ~60–80% in transformants bearing the gpr1-silencing construct (Fig. 6a). gpr1-silenced transformants showed an ~50% reduced growth rate compared to the parental strain upon cultivation on PDA plates (Fig. 6b). They sporulated continuously on solid medium and formed flat colonies with only little aerial mycelium and with a significant number of hyphae growing submerged in the agar. In liquid cultures in PDB or SM with 1% glycerol for 36 h, no conidiation occurred. Analysis of conidial germination after 9 h of incubation in PDB at 28°C revealed that 17% of the wild type conidia, but only 5% of the conidia of gpr1-silenced mutants had formed germ tubes; after 11 h, 67% of the wild type conidia and 34% of the gpr1-sil conidia had germinated.
Fig. 6.
(a) Real-time RT-PCR analyses of gpr1 gene expression in four selected gpr1-silenced transformants. mRNA levels of gpr1 in the parental strain were assigned the factor 1. (b) Colony morphology of the gpr1-silenced transformant gpr1sil-8 compared to the parental stain T. atroviride P1 upon growth on solid medium (PDA) in the dark after 4 days at 28°C
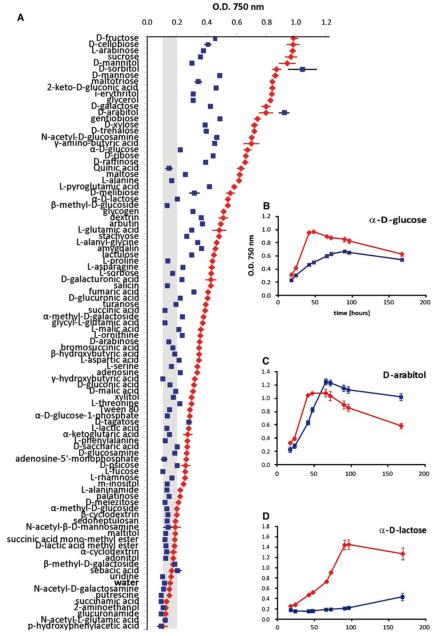
To further analyze the role of Gpr1, we used Biolog Phenotype Arrays to find specific carbon utilization properties. The carbon-source utilization profile of T. atroviride P1 has been characterized previously (Seidl et al. 2006). The authors divided the tested carbon sources into four groups by joining cluster analysis where clusters I, II, and III contain carbon sources which allow fast, moderate, and slow growth, respectively, while cluster IV contains carbon sources where the fungus grows very poorly or does not grow at all.
The gpr1-silenced mutant exhibited reduced growth on the majority of the tested carbon sources (Fig. 7) and determination of the respective growth rates between 18 and 66 h of incubation revealed a 30–70% reduction. From the carbon sources belonging to clusters I, II, and III and therefore allowing explicit growth of the parental strain, α-D-lactose and lactulose did not allow any detectable growth of the gpr1-silenced mutant during this period. On D-raffinose and maltose the mutant only grew poorly (8 and 17% of the parental strain, respectively), whereas on D-mannitol, D-arabitol, L-serine, salicin, and Tween 80 the mutant showed a similar growth rate as the parental strain.
Fig. 7.
(a) Analysis of biomass formation of the T. atroviride gpr1-silenced mutant gpr1sil-8 (filled square) in comparison to the parental strain (filled diamond) after incubation for 66 h on 95 carbon sources and water using the BIOLOG microplate assay. (b–d) Growth curves of T. atroviride parental strain (filled diamond) and the gpr1-silenced mutant (filled square) on some individual carbon sources determined after 18, 24, 42, 48, 66, 72, 96 and 168 h of incubation. The values given represent the average of three analyses
Discussion
Analysis of G protein signalling in T. atroviride revealed that the Tga1 and Tga3 Gα subunits play important roles during vegetative growth and conidiation and in regulating mycoparasitism-related genes involved in infection structure formation and chitinase as well as antifungal metabolite production, and for both an interaction with the cAMP pathway was found (Rocha-Ramirez et al. 2002; Reithner et al. 2005; Zeilinger et al. 2005). Based on these findings and to gain further insights into the complex biochemical processes that allow Trichoderma to sense numerous environmental signals, we aimed to identify upstream GPCRs from T. atroviride.
