Abstract
Prior studies of mRNA expression, protein expression, and pathway metabolite levels have implicated dysregulation of the kynurenine pathway in the etiology of schizophrenia and bipolar disorder. Here we investigate whether genes involved in kynurenine pathway regulation might interact with genes that respond to kynurenine metabolites, to enhance risk for these psychiatric phenotypes. Candidate genes were selected from prior studies of genetic association, gene expression profiling and animal models. A single nucleotide polymorphism (SNP) in each of six genes, TDO2, HM74, HM74A, MCHR1, MCHR2 and MC5R, was tested for association with phenotype (475 Caucasians, 88 African Americans with schizophrenia; 97 Caucasians, 3 African Americans with bipolar disorder; 191 Caucasian, 49 African American controls). An A allele in HM74 was significantly associated with schizophrenia and with schizophrenia plus bipolar disorder combined, odds ratios (OR) of 1.48, p = 0.011 and 1.50, p= 0.007, respectively. Augmentation of disease risk was found for the complex genotype HM74[A,any] + MCHR1[T,any] + MCHR2[C,any] which conferred an OR maximal for the combined diagnostic category of schizophrenia plus bipolar disorder (1.70, p= 0.003), carried by 30% of the cases. TDO2[CC] + MC5R[G,any] + MCHR2[GC] conferred an OR maximal for schizophrenia alone (4.84, p= 0.005), carried by 8% of schizophrenia cases. The combined risk posed by these related, complex genotypes is greater than any identified single locus and may derive from co-regulation of the kynurenine pathway by interacting genes, a lack of adequate melantropin-controlled sequestration of the kynurenine-derived pigments, or the production of melanotropin receptor ligands through kynurenine metabolism.
Keywords: psychosis; gene-gene interactions; metabolic; niacin receptor; tryptophan 2,3-dioxygenase
1. Introduction
The heritability of schizophrenia and bipolar disorder is generally accepted to be complex in nature, a characterization resulting from the observed non-Mendelian modes of inheritance, incomplete penetrance of genetic risk, polygenetic pattern of inheritance (Gottesman and Shields, 1967) and apparent heterogeneity of genetic causation (Allen et al., 2008). Evidence for shared susceptibility loci in schizophrenia and bipolar disorder has been found in several genome-wide association studies (Berrettini, 2000). The incomplete genetic penetrance observed for both phenotypes results from modulatory genetic background effects and from environmental influences that alter the neuro-developmental course (Dean and Murray, 2005).
Here we present genetic association data concerning a model for schizophrenia and bipolar disorder that incorporates most of these features, specifically, polygenic contribution, heterogeneity of genetic cause coupled with modulatory genetic background effects and a well-defined role for environmental interaction. The focal point of this model is the kynurenine pathway, the genes that influence its regulation and those that respond to its products (Figure 1). Changes in this pathway in schizophrenia and bipolar disorder have been identified at the mRNA expression, protein, and metabolite level in this laboratory and several others around the world (Schwarcz et al., 2001; Erhardt et al., 2003; Miller et al., 2004, 2006, 2008; Myint et al., 2007; Barry et al., 2008).Whether these changes represent biomarkers of disease or whether they represent disease causation is the question raised by observations that psychoses of several different etiologies involve activation of this pathway (Miller et al., 2004). To begin to address this issue, we asked whether kynurenine pathway-related genes (Figure 1) that have demonstrated expression differences between cases and show association with schizophrenia and/or bipolar disorder at the gene sequence level, specifically, tryptophan dioxygenase (TDO2) and the duplicated niacin receptor genes (HM74 and HM74A). TDO2 was shown to be elevated at the mRNA and protein levels in the postmortem frontal and anterior cingulate cortex of schizophrenia cases as compared to controls (Miller et al., 2004; Miller et al., 2006) and at the protein level in the anterior cingulate cortex of bipolar cases as compared to controls (Miller et al., 2006). The mRNA for the niacin receptors HM74 and HM74A (Figure 1) was found to be increased in the anterior cingulate cortex of schizophrenia and bipolar cases as compared to controls, but this increase in mRNA did not correspond to an increase in protein; rather, the protein for HM74 was unchanged and the protein for HM74A was decreased in schizophrenia cases as compared to controls (Miller and Dulay, 2008).
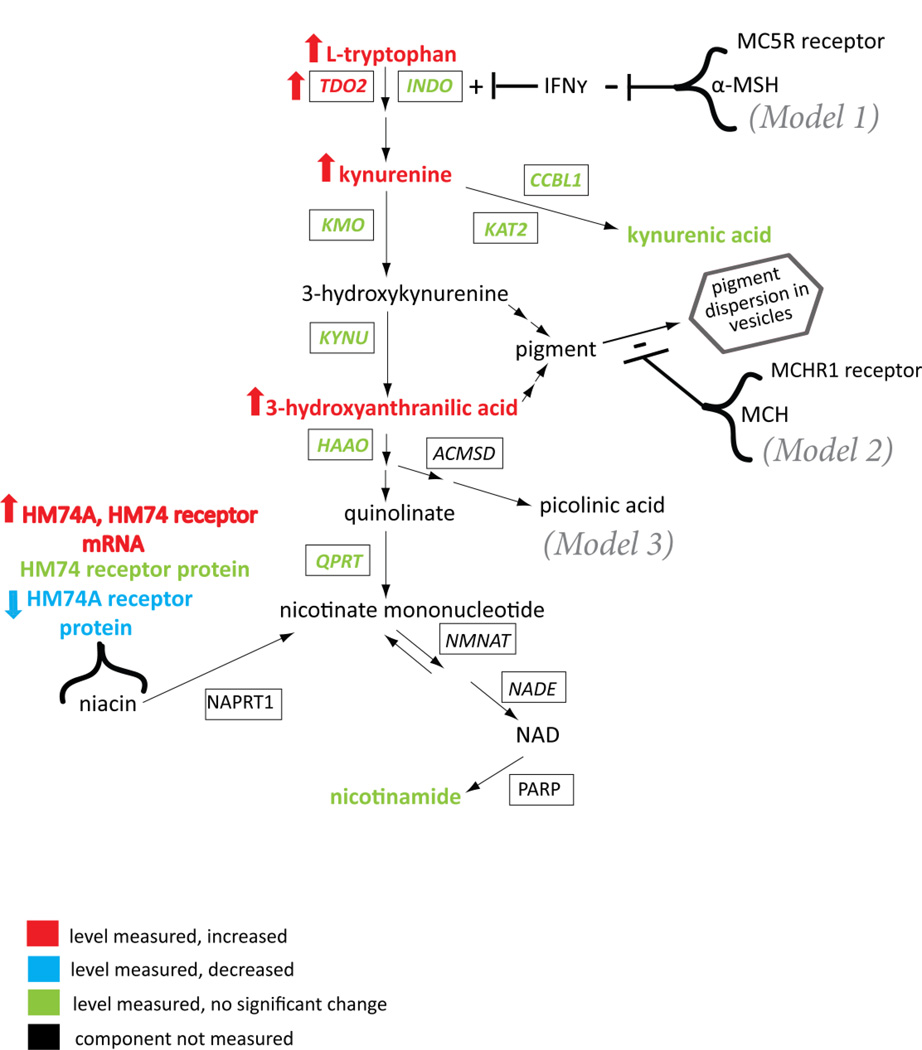
Figure 1.
