Abstract
Competence protein A (ComA) is a response regulator protein involved in the development of genetic competence in the Gram-positive spore forming bacterium Bacillus subtilis, as well as the regulation of the production of degradative enzymes and antibiotic synthesis. ComA belongs to the NarL family of proteins which are characterized by a C-terminal transcriptional activator domain that consists of a bundle of four helices, where the second and third helices (α8 and α9) form a helix-turn-helix DNA binding domain. Using NMR spectroscopy, the high resolution three-dimensional solution structure of the C-terminal DNA-binding domain of ComA (ComAC) has been determined. In addition, surface plasmon resonance and NMR protein-DNA titration experiments allowed for the analysis of the interaction of ComAC with its target DNA sequences. Combining the solution structure and biochemical data, a model of ComAC bound to the ComA recognition sequences on the srfA promoter has been developed. The model shows that for DNA binding, ComA uses the conserved helix-turn-helix motif present in other NarL family members. However, the model also reveals that ComA may use a slightly different part of the helix-turn-helix motif and there appears to be some associated domain re-orientation. These observations suggest a basis for DNA binding specificity within the NarL family.
Keywords: Competence, ComA, NMR, two-component, response regulator
Introduction
The integration of newly acquired DNA is implicated in the continued development of the pathogenic nature of bacteria and has been shown to be responsible for the development of antibiotic resistance of bacteria 1; 2; 3; 4; 5. The development of genetic competence in the Gram-positive bacterium Bacillus subtilis is regulated by the ComP-ComA two-component system 6; 7; 8; 9; 10. ComP is a transmembrane histidine kinase that autophosphorylates in response to the extracellular signaling molecule ComX 11; 12. Phosphorylated ComP then activates the response regulator protein ComA by transferring a phosphoryl group. Upon phosphorylation, ComA functions as a transcriptional activator of a variety of genes/operons. Among these, the srfA operon 7; 13; 14 ultimately controls the expression of the late com genes. The late com genes express the protein machinery that bind to and take up exogenous DNA 15; 16; 17; 18.
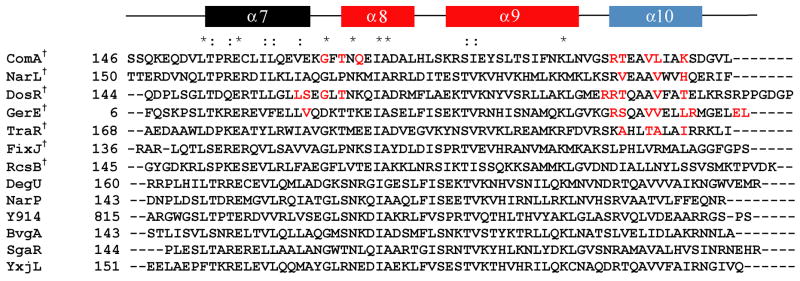
Structurally, ComA consists of two domains, an N-terminal regulatory domain and a C-terminal variable effector domain. The N-terminal domain contains a highly conserved aspartic acid residue at position 55, which is the site of phosphorylation by ComP 19; 20. Through sequence alignment and secondary structure prediction of the C-terminal effector domain, it was determined that ComA belongs to the NarL family of response regulator proteins (Figure 1) 14; 20. This family is characterized by a C-terminal transcriptional activator domain that consists of a grouping of four alpha helices, in which the second and third helices form a helix-turn-helix DNA binding domain, making the C-terminal domain responsible for the interaction with and activation of the target DNA sequence. The srfA operon is the most characterized binding target of ComA due to its critical role in the development of competence 7; 10; 13; 14; 16; 21. However, in addition to the srfA operon, other operons have been identified to be directly regulated by ComA. These include degQ 22, which encodes a regulator of degradative enzyme production, and rapA 23, rapC 24, rapE 25, and rapF, which encode transition phase regulatory proteins 26; 27; 28. Whole genome DNA microarray analysis identified these genes and also identified at least 89 genes in 35 operons to be affected by ComA independent of its role in competence 29. Original analysis of the binding site of ComA to the srfA operon identified two regions of dyad symmetry critical to the positive control of srfA transcription. This resulted in a proposed consensus sequence containing an inverted repeat with two 6 base pair half sites with a four base pair spacer, 5′-TTGCGGnnnnCCGCAA-3′ 10; 13; 14. More recently, alignment of the ComA DNA binding sites of the targets known to be directly activated by ComA identified a third half site important for transcriptional activation 30.
Figure 1.
Sequence alignment of the C-terminus of competence protein A (ComAC) with other members of the NarL family of proteins. The secondary structure prediction is shown on top. Red coloring of the secondary structure represents the helix-turn-helix domain and blue represents the helix involved in the dimerization interface. Residues in red highlight those residues identified as being specifically involved in the dimerization interface. *denotes conserved residues, : denotes conserved substitutions, †denotes proteins with known structures. The known structures include: ComAC (current work), NarL (PDB 1JE8) 35, DosR (PDB 1ZLJ) 36, GerE (PDB 1FSE)37, RcsB (PDB 1P4W)38, FixJC (1X3U)39, and TraR (1H0M and 1L3L)40.
Despite a myriad of previous studies, a structural model of the ComA-DNA binding interaction has remained elusive. To address this, we solved the high resolution NMR solution structure of the C-terminal DNA binding domain of ComA (ComAC, residues 146–214). In addition, DNA binding studies using SPR and NMR protein-DNA titration experiments were performed to analyze the interaction of ComAC with its DNA targets. Combining the ComAC structure with biochemical data allowed a model to be developed of ComAC binding to the target DNA sequences. Subsequent principal component analysis of the structural ensemble revealed an anti-correlated migration of the helix-turn-helix domains, while the protein dimerization interface possessed cooperative migration. This ‘scissor’ like motion may assist in DNA binding.
Results
Characterization of ComA and its domains
Oligomerization state of ComA
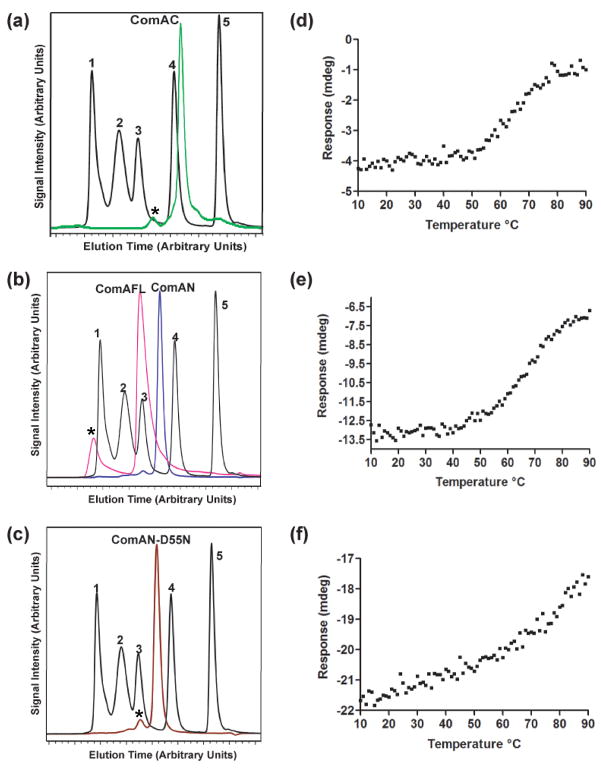
The N-terminal domain of ComA (ComAN) comprises residues 1–126 of the full-length protein while the C-terminal domain of ComA (ComAC) comprises residues 146–214 of the full-length protein. The multimerization state of ComAC as well as ComAN, ComAN-D55N, and ComAFL (full-length ComA) were determined using size exclusion chromatography (Figure 2). At all concentrations (10 μM-1 mM), ComAC eluted at a molecular weight consistent with the formation of a dimer in solution. At all concentrations (10 μM–500 μM), ComAFL was found to be a dimer, corroborating previous results 30. ComAN, as well as ComAN- D55N, was also found to exist as a dimer in solution at all concentrations (10 μM–500 μM).