As the genome sequence of T. atroviride was not available, we searched the proteome of its close relative T. reesei for GPCR-like proteins and identified a total of 55 sequences in its genome database (Table 2). The identified proteins include putative orthologues of fungal Ste2- and Ste3-like pheromone receptors previously reported to be required for the mating responses in S. cerevisiae, a homologue of the previously characterized carbon sensors GprD from A. nidulans (Han et al. 2004) and Gpr-4 from N. crassa (Li and Borkovich 2006), three putative nitrogen sensors, four CRL proteins grouping with the previously characterized N. crassa GPR-1 receptor (Krystofova and Borkovich 2006), and four RGS domain-containing GPCRs. Furthermore, we found six proteins grouping to classes VII and VIII, respectively, which were first described by Kulkarni et al. (2005) and consist of fungal GPCRs related to different mammalian GPCRs including the rat growth hormone releasing factor and the steroid receptor mPR. Interestingly, we did not detect a member of class IX, which includes the previously characterized N. crassa NOP-1 opsin (Bieszke et al. 1999; Bieszke et al.2007), in the T. reesei genome.
Based on the results on GPCRs present in the T. reesei genome, putative GPCR-encoding genes from the mycoparasite T. atroviride were isolated. As the information on functional fungal GPCRs other than pheromone receptors is very limited, the class of CRL proteins was chosen as it is not present in the genomes of the ascomycete yeasts S. cerevisiae and S. pombe but it includes proteins from ascomycete filamentous fungi like N. crassa GPR-1 which was shown to be actually expressed and functional. Furthermore, it is a rather big class represented by four members in the T. reesei genome.
Phylogenetic and topological analyses confirmed that all four encoded proteins from T. atroviride contain a distribution of the seven transmembrane helices characteristic for members of the CRL class and contain a Dicty_CAR domain (pfam05462). Furthermore, they share sequence homology to CRL proteins of other ascomycetes primarily in the seven transmembrane domains as well as the three extracellular loops which are important for ligand binding, whereas sequence similarity is minor in the third intracellular loop and the C-terminal tail which are interacting with the Gα subunits. These findings are in accordance to those from Krystofova and Borkovich (2006), who speculated that these GPCRs may therefore share functional similarity e.g. by responding to similar ligands. Although the related protein GprH of A. nidulans has already been described some years ago (Han et al. 2004), no function has yet been assigned to it. Until now, the only CRL protein of a filamentous fungus has been functionally characterized in N. crassa, where GPR-1 is required for female sexual development and is suggested by epistatic analysis to couple to the GNA-1 Gα subunit. Interestingly, in contrast to T. atroviride gpr1-sil mutants, no obvious defects during asexual growth were found in Δgpr-1 strains (Krystofova and Borkovich 2006). In the basidiomycete C. neoformans, Gpr4, a GPCR with homology to A. nidulans GprH and the D. discoideum cAR1 cAMP receptor, was found to act upstream of the Gα subunit Gpa1 in sensing amino acids to activate the cAMP-PKA signalling pathway. Furthermore, Gpr4 was reported to be rapidly internalized in response to methionine, a mechanism leading to GPCR signalling desensitization (Xue et al.2006). Nevertheless, recent analysis showed that C. neoformans Gpr4 is more similar to carbon-sensing GPCRs like Gpr1p from S. cerevisiae (Li et al. 2007).
GPCR-encoding genes like those coding for pheromone receptors were reported to be transcribed at low to undetectable levels in e.g. N. crassa, Sordaria macrospora (Pöggeler and Kück 2001), U. maydis and U. hordei (Urban et al. 1996; Anderson et al. 1999). This weak transcription of GPCR-encoding genes was also confirmed by analyses of N. crassa GPR-1 and the predicted carbon-sensing receptor GPR-4, both could not be detected using Northern analysis (Krystofova and Borkovich 2006; Li and Borkovich 2006).
Because of these findings, we decided to employ highly sensitive real-time RT-PCR for the transcriptional characterization of the T. atroviride gpr1, gpr2, gpr3, and gpr4 genes.
