Abstract
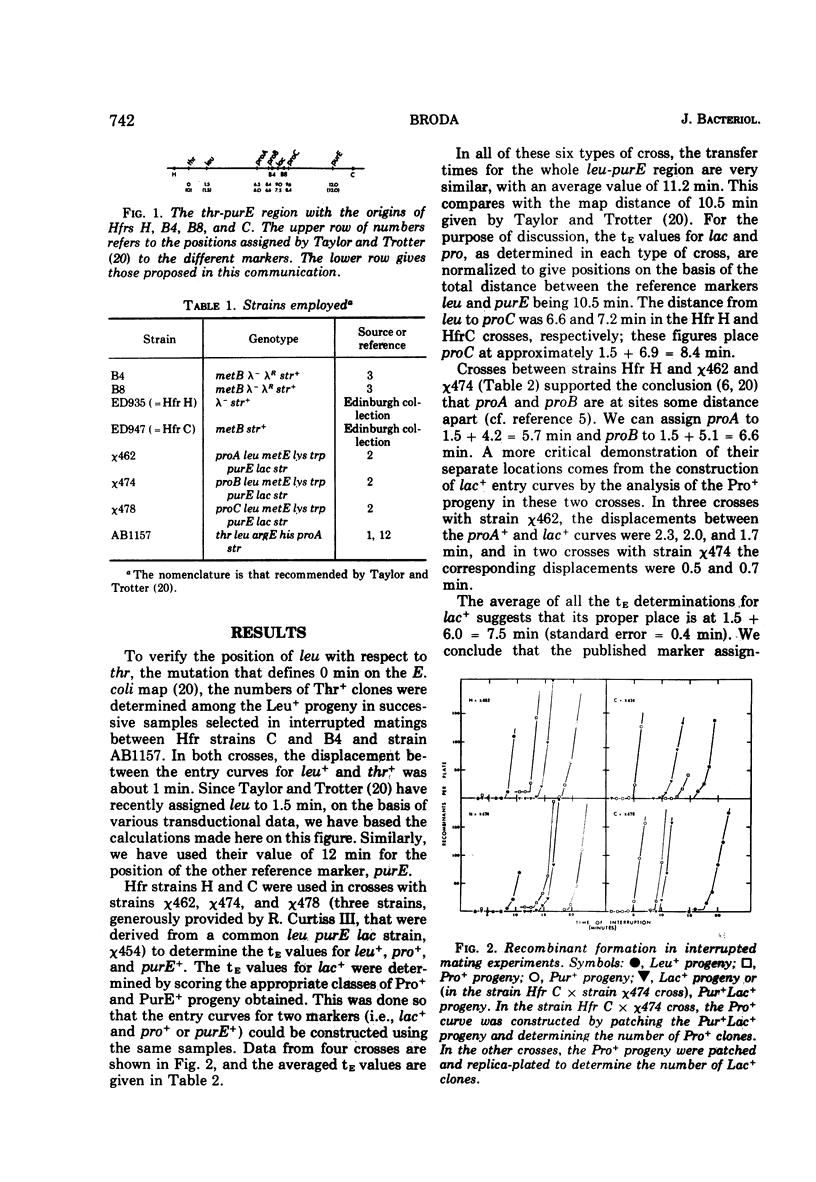
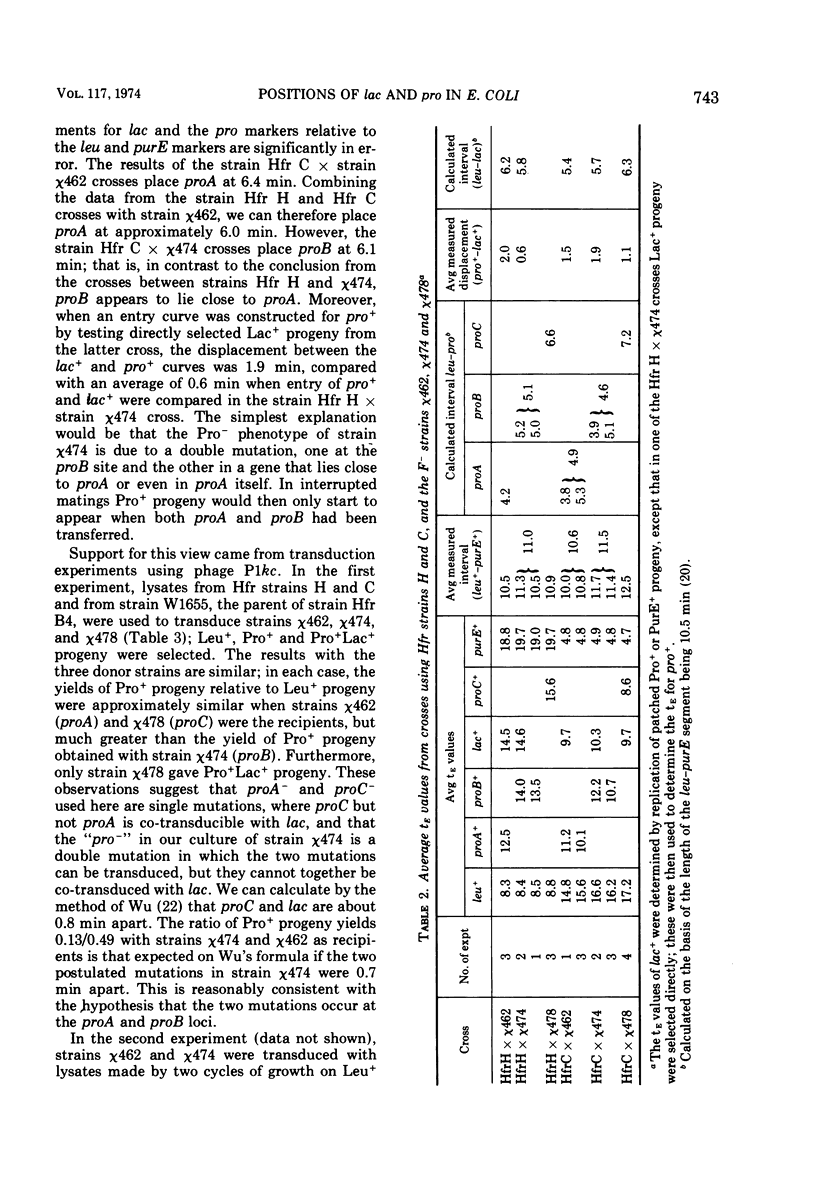
Interrupted mating experiments were performed with Hfr strains H and C and three leu lac purE recipient strains derived from a common parent and carrying, respectively, the proA−, proB−, and proC− mutations. It was concluded that if leu is placed at 1.5 min and purE at 12 min from thr, the origin on the Taylor-Trotter map, lac is at about 7.5 min and the pro genes are at about 6.0, 6.6, and 8.4 min, respectively. Both conjugational and transductional data suggest that the strain carrying the proB− mutation also carries a second mutation close to the proA site which independently confers a Pro− phenotype. The times before the onset of transfer of chromosomal deoxyribonucleic acid by both Hfr strains B4 and B8 were approximately 3 min.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg C. M., Curtiss R., 3rd Transposition derivatives of an Hfr strain of Escherichia coli K-12. Genetics. 1967 Jul;56(3):503–525. doi: 10.1093/genetics/56.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broda P., Collins J. F. Gross map distances and Hfr transfer times in Escherichia coli K-12. J Bacteriol. 1974 Feb;117(2):747–752. doi: 10.1128/jb.117.2.747-752.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CURTIS S. R., 3rd CHROMOSOMAL ABERRATIONS ASSOCIATED WITH MUTATIONS TO BACTERIOPHAGE RESISTANCE IN ESCHERICHIA COLI. J Bacteriol. 1965 Jan;89:28–40. doi: 10.1128/jb.89.1.28-40.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condamine H. Mutants des voies de biosynthèse et de dégradation de la proline chez E. coli K 12. Ann Inst Pasteur (Paris) 1971 Jan;120(1):9–22. [PubMed] [Google Scholar]