Figure 2.
Characterization of ComA. Size exclusion chromatography of: A) ComAC (green), B) ComAFL (pink), ComAN (blue), and C) ComAN-D55N (brown). Standards are in black: 1 denotes thyroglobulin (670 kDa), 2 denotes bovine γ-globulin (158 kDa), 3 denotes chicken ovalbumin (44 kDa), 4 denotes equine myoglobin (17.5 kDa), and 5 denotes vitamin B12 (1,350 Da). *Identifies peaks resulting from aggregation at the highest concentration. Thermal denaturation profiles of ComA: D) ComAC E) ComAFL, and F) ComAN, by monitoring the A222 nm over a temperature range of 10 °C to 90 °C.
Thermostability of ComA and its domains
The thermal denaturation midpoint (Tm) was monitored as a change in molar ellipticity at 222 nm as the temperature was increased in 1° increments from 10 to 90 °C for ComAFL, ComAN and ComAC. The denaturation profile (Figure 2) of both ComAFL and ComAC demonstrated a single transition, at 63 °C. ComAN on the other hand does not appear to have a distinct transition point. Comparison of the profiles of ComAN and ComAC indicates that the C-terminus is responsible for the stabilization of the full-length protein, while the N-terminus works to destabilize the full-length protein.
ComAC dimer subunit exchange
A combination of column chromatography and MALDI-TOF mass spectrometry was used to determine whether subunit exchange occurs between the individual subunits comprising the ComAC dimer complex, an essential aspect to NMR solution structure determination. Briefly, a sample of half His-tagged-ComAC and half untagged-ComAC were combined and applied to a Ni-NTA column and eluted as in the original protocol. Eluted fractions containing protein were combined and analyzed by MALDI-TOF mass spectrometry. Both the His-tagged-ComAC and untagged-ComAC were recovered in the elution confirming ComAC undergoes subunit exchange (Figure S1).
Structural determination of ComAC
Heteronuclear multidimensional NMR methods were used to assign 1H, 13C, and 15N backbone and side chain resonances, provide sequential connectivities and define distance relationships between protons, both inter- and intra- molecular, for the development of a high resolution solution structure of ComAC 31. The 1H-15N HSQC spectrum of ComAC shows good spectral dispersion; all NH resonances were assigned with the exception of Gln148 (Figure S2). Ultimately, 97% of backbone resonance assignments (HN, N, Cα, C′) were achieved while 85% of the total assignable side chain atoms were assigned. The dimerization interface was determined by performing 12C/13C isotope edited experiments using three-dimensional 13C- NOESY-HSQC experiments with an 120-ms mixing time 32.
Statistics for the ten lowest energy, water refined structures are shown in Table 1. These structures were determined with 2,316 unambiguous restraints, 1,323 ambiguous restraints with 700 intraresidue, 791 sequential, 1,050 medium range, and 1,098 long range. The ten structure ensemble had zero NOE and/or hydrogen bond violations and dihedral violations per structure. They showed an average Cα r.m.s.d. (Å) of 0.20 ± 0.04 for secondary structure (backbone), 0.56 ± 0.06 secondary structure (heavy), 0.39 ± 0.11 backbone and 0.90 ± 0.06 heavy. The achieved energies of −1416 ± −9 (kcal mol−1) for van der Waals and −5325 ± −98 (kcal mol−1) for electrostatic, in addition to the Ramachandran analysis with 99.9% in the generously allowed or better conformation indicate that the reported structure for ComAC is of excellent quality (Figure 3).
Table 1.
ComAC NMR and refinement statistics.
| NMR distance and dihedral constraints | ComAC dimera |
|---|---|
| Distance constraints | |
| Total NOE | 3,639 |
| Intramolecular | 3,150 (~87%)b |
| Intermolecular | 489 (~13%)b |
| Unambiguous | 2,316 |
| Intraresidue | 700 |
| Sequential (|i – j| = 1) | 791 |
| Medium range (|i – j| <4) | 1050 |
| Long range (|i – j| >5) | 1098 |
| Ambiguous | 1323 |
| Hydrogen bonds | 103 |
| Total dihedral angle restraints | |
| ϕ | 172 |
| ψ | 172 |
| Structure statistics | |
| Energies (kcal mol−1) | |
| van der Waals | −1416 ± −9 |
| Electrostatic | −5325 ± −98 |
| Average violations per structure | |
| NOEs and/or hydrogen bonds | 0 |
| Dihedrals | 0 |
| Violations (mean and S.D.) | |
| Distance constraints (Å) | 0.035 ± 0.005 |
| Hydrogen bonds (Å) | 0.054 ± 0.002 |
| Dihedral angle constraints (degrees) | 0.672 ± 0.024 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.00470 ± 0.00007 |
| Bond angles (degrees) | 0.615 ± 0.008 |
| Impropers (degrees) | 1.60 ± 0.04 |
| Average pairwise r.m.s.d.c (Å) | |
| Secondary structure (backbone) | 0.20 ± 0.04 |
| Secondary structure (heavy) | 0.56 ± 0.06 |
| Backbone | 0.39 ± 0.11 |
| Heavy | 0.90 ± 0.06 |
| Ramachandran analysis | |
| Most favored | 84.2% |
| Additional allowed | 10.3% |
| Generously allowed | 5.4% |
| Disallowed | 0.1% |
Statistics for all dimer residues
Percentage of total NOE’s
Pairwise r.m.s.d. was calculated among the 10 lowest energy refined structures.