For transcriptional analyses in T. atroviride, the actin-encoding gene act1 was frequently used as reference during the last decades. Nevertheless, preliminary results from real-time RT-PCR analyses showed significant variation of act1 gene transcription (Kratochwill and Brunner, unpublished), which is in accordance with a report from Gomez et al. (1997) on the regulation of actin gene transcription during interaction of different T. harzianum isolates. For these reasons, we tested four additional “housekeeping” genes for their constant transcription under varying growth conditions of T. atroviride to exclude carbon source- and surface-dependent differences in their expression. As their cDNA abundances again showed growth-dependent variations, reflecting previous findings (e.g. Zhang and Snyder 1992; Bhatia et al. 1994; Thellin et al. 1999; Bustin 2000; Schmittgen and Zakrajsek 2000; Suzuki et al. 2000), the most stable internal control gene for each condition was identified as described by Vandesompele et al. (2002). act1 turned out to be the best suited reference for real-time investigations of samples from liquid media, whereas the use of tef1 provided stable and reproducible results when comparing samples from growth on PDA plates with growth in liquid PBD. For analysing samples derived from in vitro biocontrol conditions, sar1 was chosen.
The gpr1, gpr2, gpr3 and gpr4 genes of T. atroviride all encode putative GPCRs belonging to the class of CRL proteins. In Trichoderma spp. exogenous cAMP was described to promote conidiation even in the dark (Nemcovic and Farkas 1998; Berrocal-Tito et al., 2000) and to promote mycoparasitic behaviour by increasing coiling (Omero et al. 1999). In T. virens, Δtac1 mutants missing the gene encoding adenylate cyclase were unable to overgrow plant pathogens like Sclerotium rolfsii, R. solani or Pythium sp., and cAMP signalling was shown to also be essential for growth, conidial germination and secondary metabolite production (Mukherjee et al. 2007). M. grisea was reported to respond to exogenously added cAMP by developing appressoria although the authors noted that the cell wall and cell membrane should be relatively impermeable to cAMP and thus any responses to extracellular cAMP seem to be due to cAMP receptors (Lee and Dean 1993). Furthermore, Galagan et al. (2003) hypothesized that class V receptors of N. crassa might be involved in cAMP sensing. In addition, by studying the response of T. atroviride pkr-1 antisense and over-expressing mutants to cAMP, Casas-Flores et al. (2006) obtained results showing that triggering of conidiation by cAMP is at least in part independent of PKA. In pkr-1 antisense transformants, which showed elevated levels of PKA activity and were unable to conidiate even with light, addition of cAMP to the medium provoked conidiation. pkr-1 overexpressing mutants, which had nearly undetectable PKA activity levels but nevertheless hypers-porulated in response to light and conidiated even in the dark, produced more conidia in the presence of cAMP. The finding that both the extracellular addition of dB-cAMP as well as sustained low PKA activity provokes sporulation encouraged the authors to suggest a second, alternate signalling pathway that uses a membrane receptor for exogenous cAMP. This cAMP receptor is supposed to exist beside the classical pathway of PKA activation by cAMP.
Analysis of cAMP-dependent transcription of the T. atroviride GPCR-encoding genes showed that expression of gpr3 and gpr4 was significantly reduced in the presence of exogenously added cAMP, whereas gpr1 and gpr2 mRNA levels were only slightly influenced.
When comparing growth on solid (PDA) to growth in liquid (PDB) potato-dextrose medium, again transcription of gpr3 and gpr4 showed the most noticeable responses in that mRNA levels were significantly lower upon growth of T. atroviride in liquid PDB compared to growth on PDA plates. Similar results were described for a pheromone receptor of C. neoformans: CPRa gene expression markedly increased when the cultures were shifted from liquid to solid media (Chang et al. 2003).
During plate confrontation assays, transcription of gpr1, gpr2 and gpr4 basically remained unaltered in the presence of a host fungus, whereas gpr3 transcription significantly decreased upon contact with R. solani. Interestingly, this down-regulation of gpr3 transcription could also be observed when T. atroviride was confronted with itself and when it touched a cellulose membrane, suggesting that the observed transcriptional regulation is the result of any physical contact and not of fungal–fungal interaction.