Schematic of the kynurenine pathway with relevant regulatory components, summarizing gene expression and metabolite changes previously identified for schizophrenia in the frontal and anterior cingulate cortex. The significant changes that had been identified for bipolar disorder (not shown) were limited to increased kynurenine and increased mRNA for the HM74 receptor (Miller et al., 2006; Miller and Dulay, 2008). TDO2, HM74A and HM74 had been analyzed in previous studies for mRNA (RT-PCR [Miller et al., 2004&2006; Miller and Dulay, 2008] and protein (via quantitative Western blots or semi-quantitative immunohistochemistry (Miller et al., 2004,2006; Miller and Dulay, 2008) and several of the remaining enzymes had been analyzed for mRNA expression only (RT-PCR, Miller et al., 2004). The metabolites had been quantified by HPLC (Miller et al., 2006, 2008). Enzymes are presented in boxes and denoted with their HUGO identifier. Not all steps are depicted, e.g. the signaling cascades for the receptors. The models (see Discussion) for the relevant points of potential interaction between the melanotropin receptors and the kynurenine pathway genes, gene products or metabolites are denoted models 1, 2, and 3. Note that the regulatory role of the niacin receptor genes for the kynurenine pathway can only be inferred from the fact that administration of niacin does not allosterically inhibit TDO2 to any significant extent, but by some other means, leads to remission of the kynurenine-mediated de-novo synthesis of NAD from tryptophan in pellagra patients (Hankes et al., 1971). Niacin treatment of schizophrenia patients has been reported to result in remission of psychosis (Hoffer et al., 1957).
Potential modifying genes selected for study were those for a class of melanotropin receptors with relevant effects (Miller et al., 1993) in an animal model of an auditory gating endophenotype common in individuals with schizophrenia (Nagamoto et al., 1991). Within this class of receptors, three (MCHR1, MCHR2, MC5R) reside in chromosomal regions showing association with schizophrenia and/or bipolar disorder in genome-wide association studies (Schwab et al., 1998; Levinson et al., 2000; Dick et al., 2003; Lambert et al., 2005; Lin et al., 2005) and in association studies of MCHR1, specifically (Severinsen et al., 2006).
Hypothetical points of interaction between the melanotropin receptors and the kynurenine pathway are highlighted in Figure 1. The first involves co-stimulation of the kynurenine pathway via MC5R inhibition (Taylor and Namba, 2001) of interferon-gamma expression (IFNγ, Model 1 in Figure 1). Decreased signaling through the MC5R receptor would lead to activation of the pathway. A second point of interaction relates to the sequestration of the potentially toxic pigment molecules generated by the pathway (Model 2 in Figure 1). The effect of peptide agonists for the MCHR1 receptor is to decrease melanosome formation and decrease the dispersion of pigment in melanosome vesicles (Baker and Ball, 1975). In this model, the capacity of pigment sequestration and dispersion would be adversely affected by a hyper-functional MCHR1 receptor. Alternatively, the pathway produces at least one key precursor for ligands of the melanotropin receptors, picolinic acid (Model 3, Figure 1), which in its reduced form (pipecolic acid) is the core residue in the most potent peptide ligands reported for MC5R (Bednarek et al., 2007).
Thus, the six candidate genes analyzed for association with schizophrenia and bipolar disorder were: TDO2 (4q31.2), HM74A (12q24.3), HM74(12q24.3), MCHR1 (22q13.1), MCHR2 (6q16.3) and MC5R (18p11.21). One single nucleotide polymorphism (SNP) per gene was selected, by taking advantage of two prior genetic association studies, one yielding positive results for HM74 (Shink et al., 2005) and the other for MCHR1 (Severinsen et al., 2006), while for untested genes, SNPs were selected that showed maximal differences in frequency between racial groups (Figure 2). A difference between ethnic populations in disease expression and severity has been noted by many researchers studying schizophrenia (Saha et al., 2006).
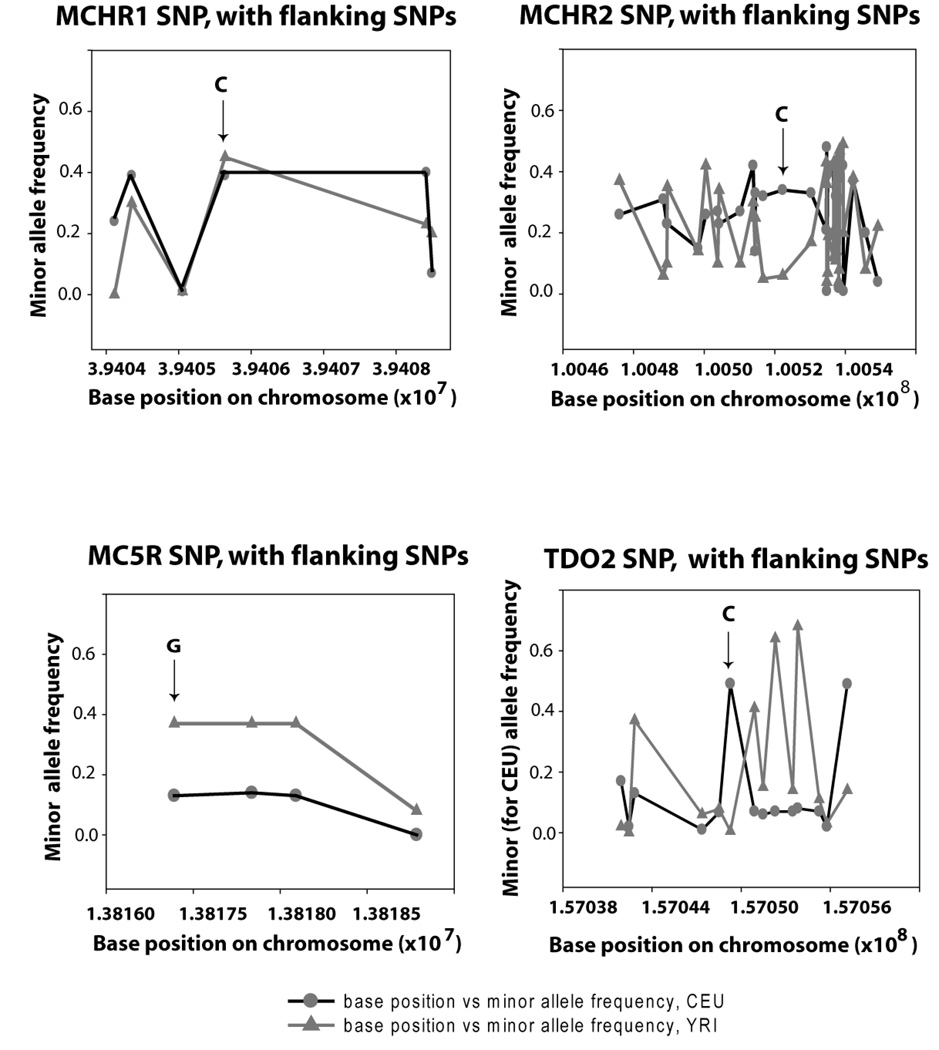
Figure 2.