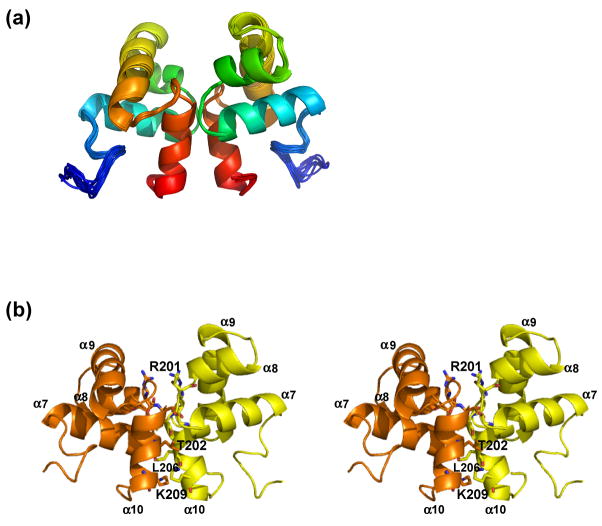
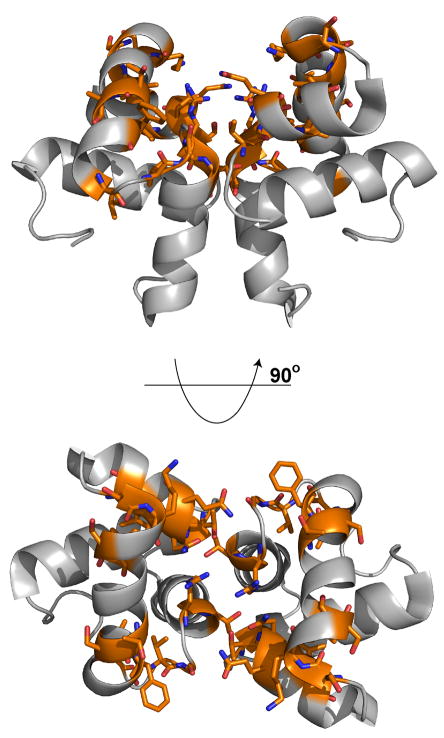
Figure 3.
Solution structure of ComAC. A) Ensemble of the top ten lowest energy structures of ComAC. B) Stereoview of ComAC dimer with subunits A and B designated as orange and yellow, respectively. Residues involved in the dimer interface are represented in stick form.
Dimerization interface of ComAC
ComAC dimerizes in a parallel fashion (Figure 3), similarly to the other proteins in the NarL family. Residues involved in the dimerization interface were determined from the 13C-NOESY-HSQC- 12C/13C isotope edited NMR experiments. These restraints included NOE’s between Val205 and Ile207’, Thr202 and Ala204’, Leu206 and Ala208’, Ile207 and Lys209’, Lys209 and Lys209’, and Ser210 and Ser210’. The amino acid residues contributing the most to the dimer interface were determined by calculating the change in surface accessibility between a monomeric subunit and the dimer complex using NACCESS 33. Upon dimerization, a surface area of 676.83 Å2 of each monomeric subunit becomes buried. This accounts for about 15% of the solvent accessible surface area per subunit. In α10, Thr202 and Leu206 contribute the most, 14.36% and 14.58% of the interface surface, respectively. These are followed by Arg201 (11.43%), Lys209 (10.17%), and Val205 (8.65%). Residues Gly168 (6.52%), Thr170 (8.21%) and Gln172 (6.19%), located in the coil between α7 and the initial part of α8, are also involved. ComAC is composed of four α-helices with the second and third helices (α8 and α9) assuming a conformation typical of helix-turn-helix DNA binding domains. This domain must then be responsible for the transcriptional activation of ComAC’s DNA targets. In the unbound ComAC structure these helices lie at an angle of 116.0° to each other.
DNA interactions
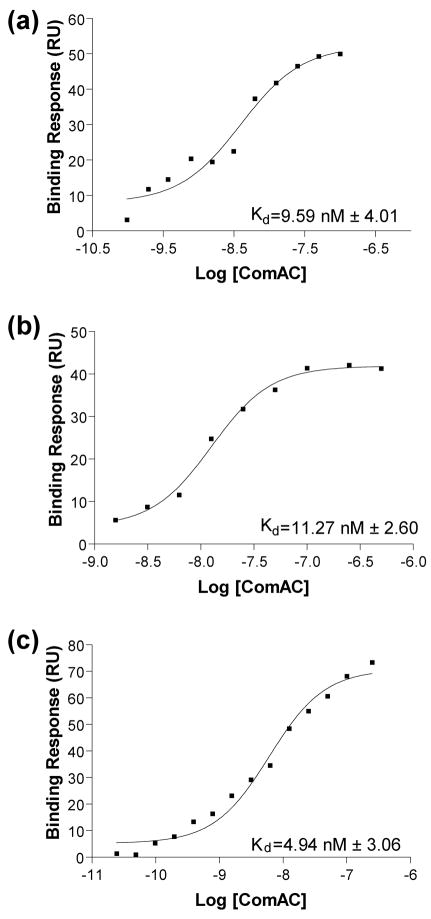
Determination of binding affinities using surface plasmon resonance
SPR analysis was used to determine the binding affinities of the wild-type srfA consensus promoter sequence as well as two different consensus sequences from reference 30. The apparent dissociation equilibrium constants (Kd), calculated using a steady-state affinity model indicated that ComAC binds to its target DNA promoter sequences with extremely low nanomolar affinity (Figure 4). ComAC bound to the wild-type srfA DNA sequence with an average Kd of 9.59 nM with a standard deviation of 4.01. ComAC bound to the consensus RE1–RE3 sequence with an average Kd of 11.27 nM with a standard deviation of 2.60 and with an average Kd of 4.94 nM with a standard deviation of 3.06 to the consensus RE3 sequence.
Figure 4.
Steady state affinity model binding curves. The apparent dissociation (equilibrium) constants (Kd), calculated using a steady-state affinity model. A) Wild-type srfA DNA sequence with an average Kd equal to 9.59 nM. B) Consensus RE1-RE3 sequence with an average Kd equal to 11.27 nM. C) Consensus RE3 sequence with an average Kd equal to 4.94 nM.
ComAC: DNA NMR titration experiments
A series of ComAC-DNA NMR titration experiments were performed using the same DNA sequences described above. Chemical shift changes, line broadening or peak disappearance were followed via 1H/15N-HSQC experiments. The residues that were found to be affected by the addition of the DNA sequences include: Thr170, Asn171, Gln172, Ile174, Ala175, Ser181, Lys182, Ser184, Ile185, Thr190, Ser191, Phe193, Leu196, Val198, Gly199, Arg201 and Thr202 (Figure 5 and S3). Those residues most affected showed peak broadening to the point of disappearance. Arg201 disappeared after titration of a ratio of DNA:protein of 0.25:1. Thr170, Asn171, Gln172, Ser181, Lys182, Thr190, Phe193, Gly199 and Thr202 disappear at a DNA:protein ratio of 0.5:1. Ala175, Ser184, Ile185, Ser191, Leu196 and Val198 disappear at a DNA:protein ratio of 0.75:1. The titration was taken to a 4:1 DNA:protein ratio.
Figure 5.
ComAC NMR solution structure with residues identified by the NMR titration studies as being affected upon the addition of DNA are highlighted in orange and stick form.