When testing the effect of different carbon sources and carbon starvation, all four T. atroviride GPCR-encoding genes showed a similar behaviour with the lowest expression on glycerol and highest mRNA levels when the fungus was cultivated without any carbon source followed by growth in the presence of the hardly utilisable chitin. These results resemble those reported for C. neoformans CPRa whose expression increased upon transfer from rich to poor medium (Chang et al. 2003), but are in contrast to N. crassa gpr-4, whose mRNA levels were highest in glycerol-grown cultures. N. crassa gpr4 was demonstrated to encode a GPCR required for carbon source-dependent asexual growth and development and it was shown to be coupled to the Gα subunit GNA-1 in a cAMP-signalling pathway (Li and Borkovich 2006).
In general, gpr3 and gpr4 showed significantly lower transcript levels compared to gpr1 and gpr2, being about 10-fold lower when T. atroviride was grown in PDB versus cultivation on PDA and under in vitro biocontrol conditions, or even about 100-fold lower upon cultivation in liquid medium with different carbon sources. Interestingly, these findings reflect the phylogenetic analysis grouping Gpr1 and Gpr2 into one branch and Gpr3 and Gpr4 in the other one (Fig. 3).
For further analyses of the T. atroviride GPCR-encoding gpr1 and gpr3 genes, a silencing method based on the synthesis of hairpin RNA was applied. Silencing of gpr1 resulted in transformants with silencing levels of 70–80%, whereas transformants with the integrated gpr3 silencing construct exhibited unaltered gpr3 mRNA levels. Low-abundant transcripts were suggested to be less susceptible to siRNA-mediated degradation than medium- and high-abundant transcripts (Hu et al. 2004). The failure to obtain silencing of gpr3 transcription may therefore be due to its low native mRNA levels. In addition, Fraser et al. (2000) reported that some genes are resistant to RNAi and in C. cinereus a lack of silencing in some transformants despite the presence of a complete silencing cassette was observed. The authors suggested that this effect could be due to a lack of expression caused by the surrounding DNA of the ectopic insertion point (Wälti et al. 2006).
gpr1-silenced mutants grow slowly with only little aerial mycelium, but instead the hyphae invade the agar and grow submerged in it. In addition, the mutants continuously produce conidia when they are cultivated on solid media and the conidia have reduced and delayed germination. The finding that silencing of gpr1 promotes constitutive sporulation even in the dark combined with the fact that exogenous cAMP induces sporulation (Nemcovic and Farkas 1998) makes it unlikely that cAMP is indeed the ligand of Gpr1. The phenotypes of the gpr1-silenced mutants are similar, although less pronounced (concerning reduction of growth rate and ratio of aerial hyphae to hyphae invading the agar), to the phenotype observed in Δtga3 mutants missing the subgroup III Gα subunit (Zeilinger et al., 2005). Whether Tga3 acts downstream of Gpr1 in the same signalling pathway needs further investigation, e.g. by applying the yeast 2-Hybrid-System and by genetic epistasis analysis.
When analysed in the Biolog system, gpr1-silenced mutants exhibited reduced growth on a variety of different carbon sources. For example, on glucose and glycerol, which are among the best carbon sources for the parental strain T. atroviride P1, the tested mutant exhibited an about 70% reduced growth rate during the first 66 h of incubation (Fig. 7). Furthermore, the mutant was not able to grow at all on the disaccharide lactose and its isomerization product lactulose, and also the disaccharide maltose and the trisaccharide raffinose did only allow sparse growth, although the sugars are well metabolized by the parental strain. Interestingly, on D-mannitol, D-arabitol, L-serine, salicin, and Tween 80 the mutant showed a similar growth rate as the parental strain suggesting that its reduced growth is carbon source-dependent and not a general effect.
Summarizing, we isolated and characterized four genes of T. atroviride P1 encoding GPCRs grouping to the class of CRL proteins. In the closely related ascomycete N. crassa this class of receptors consists of three proteins (Krystofova and Borkovich 2006), but with the exception of GPR-1 no information is available on their function. Furthermore, it is still unclear if these GPCRs respond to the same ligands or couple to the same downstream signalling components. Further characterization of T. atroviride Gpr1 showed that this GPCR affects conidiation, conidial germination and vegetative growth on a multitude of carbon sources, supporting the fundamental role of G protein signalling in Trichoderma.
Acknowledgments
This work was supported by grants from the Fonds zur Förderung Wissenschaftlicher Forschung (FWF P18109 and L225-B03). The authors thank Mariella Pauls for her assistance in performing Biolog analysis.