Variation of SNP minor allele frequencies as reported in the Hapmap database (http;//www.hapmap.org) for genes under study. The frequency class of allele (minor or major) was specified based on the Caucasian frequency. The minor allele for the selected SNP (marked by arrows) is noted for each gene. The circles represent minor allele frequencies for Caucasians (CEU) reported in Hapmap, and the triangles represent minor allele frequencies for Sub-Saharan Africans (YRI) reported in Hapmap. Not shown are the minor allele frequency distributions for HM74 and HM74A, since much of the data that has been collected for these genes are potentially confounded by the duplicated nature of the relevant sequences. The report for the HM74 SNP, denoted rs2454727 in the database, links to NCBI: http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?rs=rs2454727) which specifies a minor allele frequency (A) of 34% for the CEU population, and 24% for the YRI population.
2. Materials and Methods
2.1 Patient population and sample acquisition
The samples as acquired were coded so as to blind the investigator to the diagnostic category and the code was broken after data submission to a 3rd party (first phase of the study) and to a co-author (SL) for the second phase of the study. The internal review boards of the University of Colorado and Johns Hopkins University approved this study. The age range of the controls at the time of evaluation was 22–96 yrs. The age distribution was such that approximately 1/4 of the study controls were less than 30 yrs of age. The sample set consisted of cases and controls of both genders, obtained from two sources: a) 323 DNA samples isolated from postmortem brain tissue obtained from the Stanley Brain Bank collection (Stanley Medical Research Institute (SMRI), Chevy Chase, MD) including 311 Caucasian individuals, 125 with schizophrenia, 97 with bipolar disorder and 89 normal controls; 12 African American individuals, 8 with schizophrenia, 3 with bipolar disorder and 1 normal control; b) 580 DNA samples isolated from immortalized lymphoblastoid cells from the University of Colorado Schizophrenia Center (Dr. Sherry Leonard), including 350 Caucasian individuals with schizophrenia, 80 African American individuals with schizophrenia, 102 Caucasian normal controls, and 48 African American normal controls. The methods for postmortem diagnosis employed by SMRI involved DSM-IV criteria employed by at least two independent psychiatrists reviewing the records, as described by Torrey et al. (2000). The diagnostic methods employed by the University of Colorado Schizophrenia Center utilized a Structured Clinical Interview for DSM-IV Axis I Disorder (Stephens et al., in press). A number of related individuals representing 39 Caucasian families with more than one member diagnosed with schizophrenia, composing 20% of the total schizophrenia Caucasian case population were present in the Colorado sample collection. In addition, 9 African American families with more than one member diagnosed with schizophrenia, were part of this collection, composing 21.5% of the African American case population. The analysis was therefore adjusted for the common genetic background shared by the related individuals, and differences in ethnic background and collection sites (see subsequent statistical analyses section).
Methods employed by the SMRI Brain Bank collection are described online: (http://www.stanleyresearch.org/programs/documents/bc_statement.pdf). The SMRI collection includes adult individuals autopsied in coroner’s offices located in Maine, Minneapolis, Seattle, and San Diego. The SMRI DNA was isolated from archived, frozen postmortem tissue.
The University of Colorado collection is described in more detail elsewhere (Stephens et al., in press).
2.2 Analyses for genetic polymorphisms in candidate genes
All experimental work and determination of allele identity (Table 1) was carried by an investigator blind to the diagnostic category of each sample. The assays were carried out by TaqMan® PCR to distinguish alleles, using kits provided by Applied Biosystems (Foster City, CA), on the Applied Biosystems 7900HT-Fast Real-time PCR System: 95 °C 10 min, [92 °C 15 sec, 60 °C 60 sec] × 47 cycles. The percent call rate was 99.86% for HM74, HM74A, and MC5R; 100% for MCHR1 and MCHR2; and 99.78% for TDO2. Samples were selected randomly for replication (approximately 4% of the sample set). The correspondence between the repeated assays and the original assays was 100%.
Table 1.
SNP characteristics
| A. The database identifiers, target sequence, and related information for each SNP analyzed in this study. | ||
|---|---|---|
| Gene | SNP rs designation |
Target sequence*, type of polymorphism, any change in translation product, and ancestral allele. |
| HM74A | N/A | CAGAGGAAGAT[G/A]ACAGGTG; nonsynonymous in sole exon; A= isoleucine instead of methionine; ancestral is likely G. |
| HM74 | rs2454727 | CAGAGGAAGAT[G/A]ACAGGTG; nonsynonymous in sole exon; A= isoleucine instead of methionine; ancestral is likely G. |
| MCHR1 | rs133073 | CCTCGCTGCTGCCCACTGGTCCCAA[C/T] G CCAGCAACACCTCTGATGGCCCCG; a synonymous polymorphism in exon 1: transition substitution in exon 1; ancestral is C. |
| MCHR2 | rs9376618 | TGCTAATGGAAAACCACTGAAGACA[C/G] TTCAATAGCCACAATGTGCAAAATA; in intron 5 of full-length mRNA; transversion substitution; ancestral is G. |
| MC5R | rs2236700 | TGGTGTCTCTGTACATACACATGTT[C/G]C TCCTGGCGCGGACTCACGTCAAGC; nonsynonymous in sole exon; G = leucine instead of phenylalanine; ancestral is G. |
| TDO2 | rs2271537 | TTTACTTCTGCTATGCTTCTATATA[A/C]TT TTCTATTGTCAAAGAAAGAAAAA; intron 4; transversion substitution; ancestral is A. |
| B. Sequence of gene-specific outer primers and common inner primers (custom synthesis) used in nested PCR to distinguish the polymorphism common to both HM74A and HM74. | ||||
| Niacin- receptor duplicated gene ID |
Gene-specific forward primer for outer PCR |
Gene specific reverse primer* for outer PCR |
Common forward primer for inner PCR |
Common reverse primer for inner PCR |
|---|---|---|---|---|
| HM74A | GCCATCATCTCT TGCCTTCT |
TGGCATGGT TATTTAAGG AGAGGT |
TCCCAACTT CTTCTCCAC TTTGATC |
CTCGTGCTG CGGTTATTA TCTG |
| HM74 | GCCTAACAGTC CACCTCCTG |
CTTCTTGGA ATGGTTATTT GAGGT |
TCCCAACTT CTTCTCCAC TTTGATC |
CTCGTGCTG CGGTTATTA TCTG |
Only the target sequences, not the primer sequences for Applied Biosystems inventoried TaqMan® SNP products are released by the company.
Note that the reverse primer sequences for these genes target 4 bp of insertion/deletion that distinguishes HM74A from HM74 (Miller and Dulay, 2008).
2.3 Statistical analyses
Separate analyses were carried out comparing schizophrenic to unaffected individuals, and comparing schizophrenic plus bipolar subjects to unaffected (the number of bipolar patients was too small to be analyzed as a separate category). Various genetic models (additive, dominant, recessive, heterosis) were investigated to assess the marginal associations of six candidate SNPs to the outcome, using a logistic link function and adjusting for potential differences in ethnic groups. Generalized estimation equations (GEE; Zeger and Liang, 1986) were applied to account for relatedness of a proportion of schizophrenia cases in the University of Colorado sample collection. We also investigated potential SNP-SNP interactions in addition to the marginal SNP effects, using GEE and logic regression, an adaptive regression methodology developed specifically for this purpose (Ruczinski et al., 2003). Model selection was carried out via permutation tests and cross-validation, which addresses the multiple comparisons problem (Ruczinski et al., 2003).