HADDOCK ComAC:DNA interaction modeling
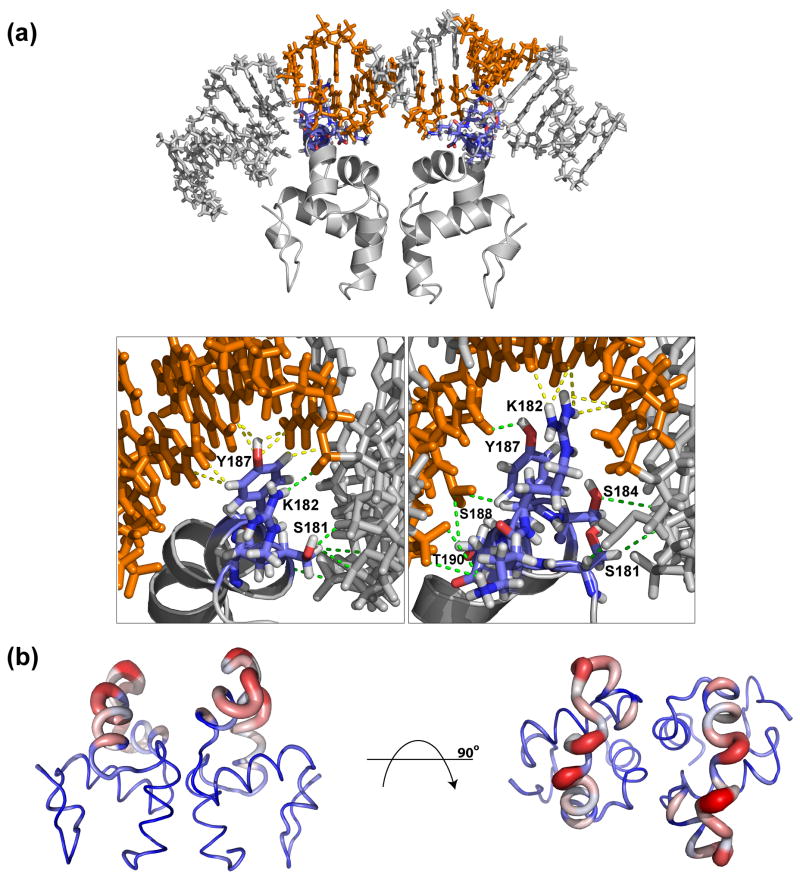
The biomolecular modeling program HADDOCK was used to generate a structural model of the interaction of ComAC with the wild-type srfA DNA sequence. The chemical shift perturbation experiments were used to define residues involved in the interaction. The resulting model and the corresponding refinement statistics can be seen in Figure 6A and Table 2, respectively. An average Cα r.m.s.d. of 1.43 ± 0.55 Å was obtained for the complex interface with an average buried surface area of 1541.36 ± 154.82 Å. PROCHECK analysis 34 of the ten lowest energy structures revealed that 97% were in the allowed regions of Ramachandran space. Comparison of the solved ComAC structure and the ComAC-srfA modeled complex indicates that the monomeric subunits maintain a similar global fold upon binding DNA. Analysis of the interaction interface from the 20 best structures, taken from the lowest energy cluster, show the position of the ComAC dimer in consecutive major grooves with helix α9 extending into the major groove to make base-pair specific interactions. At the dimerization interface, 61% of the intersubunit contacts were conserved upon binding DNA.
Figure 6.
Analysis of ComAC DNA interaction. A) Structural model of the ComAC and DNA interaction. Residues and nucleotide bases involved in the interaction are highlighted in blue and orange, respectively. Insets show a detailed look at the interaction. Green dashes designate backbone interactions and yellow dashes designate major groove interactions. B) Representation of Cα r.m.s.d. values of unbound ComAC and ComAC bound to DNA. Blue represents the lowest difference, white intermediate and red the greatest difference.
Table 2.
HADDOCK Refinement Statistics for ComAC DNA binding model.
| Violations (mean and SD) | |
| Distance constraints (Å) | 0.43 ± 0.02 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.745 ± 0.000008 |
| Bond angles (°) | 7.253 ± 0.0005 |
| Impropers (°) | 0.624 ± 0.007 |
| Average pairwise r.m.s.d.a (Å) | |
| Interface | 1.43 ± 0.55 |
| Buried surface area | 1541.36 ± 154.82 |
| Ramachandran analysis | |
| Favored | 82.1% |
| Allowed | 14.8% |
| Generously allowed | 0.1% |
| Disallowed | 3.0% |
Pairwise r.m.s.d. calculated based on all heavy atoms at the interface.
Analysis of the most populated hydrogen bond interactions between the protein and DNA show extensive hydrogen bonds. Phosphate backbone contacts include the HE# group of Gln172, the HA, HB# and HG groups of Ser181, the HB# and HG groups of Ser184, the HH group of Tyr187, the HB# group of Ser188 and the HG1 group of Thr190. Major groove hydrogen bond contacts primarily include the HH2# group of Arg183 and HE group of Tyr187. In addition, van der Waals interactions with the major groove include the CZ group of Arg183 and the CE group of Tyr187. Calculation of the Cα r.m.s.d. of the unbound ComAC structure and the ComAC-srfA modeled complex indicate that the greatest deviations occur in the helix-turn-helix DNA binding domain, α8 and α9 (Figure 6B). In α8 residues Gln172, Ile174, Asp176, Ala177, Leu178 and His179 showed the highest variation and α9 residues Ser181, Lys182, Arg183, Tyr187, Ser188 and Leu189 showed the highest variation. This indicates that the helix-turn-helix DNA binding domain is the most affected when ComAC interacts with its DNA binding partner.
Discussion
The competence transcription factor ComA is responsible for the transcriptional activation of several DNA targets 30. These DNA targets include srfA, degQ, rapA and rapC promoter regions; the products of these genes control everything from competence development, production of degradative enzymes and antibiotic synthesis to sporulation. ComA is specifically involved in the transcriptional activation of the srfA operon making it an essential player in the development of genetic competence in B. subtilis. Here we determined the high resolution solution structure of the C-terminal DNA binding domain of ComA. This domain is present in solution in dimeric form with each monomeric unit containing a helix-turn-helix DNA-binding motif (α8-α9). Each monomer consists of a grouping of four helices. The dimer is held together by interactions running along the length of α10.
ComA is a member of the NarL family of response regulators. Within this family six structures have been solved including (Figure 1): full-length NarL, the C-terminus of NarL (NarLC) in complex with DNA 35, the C-terminus of DosR (DosRC) 36, the C-terminus of GerE (GerEC) 37, the C-terminus of RcsB (RcsBC) 38, the C-terminus of FixJ (FixJC) 39 and the full-length TraR 40. ComAC has a sequence identity of 24%, 24%, 23%, 23%, 26% and 22% with each protein, respectively. As is common in many proteins with helix-turn-helix DNA-binding domains, ComAC forms a dimer in solution. This is also the case for the family members NarLC, DosRC, GerEC, and TraR. Two particularly well studied structures within the NarL family are NarLC and DosRC, therefore comparisons between ComAC and these proteins will be made where useful.
