One of the limitations of interdisciplinarity is the misunderstanding regarding specific words and concepts from one discipline to the other (1). One way to circumvent this aspect is to incorporate the vocabulary of other disciplines, when it corresponds to the appropriate concept. Gene environment interaction is a popular topic, for which there has been to date, more reviews than established findings. There has been numerous attempts to represent what types of interactions could occur (2). Geneticists have proposed the term candidate genes to infer there was a specific hypothesis, usually regarding the function of the gene, justifying its study for a given disease, whereas genomewide comparisons have been called “agnostic” (3), which etymologically means without knowledge. In that context, testing a candidate gene-environment interaction is to test a hypothesis, based on knowledge (4).
It can be done basically two ways (table 1). The first is to study gene(s) in relation to a known environmental factor, such as coal dust (5), occupational allergens, farming or smoking. In the case of smoking for example, we will be interested in genes playing a role in the antioxidant defence, because of the known oxidant role of constituents of tobacco smoke. For farming, because of the general hypotheses related to microbial agents frequently encountered in that setting, we will be interested in various innate immunity genes (6), and in general in various genes in given pathways.
Table 1.
Candidate interactions - Using various types of knowledge to formulate hypotheses
| Previous knowledge | Question |
|---|---|
|
E=> Disease Broad mode of action of E (example 1 E coal mine dust, action oxidant, inflammation, fibrois; example 2 smoking: oxidant, inflammation, example 3 allergen) – In general litterature from epidemiology G=> Effect on relevant pathway (example 1 GST, IL18, MMPs, several genes in a known pathway; 2: GST, NQO1, IL18; 3 receptor of allergen, HLA) |
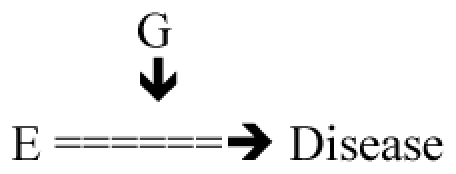 Test: E × G = ? D |
|
G => Disease Known function of gene G (example 1: G CD14, receptor of endotoxin in asthma and allergy; example 2 HLA in sarcoidosis) – In general litterature from genetics and biology E=> Role in that function (example 1: endotoxin; 2: antigen) |
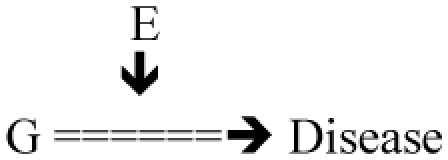 Test: G × E = ? D (formally identical from a statistical point of view to the previous one) |
| G => Disease Unknown role of G (i.e. gene of unknown function found by GWAS) G => Subphenotype of disease (Di) E => Subphenotype of disease (Di) In case of interaction, it will inform on the function of G. In the case of lack of interaction, it will provide interesting (negative) findings increasing knowledge This last strategy is likely relevant for complex disease in order to disentangle the etiological heterogeneity by considering the existing knowledge in phenotypic heterogeneity. Litterature will come from clinical and epidemiology fields. |
independence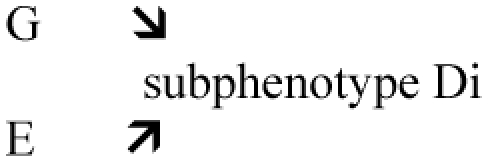 Or interaction  Test: G × E = ? Di |
| E => G expression (epigenetics, i.e. modifying DNA methylation) Previous knowledge may concern the role of E or of G on disease |
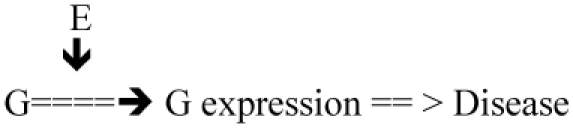 Test: G expression = ? f (E) Test: D = ? f (G expression) |
The second way is to take the complementary perspective, starting by knowledge regarding the potential role of environmental factors in relation to the known function of a specific gene. If we want to study the role of the CD14 gene, which is the receptor of endotoxin, an essential component of the Gram- bacteria membrane, we will then be interested in endotoxin exposure in the environment, i.e. something quite specific and detailed analysis on that exposure is relevant (7).
Further, it is also possible to define other types of candidate interactions, based on other types of knowledge, such as in relation to the known effect of a given environment or a given gene on a specific subphenotype. After the initial discovery by genome wide association study (GWAS) that 17q variants were related to asthma, it was shown that the association of this “new” gene (17q variant) was restricted to early onset asthma. Considering that it is known that early exposure to environmental tobacco smoke is related to early onset asthma, testing whether ETS exposure and 17q variant had interactive effects on early onset asthma was testing a candidate gene-environment interaction, even though the gene variant was initially discovered agnostically (8). Specific environmental exposures like diet can induce genetic alterations such as (somatic, acquired) mutations, as often in cancer; while this is statistically a main effect, biologically it is a physico-chemical environment-gene interaction (9). Furthermore, increasing evidence suggests that epigenetic mechanisms, such as DNA methylation modify gene expression (10). Alterations in DNA methylation may partly be due to environmental factors (11). Instances to test candidate interactions based on various types of knowledge will increase. A key aspect is to clearly specify the preexisting knowledge which justify the analytic plan. The use of the term interaction often confuses statistical interaction (product term in a model) with biological interaction and it has even been proposed to avoid that term in gene environment interactions (12). Testing a candidate interaction is based on the a plausible biological basis formulated prior to its statistical testing.