All p values are reported as uncorrected p values, with the threshold for Bonferroni correction made clear. For any particular single gene model assumed (dominant, recessive, etc), we carried out six hypothesis tests. An upper bound on the family wise error rate (FWER, i.e. the probability of at least one false rejection) can be obtained by a Bonferroni correction. Statistical significance for any of the six SNPs tested was declared if a p-value of 0.05/6 = 0.0083 or less was achieved in the final, combined data set. We did not attempt to achieve a FWER of 5% across all genetic models, since in our assessment this was not particular meaningful (we were testing the same SNPs under different assumptions for the underlying genetic model).
The distributions of the homozygotes and heterozygotes for the 6 SNPs analyzed in this work were tested for Hardy-Weinberg Equilibrium in the controls of this sample set (Table 2).
Table 2.
Table of Hardy-Weinberg expected and observed distributions for the controls in the sample set
| Locus | Common homozygotes Expected Observed |
Heterozygotes Expected Observed |
Rare homozygotes Expected Observed |
X2 | |||
|---|---|---|---|---|---|---|---|
| HM74A | 102.48 | 110 | 108.04 | 93 | 28.48 | 36 | 4.63* |
| HM74 | 108.58 | 111 | 105.65 | 101 | 25.68 | 28 | 0.46 |
| MCHR1 | 82.84 | 84 | 116.33 | 114 | 40.84 | 42 | 0.10 |
| MCHR2 | 113.44 | 118 | 103.13 | 94 | 23.44 | 28 | 1.88 |
| MC5R | 154.89 | 158 | 74.22 | 68 | 8.89 | 12 | 1.67 |
| TDO2 | 77.14 | 81 | 116.71 | 109 | 44.14 | 48 | 1.04 |
p<0.05
Population stratification was previously tested and ruled out for the Colorado DNA collection (Stephens et al., in press) by studying a series of 176 SNPs regarded as being informative for ancestry (Enoch et al., 2006). Stratification data was not available for the SMRI DNA collection.
3. Results
3.1 Association between disease and genotype at a single locus
We observed a significant association of HM74 with schizophrenia as well as schizophrenia and bipolar disease combined, after adjusting for differences in study sites and ethnic differences (Table 3). Testing a heterosis model (H), we observed a strong association of HM74 heterozygotes with schizophrenia (OR 1.50, p = 0.006; Table 3), which surpassed the Bonferroni threshold for significance of p= 0.0083 for the 6 comparisons carried out under this particular genetic model. The locus also fit a dominant (D) genetic model (HM74[A,any]). The genotype for the HM74 dominant risk locus [A] was carried by 63% of the cases with schizophrenia, 70% of the patients with bipolar disorder and 54% of controls.
Table 3.
Odds ratios, 95% confidence intervals, and p-values from GEE models for the associations of the six candidate SNPs to the outcome using a logistic link function and adjusting for differences in sites and race. Results are shown for schizophrenia (Scz) and for the combined diagnostic category of schizophrenia and bipolar disorder (Scz+Bipolar), using additive, dominant, recessive, and heterosis models (ad, D, R, and H, respectively1).
| Scz OR (95% CI) |
P* | Scz+Bipolar OR (95%CI) |
P* | |
|---|---|---|---|---|
| HM74A, ad | 0.94 (0.74–1.19) | 0.574 | 0.92 (0.73–1.15) | 0.459 |
| HM74, ad | 1.23 (0.97–1.55) | 0.093 | 1.26 (1.01–1.59) | 0.044 |
| MCHR1, ad | 1.00 (0.80–1.25) | 0.994 | 0.98 (0.80–1.21) | 0.875 |
| MCHR2, ad | 1.02 (0.83–1.27) | 0.823 | 1.00 (0.81–1.25) | 0.964 |
| MC5R, ad | 1.18 (0.90–1.54) | 0.232 | 1.11 (0.85–1.44) | 0.446 |
| TDO2, ad | 1.14 (0.91–1.41) | 0.252 | 1.16 (0.94;1.43) | 0.174 |
| HM74A, D | 1.05 (0.78–1.43) | 0.742 | 1.00 (0.75–1.35) | 0.973 |
| HM74, D | 1.48 (1.10–2.00) | 0.011 | 1.50 (1.12–2.00) | 0.007 |
| MCHR1, D | 1.08 (0.79–1.49) | 0.613 | 1.00 (0.73–1.36) | 0.992 |
| MCHR2, D | 1.00 (0.73–1.36) | 0.990 | 1.03 (0.76–1.39) | 0.843 |
| MC5R, D | 1.29 (0.94–1.76) | 0.113 | 1.19 (0.88–1.61) | 0.255 |
| TDO2, D | 1.01 (0.70–1.45 | 0.968 | 1.04 (0.73–1.49) | 0.811 |
| HM74A, R | 0.69 (0.44–1.06) | 0.093 | 0.70 (0.46–1.08) | 0.110 |
| HM74, R | 0.93 (0.60–1.45) | 0.749 | 1.04 (0.68–1.59) | 0.845 |
| MCHR1, R | 0.88 (0.60–1.28) | 0.488 | 0.95 (0.65–1.37) | 0.773 |
| MCHR2, R | 1.11 (0.72–1.72) | 0.631 | 0.95 (0.62–1.47) | 0.834 |
| MC5R, R | 0.92 (0.46–1.83) | 0.819 | 0.86 (0.43–1.69) | 0.659 |
| TDO2, R | 1.40 (0.97–2.01) | 0.069 | 1.42 (1.00–2.02) | 0.050 |
| HM74A, H | 1.25 (0.93–1.69) | 0.145 | 1.18 (0.88–1.58) | 0.260 |
| HM74, H | 1.50 (1.12–1.99) | 0.006 | 1.44 (1.09–1.90) | 0.011 |
| MCHR1, H | 1.16 (0.87–1.54) | 0.321 | 1.03 (0.78–1.36) | 0.839 |
| MCHR2, H | 0.95 (0.70–1.29) | 0.742 | 1.05 (0.78–1.42) | 0.739 |
| MC5R, H | 1.33 (0.96–1.84) | 0.082 | 1.25 (0.91–1.71) | 0.168 |
| TDO2, H | 0.79 (0.58–1.07) | 0.128 | 0.80 (0.60–1.07 | 0.138 |
HM74A, D= [A,any]; HM74, D= [A,any]; MCHR1, D= [C,any]; MCHR2, D= [C,any]; MC5R, D= [G,any]; TDO2, D=[C,any]. The recessive model (R) for each involves the homozygote of the allele specified.
A p value of 0.0083 is the threshold required for Bonferonni significance, given 6 SNP comparisons.
TDO2 showed a trend for association with disease, but the p value did not reach the threshold required for multiple comparisons. None of the single melanotropin loci showed a trend for risk of disease. Table 4 illustrates the data from the two sample populations (Stanley Medical Research Institute (SMRI) DNA collection and the University of Colorado DNA collection) shown separately to illustrate the significance level reached in each study.
Table 4.