The C-terminal domains ComAC, NarLC and DosRC are each composed of four α-helices with the second and third helices comprising the helix-turn-helix DNA binding domain. While the overall architecture is similar, slight differences in helical orientation are evident. In the unbound ComAC structure these helices lie at an angle of 116.0° with respect to each other. This angle increases to 119.6° upon binding to the srfA DNA sequence. In the unbound DosRC, they lay at an angle of 106.7° with respect to each other, but upon binding DNA this angle decreases to 102.0°. Two structures of NarL have been solved, unbound full-length (monomer) and the C-terminus bound to DNA (dimer). In the full-length, unbound structure of NarL, these helices lie at an angle of 98.8° with respect to each other, which increases to 108.7° when bound to its DNA target. The functionality of the angle between the second and third α-helices is presumably due to the binding of inverted repeats in their DNA targets, such as 5′-TTGCGGnnnnCCGCAA-3′ in the case of ComAC 30. The difference seen between ComAC, NarLC and DosRC is most likely due to the specific nature of these proteins binding their DNA targets in a slightly different manner.
Another shared feature of the NarL family is the organization of the dimer interface. The interface is formed mainly by α10 from each of the two monomeric subunits, along with the loop between α7 and α8 and the initial part of helix α8 (Figure 1). The angle between the two subunits in the unbound structures, measured at the α10 helices, is 38.2° in ComAC and 28.8° in DosRC. Upon DNA binding this angle decreases to 36.1° in ComAC and slightly increases to 30.5° in DosRC. A significant hydrophobic patch comprises the dimerization interface, primarily residues 204AVLIA208 of ComAC, 206AAVWV210 of NarLC and 200AAVFA204 of DosRC (Figure 1). ComAC has a buried surface area of about 677 Å2 per monomer upon dimerization, accounting for 15% of the solvent-accessible surface per subunit, while NarLC’s and DosRC’s interface accounts for about 10% of the solvent-accessible surface per subunit 35; 36. ComAC’s primary dimer interface residues are Thr202 (Thr198 in DosRC, Val204 in NarLC), Val205 (Val202 in DosRC and Val208 in NarLC), Leu206 and Lys209. The most divergent residue in the ComAC interface compared to DosRC and NarLC is Lys209. The equivalent residue is Thr205 in DosRC and His211 in NarLC, although these residues do share polar characteristics. Therefore, it appears that the dimerization interface is mostly conserved across these proteins.
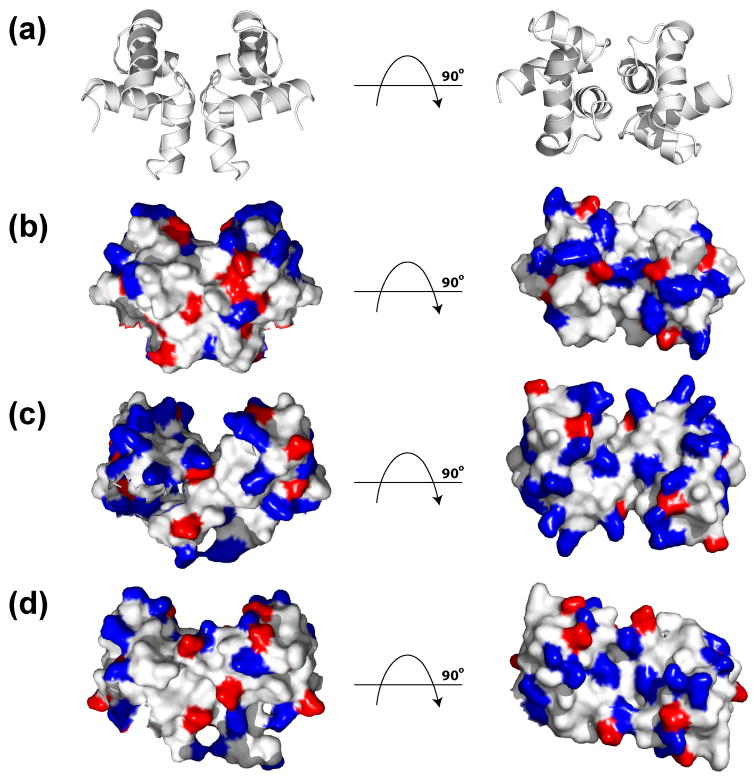
As expected these proteins show similar electrostatic surface characteristics surrounding their DNA binding helix-turn-helix motifs. A positive electrostatic surface at the beginning of α10 is seen in all three proteins and can be attributed to residues Arg201, Arg203, and Arg197 for ComAC, NarLC and DosRC, respectively (Figure 8). Indeed, across the NarL family members listed in Figure 1, a positively charged residue at this position is highly conserved (85%). Consequently, this region likely contributes to the general DNA binding mechanism for the NarL family.
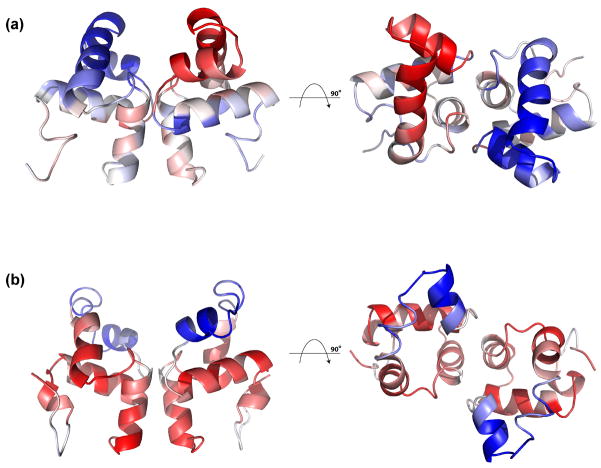
Figure 8.
Highlighting of electrostatic positive and negative amino acid residues. A) Solution structure of ComAC showing orientation of: B) ComAC, C) NarLC, and D) DosRC. Amino acid residues with positive electrostatic character (R, K, H) are highlighted in blue, while amino acid residues with negative electrostatic character (D, E) are highlighted in red.
Interestingly, there are slight differences in the helix-turn-helix DNA binding motif’s electrostatic character among ComAC, NarLC and DosRC. For NarLC, the region of greatest positive electrostatic character is in the middle of α9 which accommodates the residues responsible for DNA binding (188KVHVK192) (Figures 1 and 8C). For DosRC, the region of greatest positive electrostatic character is at the beginning of α9 (179KTVKN183), again containing the residues found to be responsible for DNA binding (Figures 1 and 8D). While NarLC and DosRC show a shift in residues responsible for DNA binding (middle vs. beginning of α9) as well as electrostatic characteristic, ComAC displays a further shift in electrostatic character to include the loop region between α8 and α9, as well as the beginning of α9 (Figures 1 and 8B). This suggests that the residues in this region (181SKRS184) are responsible for direct contact with its DNA targets and represents yet another contribution to the recognition of different DNA targets within the NarL family.