Footnotes
Nucleotide sequence data reported are available at GenBank under the accession numbers EF581847, EF581848, EF581849, EF581850, EF591763, DQ284755, DQ384865, EF534712, and EU391628.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Anderson CM, Willits DA, Kosted PJ, Ford EJ, Martinez-Espinoza AD, Sherwood JE. Molecular analysis of the pheromone and pheromone receptor genes of Ustilago hordei. Gene. 1999;240:89–97. doi: 10.1016/s0378-1119(99)00428-x. [DOI] [PubMed] [Google Scholar]
- Berrocal-Tito GM, Rosales-Saavedra T, Herrera-Estrella A, Horwitz BA. Characterization of blue-light and developmental regulation of the photolyase gene phr1 in Trichoderma harzianum. Photochem Photobiol. 2000;71:662–668. doi: 10.1562/0031-8655(2000)071<0662:coblad>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Bhatia P, Taylor WR, Greenberg AH, Wright JA. Comparison of glyceraldehyde-3-phosphodehydrogenase and 28S-ribosomal RNA gene expression as RNA loading controls for northern blot analysis of cell lines of varying malignant potential. Anal Biochem. 1994;216:223–226. doi: 10.1006/abio.1994.1028. [DOI] [PubMed] [Google Scholar]
- Bieszke JA, Spudich EN, Scott KL, Borkovich KA, Spudich JL. A eukaryotic protein, NOP-1, binds retinal to form an archaeal rhodopsin-like photochemically reactive pigment. Biochemistry. 1999;38:14138–14145. doi: 10.1021/bi9916170. [DOI] [PubMed] [Google Scholar]
- Bieszke JA, Li L, Borkovich KA. The fungal opsin gene nop-1 is negatively-regulated by a component of the blue light sensing pathway and influences conidiation-specific gene expression in Neurospora crassa. Curr Genet. 2007;52:149–157. doi: 10.1007/s00294-007-0148-8. [DOI] [PubMed] [Google Scholar]
- Birnbaumer M, Antaramian A, Themmen AP, Gilbert S. Desensitization of the human V2 vasopressin receptor. Homologous effects in the absence of heterologous desensitization. J Biol Chem. 1992;267:11783–11788. [PubMed] [Google Scholar]
- Blumer KJ, Reneke JE, Thorner J. The STE2 gene product is the ligand-binding component of the alpha-factor receptor of Saccharomyces cerevisiae. J Biol Chem. 1988;263:10836–10842. [PubMed] [Google Scholar]
- Bölker M. Sex and crime, heterotrimeric G proteins in fungal mating and pathogenesis. Fungal Genet Biol. 1998;25:143–156. doi: 10.1006/fgbi.1998.1102. [DOI] [PubMed] [Google Scholar]
- Bölker M, Urban M, Kahmann R. The a mating type locus of U. maydis specifies cell signaling components. Cell. 1992;68:441–450. doi: 10.1016/0092-8674(92)90182-c. [DOI] [PubMed] [Google Scholar]
- Borkovich KA, Alex LA, Yarden O, Freitag M, Turner GE, Read ND, Seiler S, Bell-Pedersen D, Paietta J, Plesofsky N, Plamann M, Goodrich-Tanrikulu M, Schulte U, Mannhaupt G, Nargang FE, Radford A, Selitrennikoff V, Galagan JE, Dunlap JC, Loros JJ, Catcheside D, Inoue H, Aramayo R, Polymenis M, Selker EU, Sachs MS, Marzluf GA, Paulsen I, Davis R, Ebbole DJ, Zelter A, Kalkman ER, O’Rourke R, Bowring F, Yeadon J, Ishii C, Suzuki K, Sakai W, Pratt R. Lessons from the genome sequence of Neurospora crassa, tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev. 2004;68:1–108. doi: 10.1128/MMBR.68.1.1-108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- Casas-Flores S, Rios-Momberg M, Rosales-Saavedra T, Martínez-Hernández P, Olmedo-Monfil V, Herrera-Estrella A. Cross talk between a fungal blue-light perception system and the cyclic AMP signaling pathway. Eukaryot Cell. 2006;5:499–506. doi: 10.1128/EC.5.3.499-506.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Miller GF, Kwon-Chung KJ. Importance of a developmentally regulated pheromone receptor of Cryptococcus neoformans for virulence. Infect Immun. 2003;71:4953–4960. doi: 10.1128/IAI.71.9.4953-4960.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan TM, Carroll AM, Valent B, Sweigard JA. Magnaporthe grisea pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. Plant Cell. 1999;11:2013–2030. doi: 10.1105/tpc.11.10.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman HG, Thorner J, Caron MG, Lefkowitz RJ. Model systems for the study of seven-transmembrane-segment receptors. Annu Rev Biochem. 1991;60:653–688. doi: 10.1146/annurev.bi.60.070191.003253. [DOI] [PubMed] [Google Scholar]