Odds ratios, p values and 95% confidence intervals, from GEE models for complex genotypes derived from candidate interactions and single loci of significance, tested for association with outcome and reported by study.
| Study 1a Scz OR, p value, (CI) |
Study 2b Scz OR, p value (CI) |
Combined Studiesa,b, Scz OR, p* value (CI) |
Combined studies, Scz plus bipolar disorder, OR, p* value, (CI) |
% Scz plus bipolar disorder with genotype1,2 |
|
|---|---|---|---|---|---|
| Complex risk genotypes, tested across study groups |
|||||
| A. TDO2[CC]3plus MC5R[G,any] plus MCHR2[GC] |
4.74, p= 0.044 (1.27–30.9) |
5.35, p= 0.047 (1.02– 28.0) |
4.84, p= 0.005 (1.60– 14.6) |
3.93,p= 0.014 (1.32–11.74) |
7%1 |
| B. HM74[A,any] plus MCHR1[T,any] plus MCHR2[C,any] |
2.20, p= 0.010 (1.18–4.24) |
1.46, p= 0.077 (0.96– 2.23) |
1.69, p= 0.004 (1.18–2.41) |
1.70,p= 0.003 (1.20–2.39) |
30%1 |
| Melanotropin receptor candidate interactions with the TDO2 or HM74 risk background |
|||||
| A. MC5R[G,any] plus MCHR2[GC] in TDO2[CC]3 |
3.74, p= 0.100 (0.90– 25.6) |
5.11, p= 0.084 (0.80–32.6) |
4.30, p= 0.016 (1.31– 14.14) |
3.30, p= 0.046 (1.02–10.7) |
19%2 |
| B. MCHR1 [T,any] plus MCHR2 [C,any] in HM74[A,any] |
2.26, p= 0.027 (1.11–4.71) |
1.22, p= 0.450 (0.73–2.05) |
1.52, p= 0.05 (1.00–2.32) |
1.52, p= 0.044 (1.01–2.28) |
47%2 |
| The single risk loci by site |
|||||
| HM74[A,any] | 1.39, p= 0.270 (0.78– 2.48) |
1.51, p= 0.023 (1.06– 2.15) |
1.48, p= 0.011 (1.10–2.00) |
1.50, p= 0.007 (1.12–2.00) |
63%1 |
| TDO2[CC]3 | 1.74, p= 0.077 (0.95–3.26) |
1.28, p= 0.29 (0.81–2.00) |
1.40, p= 0.069 (0.98–2.01) |
1.42, p= 0.050 (1.00– 2.02) |
33%1 |
The SMRI sample collection. For the analyses limited to this study site, African Americans were excluded as they were too few in number in this collection.
The University of Colorado Health Sciences Schizophrenia Center sample collection. Population stratification has been tested and ruled out for this sample collection (Stephens et al., in press).
A p value of 0.0083 is the threshold required for Bonferonni significance, given 6 SNP comparisons.
3.2 Candidate interactions tested between kynurenine pathway and melanotropin genes
TDO2 was found to interact significantly with the melanotropin receptor polymorphisms. The interaction was suggested by the data for individuals with the TDO2[CC] genotype, where an OR 4.30, p 0.016 for schizophrenia was conveyed by MC5R[G,any] plus MCHR2[GC] versus those without this melanotropin genotype (data line 3, Table 4). Across the study population, TDO2[CC] plus MC5R[G,any] plus MCHR2[GC] yielded an OR of 4.84, p= 0.005,when comparing schizophrenia patients to controls (data line 1, Table 4). This genotype is carried by 8% of the patients with schizophrenia, 4% of the patients with bipolar disorder and 1.6% of controls.
In addition, an interaction was present between HM74 and melanotropin receptors. In individuals with the HM74 risk genotype [A,any], the melanotropin genotypes MCHR1[T,any] plus MCHR2[C,any] conveyed an OR of 1.52 for schizophrenia, p = 0.049, and an OR of 1.52 for the combined diagnostic group, p = 0.044 (data line 4, Table 4). Across the study population, the HM74[A,any] plus MCHR1[T,any] plus MCHR2[C,any] genotype yielded an OR of 1.69, p= 0.004 when comparing the schizophrenia group to controls and an OR of 1.70, p= 0.003, for the combined diagnostic category of schizophrenia plus bipolar disorder (data line 2, Table 4). This genotype is carried by 29% of the patients with schizophrenia, 35% of the patients with bipolar disorder and 19% of controls.
A combination of both complex risk genotypes i.e., (HM74[A,any] plus MCHR1[T,any] plus MCHR2[GC] plus MC5R[G,any] plus TDO2[CC], was present in 16 individuals with schizophrenia, 1 individual with bipolar disorder and no controls.
3.3 Logic regression analysis
The logic regression method identified an interaction between diagnosis and the TDO2 and HM74 loci, adjusting simultaneously for potential differences in race and study site. The employed model selection methods addressed the multiple comparisons problem. As seen for the marginal analyses (Table 3), subjects who had two C alleles in TDO2 or at least one A allele in HM74 were at a higher risk for disease (Table 5). In addition, comparing subjects with and without genotype for TDO2 CC among those with genotype GG for HM74 (i.e. not dominant) from the same race and the same study site, the odds for the subject with TDO2[CC] are 2.93 times higher to have schizophrenia. The results for the schizophrenia plus bipolar analysis were in essence the same, except all parameter estimates achieved a higher statistical significance (Table 5).
Table 5.
Odds ratios, confidence intervals, and p-values for the significant variables identified by logic regression. We compared schizophrenic subjects only to controls (columns 1–2), and schizophrenic plus bipolar subjects to controls (columns 3–4), using generalized estimation equations to account for family structure. Adjusting for potential differences in race and study sites, we fitted a main effects models (lines 1–2), a model with main effects and an interaction (lines 3–5), and a model with the Boolean term derived from logic regression (line 6).
| Scz OR (95% CI) |
P** | Scz+Bipolar OR (95%CI) |
P** | |
|---|---|---|---|---|
| TDO2[CC] | 1.4 (0.97–2.02) | 0.07363 | 1.43(1.00–2.04) | 0.05174 |
| HM74[A,any] | 1.48 (1.09–2.00) | 0.01080 | 1.50(1.12–2.01) | 0.00651 |
| TDO2[CC] | 2.93 (1.60–5.39) | 0.00053 | 3.00(1.65–5.46) | 0.00034 |
| HM74[A,any] | 1.91 (1.35–2.69) | 0.00024 | 1.94(1.39–2.72) | 0.00010 |
| TDO2[CC] plus HM74[A,any] |
0.31* (0.15–0.64) | 0.00143 | 0.31(0.15–0.62) | 0.00110 |
| TDO2[CC] or HM74[A,any] | 1.96 (1.42–2.71) | 0.00004 | 2.00(1.46–2.74) | 0.00001 |
The OR for the interaction being less than 1 means the effects of HM74[A,any] and TDO2[CC] are not additive, i.e. that adding the individual effects would significantly overestimate the odds of disease when both HM74[A,any] and TDO2[CC] are true.
Note that permutation testing with cross-validation is not confounded by multiple comparisons. It establishes the presence or absence of signal in the data, and assesses the correct model size. The p-values are informative for quantifying the effect size relative to standard error.