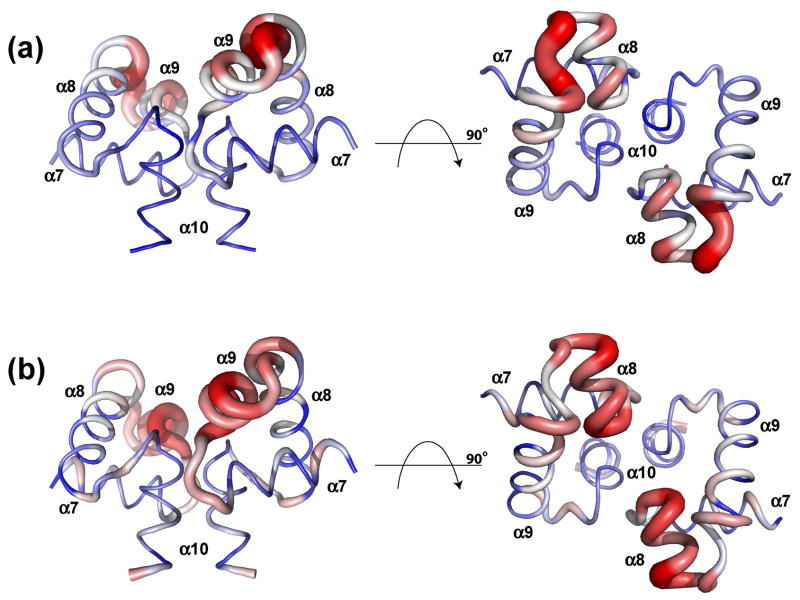
As seen in Figure 7 the greatest Cα r.m.s.d. difference between NarLC and ComAC is present at the N-terminal end of helix α9 and slightly in helix α8, while for DosRC and ComAC the greatest Cα r.m.s.d. difference occurs in helix α8. In both cases this region comprises the DNA binding helix-turn-helix motif. When comparing the overall dimeric structure of ComAC with the dimeric structures of NarLC and DosRC over all residues, the Cα r.m.s.d. values of 4.183 and 4.414 Å are observed. The Cα r.m.s.d. values between NarLC and DosRC themselves are considerably lower. However an alignment of the Cα atoms over only one subunit lowers the Cα r.m.s.d. considerably; 1.608 and 1.613 Å Cα r.m.s.d.’s observed for DosRC-ComAC and NarLC-ComAC, respectively. This suggests a slight difference in the orientation of the two ComAC monomeric subunits with respect to one another, compared to the other proteins. It is important to note that despite many structural similarities, proteins within the NarL family do not appear to recognize one another’s DNA targets. It is plausible that subunit orientation within the family, along with the shifting of recognition residues within the helix-turn-helix motif discussed above, helps drive DNA specificity (Figure 7).
Figure 7.
Calculation of Cα r.m.s.d. variation of ComAC and its homologues in their DNA bound state. The Cα r.m.s.d. differences calculated between ComAC and A) NarLC, B) DosRC. Blue represents the lowest difference, white intermediate and red the greatest difference in addition to the increasing size of the cartoon.
Further studies here support this hypothesis. SPR, NMR chemical shift perturbation studies, and molecular modeling were used to examine how ComAC can interact with its target DNA sequences. SPR experiments determined the binding constants of three different DNA sequences that contained three recognition elements (RE). Consensus sequences have been determined for each of the three recognition elements. The DNA sequence designated “consensus RE3” contains a consensus sequence at position 3 and the consensus RE1-RE3 sequence contains a consensus sequence in all three RE sequences. Similar binding constants, in the low nanomolar range, were obtained for both. Consensus RE3 has a Kd of 4.94 nM ± 3.06 and consensus RE1-RE3 has a Kd of 11.26 nM ± 2.60. In addition, ComAC bound to the wild-type srfA DNA sequence with an average Kd of 9.59 nM with a standard deviation of 4.01. Using a DNase footprinting analysis experiment, NarLC showed a similar binding affinity for its DNA target as the full-length NarL protein, with a Kd of 0.15 nM 35. No binding affinities for DosR and its target DNA sequences could be found in the current literature.
NMR titration experiments between ComAC and the ComA consensus sequence in the srfA promoter (5′-GAATCTTTCGGCATCCCGCATGAAA-3′) identified residues essential for DNA binding. These residues include Thr170, Asn171, Gln172, Ile174, Ala175 (in α8); Ser181, Lys182 (in α8-α9 loop); Ser184, Ile185, Thr190, Ser191, Phe193, Leu196 (in α9), Val198, Gly200, Arg201 and Thr202 (in α9-α10 loop and beginning of α10). Both NarLC and DosRC have the following residues in corresponding positions involved in their DNA binding interfaces: Thr170, Asn171, Ser181, Ser184, Thr190 and Thr202. In addition, NarLC also has residues corresponding to Gln172, Val198 and Gly200 while DosRC also has a residue corresponding to Arg201. These residues all lie in the helix-turn-helix DNA binding domain of these proteins. Just as there are similarities between the regions responsible for DNA binding between these proteins, these data seem to suggest a greater role of α8 and the loop between α8-α9 in DNA recognition for ComAC when compared to DosRC and NarLC.
Principal components analysis (PCA) is a standard tool in the field of multivariate analysis which extracts from a set of interrelated variables a much smaller set of variables that retains most of the variation contained in the full set. A correlation matrix consisting of elements of the average of the pairwise products of displacements from their average position of landmarks (such as the Cα positions in a protein), PCA can be very helpful in identifying, from an NMR structural ensemble, correlations in conformational rearrangements within a protein 41. Here we performed principal components analysis on the ComAC NMR structural ensemble. Structural PCA plots of the refined NMR structures of the unbound and DNA bound model of ComAC are shown (Figures 9A and 9B, respectively). Most important are the relative motions of structural elements in the unbound state, since this impacts the binding event. On average, within the dimeric NMR structure ensemble, (i) α8 and α9 move in the same direction within each monomer, but in an opposite sense to α8′ and α9′ from the other monomer; (ii) α10 and α10′ move in the same direction within the dimer (Figure 9A).
Figure 9.
THESEUS PCA analysis of the unbound and bound structure ensembles. PCA plots of the maximum likelihood correlation matrix for the structure ensembles of (A) unbound ComAC, first principal component and (B) bound ComAC, second principal component. Regions colored similarly (red or blue) are self-correlated, whereas regions colored differently (red versus blue) are anticorrelated.
The anti-correlated motion of α8/α9 and α8′/α9′ is particularly interesting as it suggests that each helix-turn-helix motif can push away from the dimeric interface (Figure 9A) to space correctly and interact more easily with adjacent recognition elements in the DNA target sequence (Figure 6A). This binding event may therefore resemble a “scissor” like motion of the helix-turn-helix motifs. Concomitant re-orientations during complex formation with target DNA sequences may also enable residues critical to DNA recognition and binding to make necessary contacts. Interestingly, as noted above for the ComAC-DNA model, the angle between α8 and α9 increases upon DNA binding. This type of motion is seen in the PCA analysis (Figure 9B). Comparative PCA investigations with other members of the NarL family were not performed for the following reasons. PCA studies require ensembles of structures. The structures of NarL/NarLC, DosRC, GerEC and TraR were solved by X-ray crystallography and ensembles of structures are not available from the Protein Data Bank. The NMR structure of FixJC suggests it is monomeric in solution and its mechanism of action is likely somewhat different than for those family members that exist functionally as dimers.