- Curtiss R., 3rd Bacterial conjugation. Annu Rev Microbiol. 1969;23:69–136. doi: 10.1146/annurev.mi.23.100169.000441. [DOI] [PubMed] [Google Scholar]
- Curtiss R., 3rd, Charamella L. J., Stallions D. R., Mays J. A. Parental functions during conjugation in Escherichia coli K-12. Bacteriol Rev. 1968 Dec;32(4 Pt 1):320–348. [PMC free article] [PubMed] [Google Scholar]
- HOWARD-FLANDERS P., SIMSON E., THERIOT L. A LOCUS THAT CONTROLS FILAMENT FORMATION AND SENSITIVITY TO RADIATION IN ESCHERICHIA COLI K-12. Genetics. 1964 Feb;49:237–246. doi: 10.1093/genetics/49.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itikawa H., Baumberg S., Vogel H. J. Enzymic basis for a genetic suppression: accumulation and deacylation of N-acetylglutamic gamma-semialdehyde in enterobacterial mutants. Biochim Biophys Acta. 1968 Jul 9;159(3):547–550. doi: 10.1016/0005-2744(68)90142-3. [DOI] [PubMed] [Google Scholar]
- Klotz L. C., Zimm B. H. Size of DNA determined by viscoelastic measurements: results on bacteriophages, Bacillus subtilis and Escherichia coli. J Mol Biol. 1972 Dec 30;72(3):779–800. doi: 10.1016/0022-2836(72)90191-x. [DOI] [PubMed] [Google Scholar]
- Kuo T. T., Stocker B. A. Suppression of proline requirement of proA and proAB deletion mutants in Salmonella typhimurium by mutation to arginine requirement. J Bacteriol. 1969 May;98(2):593–598. doi: 10.1128/jb.98.2.593-598.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOW B., WOOD T. H. A QUICK AND EFFICIENT METHOD FOR INTERRUPTION OF BACTERIAL CONJUGATION. Genet Res. 1965 Jul;6:300–303. doi: 10.1017/s001667230000416x. [DOI] [PubMed] [Google Scholar]