4. Discussion
4.1 Genotypes associated with phenotype
The risk conveyed by two complex genotypes for schizophrenia and bipolar disorder exceeded that of the single loci, suggesting that the gene-gene interactions augment the probability of disease (Table 4). The TDO2/MC5R/MCHR2 genotype was associated with an OR of 4.8 for the diagnosis of schizophrenia and the HM74/MCHR1/MCHR2 risk genotype was associated with an OR of 1.7 for the combined diagnostic category, together pertaining to approximately 40% of the patient population.
Of note, the risk alleles in HM74 and MCHR1 match those previously identified by others, i.e. the A allele in rs2454727 of HM74 (Shink et al, 2005) and the T allele in rs133073 of MCHR1 (Severinsen et al., 2006). Furthermore, the complex genotype results for the other loci are consistent with chromosomal markers previously identified as associated with schizophrenia and/or bipolar disorder in genome-wide linkage and association studies. To summarize the prior reports, significant outcomes were obtained for: a) markers within 1cM of MCHR2 (bipolar disorder, p≤ 0.05, Dick et al., 2003; Lambert et al., 2005; and schizophrenia, unweighted MLS p<0.01, Levinson et al., 2000); b) markers within 1MB of MC5R (bipolar disorder, p<0.001, Lin et al., 2005; schizophrenia, p= 0.02, Schwab et al., 1998); and c) for a broad region on chromosome 4q (133MB-187MB) spanning the location of TDO2 (schizophrenia, p<0.004; Vawter et al., 2006) and for a marker within 1.5 MB of TDO2 (schizophrenia, p= 0.01, Hovatta et al., 1999).
4.2 Interpreting the genotype risks
4.2.1 Kynurenic acid as a mediator of pathophysiology
The psychotomimetic effects potentially mediated by kynurenine pathway intermediates, include antagonism of the NMDA receptor by kynurenic acid as proposed by Schwarcz (2001) and reviewed by Coyle (2006). In addition, Hilmas et al. (2001) have demonstrated kynurenic acid inhibition of the α-7 nicotinic receptor. Antagonism of this receptor in the rat (Shepard et al., 2003) elicits the auditory gating endophenotype associated with schizophrenia (Freedman et al., 2003). Activators of the pathway would be expected to elevate kynurenic acid levels and this may underlie the gene-gene interaction found for TDO2 and MC5R (Model 1, Figure 1).
4.2.2 Neurotoxicity of pathway intermediates and pigment products
Neurotoxic intermediates are also of interest, as the known neurotoxins produced by kynurenine pathway activation include quinolinic acid and 3-hydroxykynurenine (Guidetti and Schwarcz, 1999). Again, these products will be generated by activation of the pathway and would potentially relate to the gene-gene interaction of two co-activators of the pathway (TDO2 and MC5R, Model 1, Figure 1). To the list of neurotoxic products should be added the pigment products of 3-hydroxykynurenine and 3-hydroxyanthranilic acid as the elevation in pathway intermediates extends through to 3-hydroxyanthranilic acid in schizophrenia (Miller et al., 2008). This interpretation of the pathophysiology relates to the gene-gene interaction of HM74A and MCHR1 (Model 2 in Figure 1, wherein the pigment products may not be adequately sequestered when the risk genotype for MCHR1 is present combined with the risk genotype for HM74A, as the ligand for HM74A (niacin) controls levels of kynurenine pathway products in human subjects (Hankes et al., 1971). The types of kynurenine pathway pigments range from the xanthomattins, a form that predominates in the eye and thought to be involved in cataract formation (Vazquez et al., 2000) to the melanin family of pigments (Vogliardi et al.,2004), to the antibiotic, cinnabarinic acid (Vazquez et al., 2000). As a general class, the toxicity of pigment molecules and their reactive precursors has been well recognized, including the well-known kernicterus induced by bilirubin pigment and the toxicity of neuromelanin when it is not adequately sequestered (Offen et al., 1997). In-vitro, the type of melanin formed can be strongly affected by the presence of the kynurenine metabolite 3-hydroxyanthranilic acid (Figure 1), which shifts the production of the classic polymerized black eumelanin to the formation of a reddish-brown pigment that is less-completely polymerized and potentially more soluble (Soddu et al., 2004). As described for heme (Slater et al., 1991), polymerization affords protection from dispersion of the toxic component of pigment, whereas enhanced solubility increases the distance across which the toxic component can act through the cytoplasm or the intercellular milieu.
None of the proposed models explain the gene-gene interactions found for the melanotropin receptor MCHR2. Functional MCHR2 is expressed in humans and primates, but not rodents (Tan et al., 2002), a fact that has hindered understanding of its physiological role. In this study, genotyping of MCHR2 was undertaken solely due to its chromosomal location being associated with disease phenotype in genome-wide association studies as discussed in Section 4.1 above.
4.4 Limitations of the study
As with many genetic association and linkage studies for schizophrenia and bipolar disorder, some of the controls fall within the age range of peak onset. Here, ¼ of the controls were less than 30 years of age at the time of diagnosis, and thus, it is certainly possible that an unknown percentage have gone on to develop a psychiatric diagnosis. However, none of the controls reported first degree relatives with schizophrenia or bipolar disorder, and thus, if some were to have been subsequently received a psychiatric diagnosis of relevance, it is likely that fewer of those cases would be genetic in origin than are cases with a family history of disease.
The actual mechanisms for the gene-gene interactions reported here cannot be ascertained from the results we present or from prior data. Therefore, three hypothetical models have been presented for the gene-gene interactions based on the known function of the genes involved, but those models are solely for the purposes of stimulating further research and do not represent established theory.
5. Conclusion
The associations and interactions we have identified in this preliminary genetic study provide the basis for a novel direction in schizophrenia and bipolar genetic research, one that is relevant to a substantial percentage of the patient population. Maximal risk for schizophrenia and bipolar disorder was found to be conveyed by a complex genotype of TDO2/MCHR2/MC5R, and, although relevant to a larger percent of the patient population, lesser risk was conveyed by a complex genotype of HM74/MCHR1/MCHR2. Obtaining the haplotype for the associated alleles will be the crucial next step for ascertaining the causative sequence and will allow a more accurate determination of risk. Follow-up studies in animal models will be necessary to decipher the mechanism of interaction between the genes for melanotropin receptors and kynurenine pathway regulators, to provide a framework for investigating interactions with other related genes, and to reveal the optimal therapeutic targets.
Acknowledgments
The authors are very grateful to Theodore and Vada Stanley for their support of SMRI, which funded a large portion of this study. This work was also supported by the Veterans Affairs Medical Research Service [SL]; and the National Institutes of Health [HL090577 to IR, MH066115 to RGR, and DA09457, MH08177, MH068582 to SL]. The study sponsors played no role in the study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the paper for publication.
Footnotes
PM, IR, RGR, MS, BS and SL have no competing financial interests. Since submission of the manuscript, CLM has transitioned to self-employment in the sole proprietorship MillerBio, a for-profit company dedicated to the development of genetic tools to study the human genome. The success of those products will be no more affected by the publication of this paper than the publication of other papers demonstrating an association between genes and human disease. Furthermore, no patents will be filed on the association between schizophrenia or bipolar disorder and the genes with the associated SNPs we report here as conveying risk for disease.