- Druzhinina IS, Schmoll M, Seiboth B, Kubicek CP. Global carbon utilization profiles of wild-type, mutant, and transformant strains of Hypocrea jecorina. Appl Environ Microbiol. 2006;72:2126–2133. doi: 10.1128/AEM.72.3.2126-2133.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Mol Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Csmpos M, Sohrmann M, Ahringer J. Functional genomic analysis if C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, Rehman B, Elkins T, Engels R, Wang S, Nielsen CB, Butler J, Endrizzi M, Qui D, Ianakiev P, Bell-Pedersen D, Nelson MA, Werner-Washburne M, Selitrennikoff CP, Kinsey JA, Braun EL, Zelter A, Schulte U, Kothe GO, Jedd G, Mewes W, Staben C, Marcotte E, Greenberg D, Roy A, Foley K, Naylor J, Stange-Thomann N, Barrett R, Gnerre S, Kamal M, Kamvysselis M, Mauceli E, Bielke C, Rudd S, Frishman D, Krystofova S, Rasmussen C, Metzenberg RL, Perkins DD, Kroken S, Cogoni C, Macino G, Catcheside D, Li W, Pratt RJ, Osmani SA, DeSouza CP, Glass L, Orbach MJ, Berglund JA, Voelker R, Yarden O, Plamann M, Seiler S, Dunlap J, Radford A, Aramayo R, Natvig DO, Alex LA, Mannhaupt G, Ebbole DJ, Freitag M, Paulsen I, Sachs MS, Lander ES, Nusbaum C, Birren B. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- Gomez I, Chet I, Herrera-Estrella A. Genetic diversity and vegetative compatibility among Trichoderma harzianum isolates. Mol Gen Genet. 1997;256:127–135. doi: 10.1007/s004380050554. [DOI] [PubMed] [Google Scholar]
- Gutkind JS. Cell growth control by G protein-coupled receptors, from signal transduction to signal integration. Oncogene. 1998;17:1331–1342. doi: 10.1038/sj.onc.1202186. (11 Reviews) [DOI] [PubMed] [Google Scholar]
- Hagen DC, McCaffrey G, Sprague GF., Jr Evidence the yeast STE3 gene encodes a receptor for the peptide pheromone a factor, gene sequence and implications for the structure of the presumed receptor. Proc Natl Acad Sci USA. 1986;83:1418–1422. doi: 10.1073/pnas.83.5.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han KH, Seo JA, Yu JH. A putative G protein-coupled receptor negatively controls sexual development in Aspergillus nidulans. Mol Microbiol. 2004;51:1333–1345. doi: 10.1111/j.1365-2958.2003.03940.x. [DOI] [PubMed] [Google Scholar]
- Hu X, Hipolito S, Lynn R, Abraham V, Ramos S, Wong-Staal F. Relative gene-silencing efficiencies of small interfering RNAs targeting sense and antisense transcripts from the same genetic locus. Nucleic Acids Res. 2004;32:4609–4617. doi: 10.1093/nar/gkh790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaziro Y, Itoh H, Kozasa T, Nakafuku M, Satoh T. Structure and function of signal-transducing GTP-binding proteins. Annu Rev Biochem. 1991;60:349–400. doi: 10.1146/annurev.bi.60.070191.002025. [DOI] [PubMed] [Google Scholar]
- Kim H, Borkovich KA. A pheromone receptor gene, pre–1, is essential for mating type-specific directional growth and fusion of trichogynes and female fertility in Neurospora crassa. Mol Microbiol. 2004;52:1781–1798. doi: 10.1111/j.1365-2958.2004.04096.x. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model, application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Krystofova S, Borkovich KA. The predicted G-protein-coupled receptor GPR–1 is required for female sexual development in the multicellular fungus Neurospora crassa. Eukaryot Cell. 2006;5(9):1503–1516. doi: 10.1128/EC.00124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni RD, Thon MR, Pan H, Dean RA. Novel G-protein-coupled receptor-like proteins in the plant pathogenic fungus Magnaporthe grisea. Genome Biol. 2005;6(3):R24. doi: 10.1186/gb-2005-6-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon A, Han KH, Seo JA, Yu JH, d’Enfert C. G-protein and cAMP-mediated signaling in aspergilli, a genomic perspective. Fungal Genet Biol. 2006;43(7):490–502. doi: 10.1016/j.fgb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lee YH, Dean RA. cAMP regulates infection structure formation in the plant pathogenic fungus Magnaporthe grisea. Plant Cell. 1993;5(6):693–700. doi: 10.1105/tpc.5.6.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Borkovich KA. GPR–4 is a G-protein coupled receptor required for carbon source dependent asexual growth and development in Neurospora crassa. Eucaryotic Cell. 2006;58:1278–1300. doi: 10.1128/EC.00109-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wright SJ, Krystofova S, Park G, Borkovich KA. Heterotrimeric G protein signaling in filamentous fungi. Annu Rev Microbiol. 2007;61:423–452. doi: 10.1146/annurev.micro.61.080706.093432. [DOI] [PubMed] [Google Scholar]
- Lu Z, Tombolini R, Woo S, Zeilinger S, Lorito M, Jansson JK. In vivo study of trichoderma-pathogen-plant interactions, using constitutive and inducible green Xuorescent protein reporter systems. Appl Environ Microbiol. 2004;70(5):3073–3081. doi: 10.1128/AEM.70.5.3073-3081.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach RL, Schindler M, Kubicek CP. Transformation of Trichoderma reesei based on hygromycinB resistance using homologous expression signals. Curr Genet. 1994;25:567–570. doi: 10.1007/BF00351679. [DOI] [PubMed] [Google Scholar]
- Mukherjee M, Mukherjee PK, Kale SP. cAMP signalling is involved in growth, germination, mycoparasitism and secondary metabolism in Trichoderma virens. Microbiology. 2007;153(Pt 6):1734–1742. doi: 10.1099/mic.0.2007/005702-0. [DOI] [PubMed] [Google Scholar]
- Neer EJ. Heterotrimeric G proteins, organizers of transmembrane signals. Cell. 1995;80(2):249–257. doi: 10.1016/0092-8674(95)90407-7. [DOI] [PubMed] [Google Scholar]
- Nemcovic M, Farkas V. Stimulation of conidiation by derivates of cAMP in Trichoderma viride. Folia Microbiol. 1998;43:399–402. [Google Scholar]
- Olesnicky NS, Brown AJ, Dowell SJ, Casselton LA. A constitutively active G-protein-coupled receptor causes mating self-compatibility in the mushroom Coprinus. EMBO J. 1999;18(10):2756–2763. doi: 10.1093/emboj/18.10.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omero C, Inbar J, Rocha-Ramirez V, Herrera-Estrella A, Chet I, Horwitz BA. G-protein activators and cAMP promote mycoparasitic behaviour in Trichoderma harzianum. Mycol Res. 1999;103:1637–1642. [Google Scholar]
- Peterbauer CK, Lorito M, Hayes CK, Harman GE, Kubicek CP. Molecular cloning and expression of the nag1 gene (N-acetyl-beta-D-glucosaminidase-encoding gene) from Trichoderma harzianum P1. Curr Genet. 1996;30(4):325–331. doi: 10.1007/s002940050140. [DOI] [PubMed] [Google Scholar]
- Peterbauer CK, Litscher D, Kubicek CP. The Trichoderma atroviride seb1-(stress response element binding) gene encodes an AGGGG-binding protein which is involved in osmotic stress response. Mol Genet Genomics. 2002;268:223–231. doi: 10.1007/s00438-002-0732-z. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöggeler S, Kück U. Identification of transcriptionally expressed pheromone receptor genes in filamentous ascomycetes. Gene. 2001;280:9–17. doi: 10.1016/s0378-1119(01)00786-7. [DOI] [PubMed] [Google Scholar]