It has often been considered by geneticists that research in complex diseases should start on “hard” data, i.e. the genetic aspects and that the study of environment is secondary (13), whereas traditionally in environmental research, genetic susceptiblity was not viewed as essential to understand the role of environment (14). Things are now changing. Besides the importance of environmental factors, the modest odds ratios observed for gene variants discovered by GWAs for numerous common diseases suggest that the pattern is extremely complex and that tackling gene-environment interactions from both sides is relevant. Now both environment specialists and geneticists see common ground, and the appropriation by all of the vocabulary used by both disciplines should facilitate collaborations. Speaking more often of candidate interactions, i.e. to borrow, as epidemiologists terms coined by geneticists is one way to construct interdisciplinary research (1) on common concepts. The development of a glossary feature initiated in this journal goes in the same direction (15, 16).
Acknowledgments
Supported by the Ga2len project (Global Allergy and Asthma European Network) EU-FOOD-CT-2004-506378
Footnotes
Publisher's Disclaimer: The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd and its Licensees to permit this article (if accepted) to be published in JECH editions and any other BMJPGL products to exploit all subsidiary rights, as set out in our licence (http://jech.bmj.com/ifora/licence.pdf)
References
- 1.Lynch J. It’s not easy being interdisciplinary. Int J Epidemiol. 2006;35:1119–1122. doi: 10.1093/ije/dyl200. [DOI] [PubMed] [Google Scholar]
- 2.Otman R. Theoretical epidemiology. Gene-Environment interaction: definition and study designs. Prev med. 1996;25:764–770. doi: 10.1006/pmed.1996.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunter DJ, Kraft P. Drinking from the fire hose – Statistical issues in genomewide association studies. New Engl J Med. 2007;357:436–439. doi: 10.1056/NEJMp078120. [DOI] [PubMed] [Google Scholar]
- 4.Kauffmann F, Nadif R. Candidate interactions. Europ Respir J. 2007;30:1289–1290. doi: 10.1183/09031936.00055507. [DOI] [PubMed] [Google Scholar]
- 5.Nadif R, Mintz M, Marzec J, Jedlicka A, Kauffmann F, Kleeberger SR. IL18 and IL18R1 polymorphisms, lung HRCT and fibrosis: a longitudinal study in coal miners. Eur Respir J. 2006;26:1100–1105. doi: 10.1183/09031936.00031506. [DOI] [PubMed] [Google Scholar]
- 6.Smit LAM, Siroux V, Bouzigon E, Oryszczyn MP, Lathrop M, Demenais F, Kauffmann F on behalf of the EGEA cooperative group. CD14 and Toll-like receptor gene polymorphisms, country living, and asthma in adults. Am J Respir Crit Care Med. 2009;179:363–368. doi: 10.1164/rccm.200810-1533OC. [DOI] [PubMed] [Google Scholar]
- 7.Eder W, Kilecki W, Yu L, von Mutius E, Riedler J, Braun-Farländer C, Nowak D, Martinez FD the Allergy and Endotoxin (ALEX) Study team. Opposite effects of CD 14/-260 on serum IgE levels in children raised in different environments. J Allergy Clin Immunol. 2005;116:601–607. doi: 10.1016/j.jaci.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Bouzigon E, Corda E, Aschard H, Dizier MH, Boland A, Bousquet J, Chateigner N, Gormand F, Just J, Le Moual N, Scheinmann P, Siroux V, Vervloet D, Zelenika D, Pin I, Kauffmann F, Lathrop M, Demenais F. Effect of 17q21 Variants and Smoking Exposure in Early-Onset Asthma. N Engl J Med. 2008;359:1985–94. doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 9.Rosenbaum PR. The case-only odds ratio as a causal parameter. Biometrics. 2004;60:233–40. doi: 10.1111/j.0006-341X.2004.00154.x. [DOI] [PubMed] [Google Scholar]
- 10.Foley DL, Craig JM, Morely R, Olsson CJ, Dwyer R, Smith K, Saffery R. Prospects for epigenetic epidemiology. Am J Epidemiol. 2009;169:389–400. doi: 10.1093/aje/kwn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore L, Bonefeld-Jorgensen EC. Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic Inuits. Environ Health Perspect. 2008;116:1547–52. doi: 10.1289/ehp.11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenland S. Interactions in epidemiology: relevance, identification, and estimation. Epidemiology. 2009;20:14–17. doi: 10.1097/EDE.0b013e318193e7b5. [DOI] [PubMed] [Google Scholar]
- 13.Schork NJ. Genetics of complex disease: approaches, problems, and solutions. Am J Respir Crit Care Med. 1997;156(4 Pt 2):S103–9. doi: 10.1164/ajrccm.156.4.12-tac-5. [DOI] [PubMed] [Google Scholar]
- 14.Schulte PA. A conceptual framework for the validation and use of biologic markers. Environ res. 1989;48:129–144. doi: 10.1016/s0013-9351(89)80029-5. [DOI] [PubMed] [Google Scholar]
- 15.Malats N, Calafell F. Advanced glossary on genetic epidemiology. J Epidemiol Community Health. 2003;57:562–564. doi: 10.1136/jech.57.8.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunasekara FI, Carter K, Blakely T. Glossary for econometrics and epidemiology. J Epidemiol Community Health. 2008;62:858–86. doi: 10.1136/jech.2008.077461. [DOI] [PubMed] [Google Scholar]