References
- Allen N, Bagade S, McQueen MB, Ioannidis JP, Kavvoura FK, Khoury MJ, Tanzi RE, Bertram L. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat. Genet. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- Baker BI, Ball JN. Evidence for a dual pituitary control of teleost melanophores. Gen. Comp. Endocrinol. 1975;25:147–152. doi: 10.1016/0016-6480(75)90185-9. [DOI] [PubMed] [Google Scholar]
- Barry S, Clarke G, Scully P, Dinan TG. Kynurenine pathway in psychosis: evidence of increased tryptophan degradation. J. Psychopharmacol. 2008 Jun 18; doi: 10.1177/0269881108089583. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Bednarek MA, MacNeil T, Tang R, Fong TM, Cabello MA, Maroto M, Teran A. Potent and selective agonists of alpha-melanotropin (αMSH) action at human melanocortin receptor 5; linear analogs of alpha-melanotropin. Peptides. 2007;28:1020–1028. doi: 10.1016/j.peptides.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Berrettini WH. Susceptibility loci for bipolar disorder: overlap with inherited vulnerability to schizophrenia. Biol. Psychiatry. 2000;47:245–251. doi: 10.1016/s0006-3223(99)00226-7. [DOI] [PubMed] [Google Scholar]
- Coyle JT. Glial metabolites of tryptophan and excitotoxicity: coming unglued. Exp. Neurol. 2006;197:4–7. doi: 10.1016/j.expneurol.2005.10.021. [DOI] [PubMed] [Google Scholar]
- Dean K, Murray RM. Environmental risk factors for psychosis. Dialogues Clin. Neurosci. 2005;7:69–80. doi: 10.31887/DCNS.2005.7.1/kdean. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Foroud T, Flury L, Bowman ES, Miller MJ, Rau NL, Moe PR, Samavedy N, El-Mallakh R, Manji H, Glitz DA, Meyer ET, Smiley C, Hahn R, Widmark C, McKinney R, Sutton L, Ballas C, Grice D, Berrettini W, Byerley W, Coryell W, DePaulo R, MacKinnon DF, Gershon ES, Kelsoe JR, McMahon FJ, McInnis M, Murphy DL, Reich T, Scheftner W, Nurnberger JI., Jr Genomewide linkage analyses of bipolar disorder: a new sample of 250 pedigrees from the National Institute of Mental Health Genetics Initiative. Am. J. Hum. Genet. 2003;73:107–114. doi: 10.1086/376562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Shen PH, Xu K, Hodgkinson C, Goldman D. Using ancestry-informative markers to define populations and detect population stratification. J. Psychopharmacol. 2006;20:4S19–4S26. doi: 10.1177/1359786806066041. [DOI] [PubMed] [Google Scholar]
- Erhardt S, Schwieler L, Engberg G. Kynurenic acid and schizophrenia. Adv. Exp. Med. Biol. 2003;527:155–165. doi: 10.1007/978-1-4615-0135-0_18. [DOI] [PubMed] [Google Scholar]
- Freedman R, Olincy A, Ross RG, Waldo MC, Stevens KE, Adler LE, Leonard S. The genetics of sensory gating deficits in schizophrenia. Curr. Psychiatry Rep. 2003;5:155–161. doi: 10.1007/s11920-003-0032-2. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Shields J. A polygenic theory of schizophrenia. Proc. Natl. Acad. Sci. U. S. A. 1967;58:199–205. doi: 10.1073/pnas.58.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidetti P, Schwarcz R. 3-Hyroxykynurenine potentiates quinolinate but not NMDA toxicity in the rat striatum. Eur. J. Neurosci. 1999;11:3857–3863. doi: 10.1046/j.1460-9568.1999.00806.x. [DOI] [PubMed] [Google Scholar]
- Hankes LV, Leklem JE, Brown RR, Mekel RC. Tryptophan metabolism in patients with pellagra: problem of vitamin B 6 enzyme activity and feedback control of tryptophan pyrrolase enzyme. Am. J. Clin. Nutr. 1971;24:730–739. doi: 10.1093/ajcn/24.6.730. [DOI] [PubMed] [Google Scholar]
- Hilmas C, Pereira EFR, Alkondon M, Rassoulpour A, Schwarcz R, Albuquerque EX. The brain metabolite kynurenic acid inhibits alpha7 nicotinic receptor activity and increases non-alpha7 nicotinic receptor expression: physiopathological implications. J. Neurosci. 2001;21:7463–7473. doi: 10.1523/JNEUROSCI.21-19-07463.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffer A, Osmond H, Callbeck MJ, Kahan I. Treatment of schizophrenia with nicotinic acid and nicotinamide. J. Clin. Exper. Psychopathol. 1957;18:131–158. [PubMed] [Google Scholar]
- Hovatta I, Varilo T, Suvisaari J, Terwilliger JD, Ollikainen V, Arajärvi R, Juvonen H, Kokko-Sahin ML, Väisänen L, Mannila H, Lönnqvist J, Peltonen L. A genomewide screen for schizophrenia genes in an isolated Finnish subpopulation, suggesting multiple susceptibility loci. Am. J. Hum. Genet. 1999;65:1114–1124. doi: 10.1086/302567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert D, Middle F, Hamshere ML, Segurado R, Raybould R, Corvin A, Green E, O’Mahony E, Nikolov I, Mulcahy T, Haque S, Bort S, Bennett P, Norton N, Owen MJ, Kirov G, Lendon C, Jones L, Jones I, Holmans P, Gill M, Craddock N. Stage 2 of the Wellcome Trust UK-Irish bipolar affective disorder sibling-pair genome screen: evidence for linkage on chromosomes 6q16-q21, 4q12-q21, 9p21, 10p14-p12 and 18q22. Mol. Psychiatry. 2005;10:831–841. doi: 10.1038/sj.mp.4001684. [DOI] [PubMed] [Google Scholar]
- Levinson DF, Holmans P, Straub RE, Owen MJ, Wildenauer DB, Gejman PV, Pulver AE, Laurent C, Kendler KS, Walsh D, Norton N, Williams NM, Schwab SG, Lerer B, Mowry BJ, Sanders AR, Antonarakis SE, Blouin JL, DeLeuze JF, Mallet J. Multicenter linkage study of schizophrenia candidate regions on chromosomes 5q, 6q, 10p, and 13q: schizophrenia linkage collaborative group III. Am. J. Hum. Genet. 2000;67:652–663. doi: 10.1086/303041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PI, McInnis MG, Potash JB, Willour VL, Mackinnon DF, Miao K, Depaulo JR, Zandi PP. Assessment of the effect of age at onset on linkage to bipolar disorder: evidence on chromosomes 18p and 21q. Am. J. Hum. Genet. 2005;77:545–555. doi: 10.1086/491602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CL, Hruby VJ, Matsunaga TO, Bickford PC. Alpha-MSH and MCH are functional antagonists in a CNS auditory gating paradigm. Peptides. 1993;14:431–440. doi: 10.1016/0196-9781(93)90128-4. [DOI] [PubMed] [Google Scholar]
- Miller CL, Llenos IC, Dulay JR, Barillo MM, Yolken RH, Weis S. Expression of the kynurenine pathway enzyme tryptophan 2,3-dioxygenase is increased in the frontal cortex of individuals with schizophrenia. Neurobiol. Dis. 2004;15:618–629. doi: 10.1016/j.nbd.2003.12.015. [DOI] [PubMed] [Google Scholar]