- Low B. Low recombination frequency for markers very near the origin in conjugation in E. coli. Genet Res. 1965 Nov;6(3):469–473. doi: 10.1017/s0016672300004341. [DOI] [PubMed] [Google Scholar]
- Low B. Rapid mapping of conditional and auxotrophic mutations in Escherichia coli K-12. J Bacteriol. 1973 Feb;113(2):798–812. doi: 10.1128/jb.113.2.798-812.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts L. M., Reeve E. C. Two mutations giving low-level streptomycin resistance in Escherichia coli K 12. Genet Res. 1970 Dec;16(3):359–365. doi: 10.1017/s0016672300002640. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Hsu M. T., Otsubo E., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. I. Structure of F-prime factors. J Mol Biol. 1972 Nov 14;71(2):471–497. doi: 10.1016/0022-2836(72)90363-4. [DOI] [PubMed] [Google Scholar]
- TAYLOR A. L., THOMAN M. S. THE GENETIC MAP OF ESCHERICHIA COLI K-12. Genetics. 1964 Oct;50:659–677. doi: 10.1093/genetics/50.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A. L., Trotter C. D. Linkage map of Escherichia coli strain K-12. Bacteriol Rev. 1972 Dec;36(4):504–524. doi: 10.1128/br.36.4.504-524.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood T. H. Effects of temperature, agitation, and donor strain on chromosome transfer in Escherichia coli K-12. J Bacteriol. 1968 Dec;96(6):2077–2084. doi: 10.1128/jb.96.6.2077-2084.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANOFSKY C., LENNOX E. S. Transduction and recombination study of linkage relationships among the genes controlling tryptophan synthesis in Escherichia coli. Virology. 1959 Aug;8:425–447. doi: 10.1016/0042-6822(59)90046-7. [DOI] [PubMed] [Google Scholar]