- Reithner B, Brunner K, Schuhmacher R, Peissl I, Seidl V, Krska R, Zeilinger S. The G protein alpha subunit Tga1 of Trichoderma atroviride is involved in chitinase formation and differential production of antifungal metabolites. Fungal Genet Biol. 2005;42(9):749–760. doi: 10.1016/j.fgb.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Rocha-Ramirez V, Omero C, Chet I, Horwitz BA, Herrera-Estrella A. Trichoderma atroviride G-protein alpha-subunit gene tga1 is involved in mycoparasitic coiling and conidiation. Eukaryot Cell. 2002;1(4):594–605. doi: 10.1128/EC.1.4.594-605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory Press; New York: 1989. [Google Scholar]
- Schmittgen TD, Zakrajsek BA. Effect of experimental treatment on housekeeping gene expression, validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods. 2000;46:69–81. doi: 10.1016/s0165-022x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- Seidl V, Druzhinina IS, Kubicek CP. A screening system for carbon sources enhancing beta-N-acetylglucosaminidase formation in Hypocrea atroviridis (Trichoderma atroviride) Microbiology. 2006;152(Pt 7):2003–2012. doi: 10.1099/mic.0.28897-0. [DOI] [PubMed] [Google Scholar]
- Seo JA, Han KH, Yu JH. The gprA and gprB genes encode putative G protein-coupled receptors required for self-fertilization in Aspergillus nidulans. Mol Microbiol. 2004;53(6):1611–1623. doi: 10.1111/j.1365-2958.2004.04232.x. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Higgins PJ, Crawford DR. Control selection for RNA quantitation. BioTechniques. 2000;29:332–337. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Davey J, Imai Y, Yamamoto M. Schizosaccharomyces pombe map3+ encodes the putative M-factor receptor. Mol Cell Biol. 1993;13(1):80–88. doi: 10.1128/mcb.13.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E. Housekeeping genes as internal standards, use and limits. J Biotechnol. 1999;75(2–3):291–295. doi: 10.1016/s0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- Tichopad A, Dilger M, Schwarz G, Pfaffl MW. Standardized determination of real-time PCR efficiency from a single reaction set-up. Nucleic Acids Res. 2003;31(20):e122. doi: 10.1093/nar/gng122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban M, Kahmann R, Bolker M. Identification of the pheromone response element in Ustilago maydis. Mol Gen Genet. 1996;251(1):31–37. doi: 10.1007/BF02174341. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):research 34.1–34.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigó R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Versele M, Lemaire K, Thevelein JM. Sex and sugar in yeast, two distinct GPCR systems. EMBO Rep. 2001;2(7):574–579. doi: 10.1093/embo-reports/kve132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wälti MA, Villalba C, Buser RM, Grünler A, Aebi M, Künzler M. Targeted gene silencing in the model mushroom Coprinopsis cinerea (Coprinus cinereus) by expression of homologous hairpin RNAs. Eukaryot Cell. 2006;5(4):732–744. doi: 10.1128/EC.5.4.732-744.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Bahn YS, Cox GM, Heitman J. G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans. Mol Biol Cell. 2006;17(2):667–679. doi: 10.1091/mbc.E05-07-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilinger S, Galhaup C, Payer K, Woo SL, Mach RL, Fekete C, Lorito M, Kubicek CP. Chitinase gene expression during mycoparasitic interaction of Trichoderma harzianum with its host. Fungal Genet Biol. 1999;26(2):131–140. doi: 10.1006/fgbi.1998.1111. [DOI] [PubMed] [Google Scholar]
- Zeilinger S, Reithner B, Scala V, Peissl I, Lorito M, Mach RL. Signal transduction by Tga3, a novel G protein alpha subunit of Trichoderma atroviride. Appl Environ Microbiol. 2005;71(3):1591–1597. doi: 10.1128/AEM.71.3.1591-1597.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Snyder SH. Nitric oxide stimulates auto-ADP-ribosylation of glyceraldehyde-3-phosphate dehydrogenase. Proc Natl Acad Sci USA. 1992;89(20):9382–9385. doi: 10.1073/pnas.89.20.9382. [DOI] [PMC free article] [PubMed] [Google Scholar]