- Miller CL, Llenos IC, Dulay JR, Weis S. Upregulation of the initiating step of the kynurenine pathway in postmortem anterior cingulate cortex from individuals with schizophrenia and bipolar disorder. Brain Res. 2006:1073–1074. 25–37. doi: 10.1016/j.brainres.2005.12.056. [DOI] [PubMed] [Google Scholar]
- Miller CL, Llenos IC, Cwik M, Walkup J, Weis S. Alterations in kynurenine precursor and product levels in schizophrenia and bipolar disorder. Neurochem. Int. 2008;52:1297–1303. doi: 10.1016/j.neuint.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Miller CL, Dulay JR. The high-affinity niacin receptor HM74A is decreased in the anterior cingulate cortex of individuals with schizophrenia. Brain Res. Bull. 2008;77:33–41. doi: 10.1016/j.brainresbull.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Myint AM, Kim YK, Verkerk R, Park SH, Scharpé S, Steinbusch HW, Leonard BE. Tryptophan breakdown pathway in bipolar mania. J. Affect. Disord. 2007;102:65–72. doi: 10.1016/j.jad.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Nagamoto HT, Adler LE, Waldo MC, Griffith J, Freedman R. Gating of auditory response in schizophrenics and normal controls. Effects of recording site and stimulation interval on the P50 wave. Schizophr. Res. 1991;4:31–40. doi: 10.1016/0920-9964(91)90007-e. [DOI] [PubMed] [Google Scholar]
- Offen D, Ziv I, Barzilai A, Gorodin S, Glater E, Hochman A, Melamed E. Dopamine-melanin induces apoptosis in PC12 cells; possible implications for the etiology of Parkinson’s disease. Neurochem. Int. 1997;31:207–216. doi: 10.1016/s0197-0186(96)00150-7. [DOI] [PubMed] [Google Scholar]
- Ruczinski I, Kooperberg C, LeBlanc ML. Logic regression. J. Computat. Graph. Stat. 2003;12:475–511. [Google Scholar]
- Saha S, Chant DC, Welham JL, McGrath JJ. The incidence and prevalence of schizophrenia varies with latitude. Acta Psychiatr..Scand. 2006;114:36–39. doi: 10.1111/j.1600-0447.2005.00742.x. [DOI] [PubMed] [Google Scholar]
- Schwab SG, Hallmayer J, Lerer B, Albus M, Borrmann M, Hönig S, Strauss M, Segman R, Lichtermann D, Knapp M, Trixler M, Maier W, Wildenauer DB. Support for a chromosome 18p locus conferring susceptibility to functional psychoses in families with schizophrenia, by association and linkage analysis. Am. J. Hum. Genet. 1998;63:1139–1152. doi: 10.1086/302046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severinsen JE, Als TD, Binderup H, Kruse TA, Wang AG, Vang M, Muir WJ, Blackwood DH, Mors O, Børglum AD. Association analyses suggest GPR24 as a shared susceptibility gene for bipolar affective disorder and schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006;141:524–533. doi: 10.1002/ajmg.b.30335. [DOI] [PubMed] [Google Scholar]
- Shepard PD, Joy B, Clerkkin L, Schwarcz R. Micromolar brain levels of kynurenic acid are associated with disruption of auditory sensory gating in the rat. Neuropsychopharacology. 2003;28:1454–1462. doi: 10.1038/sj.npp.1300188. [DOI] [PubMed] [Google Scholar]
- Shink E, Harvey M, Tremblay M, Gagné B, Belleau P, Raymond C, Labbé M, Dubé MP, Lafrenière RG, Barden N. Analysis of microsatellite markers and single nucleotide polymorphisms in candidate genes for susceptibility to bipolar affective disorder in the chromosome 12Q24.31 region. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2005;135:50–58. doi: 10.1002/ajmg.b.30165. [DOI] [PubMed] [Google Scholar]
- Slater AF, Swiggard WJ, Orton BR, Flitter WD, Goldber DE, Cerami A, Henderson GB. An iron-carboxylate bond links the heme units of malaria pigment. Proc. Natl. Acad. Sci. U.S.A. 1991;88:325–329. doi: 10.1073/pnas.88.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soddu G, Sanjust E, Murgia S, Rescigno A. Interference of some tryptophan metabolites in the formation of melanin in vitro. Pigment Cell Res. 2004;17:135–141. doi: 10.1046/j.1600-0749.2003.00120.x. [DOI] [PubMed] [Google Scholar]
- Stephens SH, Logel J, Barton A, Franks A, Schultz J, Short M, Dickenson J, James B, Fingerlin TE, Wagner B, Hodgkinson C, Graw S, Ross RG, Freedman R, Leonard S. Association of the 5’-upstream regulatory region of the α7 nicotinic acetylcholine receptor subunit gene (CHRNA7) with schizophrenia. Schizophrenia Res. doi: 10.1016/j.schres.2008.12.017. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CP, Sano H, Iwaasa H, Pan J, Sailer AW, Hreniuk DL, Feighner SD, Palyha OC, Pong SS, Figueroa DJ, Austin CP, Jian MM, Yu H, Ito J, Ito M, Ito M, Guan XM, MacNeil DJ, Kanatani A, Van der Ploeg LH, Howard AD. Melanin-concentrating hormone receptor subtypes 1 and 2: species-specific expression. Genomics. 2002;79:785–792. doi: 10.1006/geno.2002.6771. [DOI] [PubMed] [Google Scholar]
- Taylor A, Namba K. In vitro induction of CD25+ CD4+ regulatory T cells by the neuropeptide alpha-melanocyte stimulating hormone (alpha-MSH) Immunol. Cell. Biol. 2001;79:358–367. doi: 10.1046/j.1440-1711.2001.01022.x. [DOI] [PubMed] [Google Scholar]
- Torrey EF, Webster M, Knable M, Johnston N, Yolken RH. The Stanley Foundation brain collection and neuropathology consortium. Schizophr. Res. 2000;44:151–155. doi: 10.1016/S0920-9964(99)00192-9. [DOI] [PubMed] [Google Scholar]
- Vawter MP, Atz ME, Rollins BL, Cooper-Casey KM, Shao L, Byerley WF. Genome scans and gene expression microarrays converge to identify gene regulatory loci relevant in schizophrenia. Hum. Genet. 2006;119:558–570. doi: 10.1007/s00439-006-0172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez S, Garner B, Sheil MM, Truscott RJ. Characterisation of the major autoxidation products of 3-hydroxykynurenine under physiological conditions. Free Radic. Res. 2000;32:11–23. doi: 10.1080/10715760000300021. [DOI] [PubMed] [Google Scholar]
- Vogliardi S, Bertazzo A, Comai S, Costa CV, Allegri G, Seraglia R, Traldi P. An investigation on the role of 3-hydroxykynurenine in pigment formation by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2004;18:1413–1420. doi: 10.1002/rcm.1497. [DOI] [PubMed] [Google Scholar]
- Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42:121–130. [PubMed] [Google Scholar]