The ComAC:srfA model outlined here suggests both similarities and differences in how members of the NarL family of response regulators interact with their DNA targets. To facilitate its protein:DNA interactions, ComAC relies predominantly on the most N-terminal residues of helix α9. In comparison, NarLC mainly utilizes residues in the center of helix α9 while DosRC’s binding region falls in between that of ComAC and NarLC. Experiments performed here identify amino acid residues in helices α8 and α9 of ComAC as being important for binding to its target DNA sequences. Principal components analysis of the ComAC structure suggests that anti-correlated movement of each helix-turn-helix motif within each monomeric subunit of the full dimer structure may facilitate critical protein:DNA interactions.
Materials and methods
Expression and purification of ComAC
DNA for transformation of Escherichia coli was isolated using the QIAprep Miniprep Spin Kit (Qiagen). The purified plasmids were transformed into competent E. coli BL21(DE3)pLysS cells (Novagen). One liter of LB broth containing 100 μg/mL kanamycin was inoculated and grown at 37 °C at 160 rpm until the cells reached an optical density at 600 nm (OD600nm) of 0.8. Isopropyl-β-D-thiogalactopyranoside (IPTG) was added to a concentration of 1 mM, the temperature was reduced to 32 °C at 120 rpm and incubation continued for 3 hours. The cells were pelleted by centrifugation (7500 rpm for 20 minutes) and resuspended with 25 mL of lysis buffer (25 mM NaH2PO4, pH 8.0, 300 mM NaCl, 5 mM imidazole). In addition to the lysis buffer, 0.25 μM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF) and 0.01% Triton X-100 were also added. The cells were sonicated 8 times (3 minutes of 8 second pulses followed by 2 seconds rest). The resulting suspension was centrifuged at 17,500 rpm for 15 minutes. The supernatant was removed and saved as the crude extract. The crude extract was applied to a Ni-NTA affinity resin (Qiagen) that was previously equilibrated using the lysis buffer. Once the column was loaded with the ComAC crude lysate it was washed with 100 mL of the lysis buffer. ComAC was eluted from the column using an imidazole gradient of 5 mM to 250 mM. Fractions containing the ComAC were pooled and concentrated to 45 mL. 100 units of thrombin were added to the concentrated sample and the cleavage reaction was allowed to occur for 2 hours at room temperature. The cleavage was stopped by the addition of 1 mL of Protease Inhibitor Cocktail (Sigma). The now cleaved sample was immediately and extensively dialyzed against 25 mM NaH2PO4, pH 6.5, 100 mM NaCl, 2 mM ethylenediaminetetraacetic acid (EDTA), 1 mM dithiothreitol (DTT). Throughout the protocol the presence of ComAC was monitored using 12% Tricine Gel Electrophoresis. Production of 1H-15N-13C samples for NMR analysis followed the above protocol with the substitution of minimal media (M9T) with 1 g of 15N-ammonium chloride and 1 g of U-13C6 glucose for expression.
Construction of ComAN and ComAN-D55N mutant expression vectors
The ComA vector template used for the expression of the 5′ region of the comA gene encoding ComAN (residues 1–126) was provided by Dr. Dave Dubnau (The Public Health Research Institute, Newark, NJ). Primers were designed to incorporate NheI and SacI restriction endonuclease cleavage sites at the 5′ and 3′ ends of the coding sequence, respectively. The fragment was inserted into the pET28a vector (Novagen). This construct resulted in a thrombin cleavable His-tag (6x) at the N-terminus of the recombinant ComAN protein. This results in the production of a protein with a molecular weight of approximately 17,000 Da. A gene encoding a mutant ComAN protein in which an asparagine residue replaces an aspartate residue at position 55 (ComAN-D55N) was also constructed. Primer sequences were purchased from Integrated DNA Technologies. The primer sequences are:
ComAN-Forward: 5′-CTA GCT AGC ATG AAA AAG ATA CTA GTG-3′
ComAN-Bkwd: 5′ GCT GTT TAA AGA GCT CAA ATT ACT ATA AAA TTT CTC 3′
ComAN-D55N-Forward: 5′-GAT CTC ATT TTA ATG AAC CTG AAT CTA GGC-3′
ComAN-D55N-Bkwd: 5′-GCC TAG ATT CAG GTT CAT TAA AAT GAG ATC-3′
Expression and purification of ComAN and ComAN-D55N
Expression and purification were performed as described for ComAC.
Expression and purification of ComAFL
The ComAFL vector was provided by Dr. Dave Dubnau (The Public Health Research Institute, Newark, NJ). Briefly, this is a pET28a vector (Novagen) and results in a C-terminal non-thrombin cleavable His-tag (6x) recombinant ComAFL protein. This produces a protein with a molecular weight of approximately 25,000 Da. Expression and purification were performed as described for ComAC.
Size exclusion liquid chromatography
Size exclusion liquid chromatography was used to determine the multimerization state of ComAC, ComAN, ComAN-D55N and ComAFL proteins. A protein molecular weight standard (BIO-RAD) containing bovine thyroglobulin (670,000 Da), bovine γ-globulin (158,000 Da), chicken ovalbumin (44,000 Da), equine myoglobin 17 (17,000 Da) and vitamin B12 (1,350 Da) was used. Analysis was performed at room temperature using a Waters Breeze HPLC system equipped with a 4.6 mm ID × 30 cm TOSOH TSK-GEL SW2000 size exclusion column. Sample volumes of 10 μL with concentrations ranging from 10 μM to 1 mM were injected with a flow rate of 0.3 mL/min for twenty minutes. A mobile phase of 25 mM NaH2PO4, pH 6.5, 100 mM NaCl was used. Chromatographs were analyzed with Waters Breeze software version 3.30.
Circular dichroism spectroscopy
Far UV spectra of ComAC, ComAN, ComAN-D55N and ComAFL at a concentration of 60 μM were acquired with an Applied Photophysics Pistar-180 spectrometer, using a buffer of 25 mM NaH2PO4, pH 6.5, 100 mM NaCl. A circular 200 μL quartz cuvette (Hellma) with a 0.02 cm path length was used for all measurements. Spectra were recorded at a wavelength of 222 nm, to monitor α-helical content. A midpoint denaturation temperature (Tm) was determined by heating samples from 10 to 90 °C at a rate of 1 °C/min. Five replicates were collected for each protein and corrected for the buffer blank.
Subunit exchange of ComAC
ComAC was expressed and purified as described above. Fractions containing ComAC were combined and excess imidazole removed by dialysis. Half of the sample was set aside (his-tag (6x) -ComAC) and did not undergo thrombin cleavage. The other half underwent thrombin cleavage as described above. All uncleaved sample was removed by running the sample over a second Ni-NTA column, resulting in a thrombin-cleaved ComAC (untagged ComAC) sample. The his-tag (6x) -ComAC and untagged ComAC were then combined and reapplied to the Ni-NTA column and eluted as in the original protocol. Fractions containing ComAC, evaluated by 12% Tricine gel electrophoresis, were combined. Positive ion MALDI-TOF MS analyses were performed using an Applied Biosystems Voyager Super DE STR. This instrument is equipped with a nitrogen laser (337 nm) to desorb and ionize the samples. A close external calibration, using two points to bracket the mass range of interest, was used. A saturated solution of α-cyano-4-hydroxycinnamic acid in 45:45:10 EtOH:H2O:formic acid (v/v/v) was used as the MALDI matrix. Spectra were obtained over a mass range of 2,000 to 20,000 Da in linear mode with an average of 100 laser shots per spectrum. For the MALDI analyses, protein samples were prepared using micro reversed phase chromatography on C4- zip tips (Millipore). The tip resin was prepared by rinsing repeatedly with 10 μl of 100% acetonitrile/0.1% formic acid. This was followed by repeated washing with 0.1% formic acid. The protein was then concentrated by repeated pipetting, 10 μl seven times. Salts were removed by extensive washing with 0.1% formic acid. The protein was eluted with 2.5 μl of 70% acetonitrile/0.1% formic acid and 0.5 μl of the protein sample were mixed with 0.5 μl MALDI matrix and deposited on a 100-well MALDI sample target.
NMR spectroscopy and structure calculations
All experiments were performed at 288.15 K on a Varian INOVA 600 MHz equipped with a Varian cryogenic probe. Protein samples concentrated to 1.0–2.0 mM in the following buffer were used: 90:10% or 1:99% H20/D2O, 25 mM NaH2PO4, pH 6.5, 100 mM NaCl, 2 mM EDTA, and 1 mM DTT. Sequential backbone assignments were made from HNCACB, CBCA(CO)NH, HNCA, HNCO and HN(CA)CO experiments 42; 43; 44; 45; 46. Side chains were assigned from H(CCO)NH, C(CO)NH, and HCCH-TOCSY experiments 42; 43; 45. Exchange protected amides were monitored by sequentially recording 100 12-min two-dimensional 1H-15N-HSQC experiments. TALOS was used to determine coupling constants for assigning backbone ψ and ϕ angles 47. NOE’s were obtained from three-dimensional 15N-NOESY-HSQC experiments with 50-, 100-, and 120-ms mixing times and three-dimensional 13C-NOESY-HSQC experiments with an 120-ms mixing time. 12C/13C isotope edited experiments using three-dimensional 13C-NOESY-HSQC experiments with an 120-ms mixing time was used to determine the amino acid residues involved in the dimer interface 32. This was done by combining 13C-labeled ComAC combined with natural abundance ComAC in a 1:1 ratio. Structures were calculated with NOE’s (inter- and intra- molecular), hydrogen bond restraints (inter- and intra- molecular), amide exchange experiments, and ψ and ϕ angles (TALOS predictions). ARIA (version 1.2) and CNS (version 1.1) programs were used to compute the solution structure starting from an extended structure with random side chain conformations 48; 49. The CNS protocols used simulated annealing with torsion angle and Cartesian space dynamics using the default parameters. Manually assigned inter- and intra- molecular NOE’s were input to ARIA as assigned and uncalibrated with respect to distance. The total number of ambiguous NOE restraints allowed for each peak on the NOESY spectra was set to 20. Distance restraints, derived from the manually assigned NOE’s, were set to 1.8–6.0 Å. The dihedral angle restraints were taken to be ±2 S.D. values or at least ±20 from the average values predicted by TALOS 47. In this study, the dihedral angles were restrained to ϕ= −70±50° and ψ= −50±50° for the helical regions. The noncrystallographic symmetry energy term was used to keep the Cα atoms of the monomers superimposable, and distance symmetry potential was used to ensure that the relative orientations of all of the Cα atoms of the monomers were symmetric 49. The spectra were processed with NMRPIPE and analyzed with NMRVIEW on LINUX workstations running Fedora Core 50; 51. Molecules were visualized and aligned using PyMol 52. The ten lowest energy structures were further water-refined with ARIA. Principal components analysis (PCA) was performed on the NMR structure ensembles by using THESEUS 41.
Surface plasmon resonance (SPR) ComAC DNA binding experiments
An ICx Nomadics SensiQ dual channel surface plasmon resonance instrument was used to perform all binding experiments. For these experiments, the double stranded DNA sequences were immobilized to the sensor surface and the protein solution injected over the surface. To achieve this, neutravidin was immobilized onto a carboxylated surface (COOH1 sensor) over each flow cell channel through amine coupling. Briefly, amine coupling was performed by activating the surface by injecting 100 μl of a mixture of NHS/EDC over both channels at a flow rate of 50 μl/min. This was followed by an injection of 100 μl of a 50 μg/mL solution of neutravidin in 10 mM sodium acetate, pH 4.5, over both channels at a flow rate of 5 μl/min. 75 μl of ethanolamine (1 M) was injected at a flow rate of 25 μl/min over both channels to block remaining activated carboxyl groups. DNA sequences used for these are experiments are listed below. Oligonucleotide sequences were synthesized by IDT DNA and HPLC purified. The 5′ end of one strand was biotinylated to facilitate immobilization of the DNA to the neutravidin immobilized to the chip. Complementary strands were annealed by combining equimolar amounts and heating to 95 °C and cooled to room temperature. Sequences used in this study were determined from reference 30. The sequences are (underlined/bold portions correspond to a consensus recognition element):
| RE1 | RE2 | RE3 | |
| Wild type srfA: | 5′-TTTCGGcatc | CCGCATgaaact | TTTCAC-3′ |
| Consensus RE3: | 5′-TTTCGGcatc | CCGCATgaaact | TTGCGG-3′ |
| Consensus RE1-RE3: | 5′-TTGCGGcatc | CCGCAAgaaact | TTGCGG-3′ |
Biotinylated double stranded DNA sequences were injected over channel 1 only at a concentration of 1 μM until a response of 100 RU’s was achieved. Steady state assay experiments were performed at room temperature. A running buffer of 10 mM HEPES, pH 7.6, 2 mM MgCl2, 0.1 mM EDTA, 200 mM KCl, was used throughout the experiment. The steady state assay was performed by injecting increasing concentrations of ComAC over both channels, 125 μl at a flow rate of 5 μl/min. Upon completion of the binding profiles, all responses were baseline corrected to an injection of buffer only and subtraction of the reference flow cell (flow cell 2). This was done to correct for non-specific binding and instrument drift. The response value at equilibrium (Kd) was determined using the accompanying ICx Nomadics Q-DAT software.
ComAC DNA NMR titration experiments
All experiments were performed at 288.15 K on a Varian INOVA 600 MHz equipped with a Varian triple resonance cryogenic probe. The ComAC protein sample was concentrated to 250 μM in a buffer of 90:10% H2O/D2O, 25 mM NaH2PO4, pH 6.5, 100 mM NaCl, 2 mM EDTA, 1 mM DTT. DNA samples were titrated into samples that provided the following molar DNA:protein ratios: 0:1, 0.25:1, 0.5:1, 0.75:1, 1:1, 2:1, and 4:1. At each titration point a two-dimensional 1H-15N-HSQC experiment was collected. Data were processed using NMRPIPE and analyzed using NMRVIEW 50; 51. Oligonucleotide sequences were synthesized by IDT DNA and HPLC purified. Complementary strands were annealed by combining equimolar amounts and heating to 95 °C and cooled to room temperature. Sequences used in this study were identical to those used in the SPR studies and determined from reference 30.
ComAC:DNA modeling
The DNA pdbs used in the modeling study were constructed through X3DNA’s webserver (w3DNA: http://w3dna.rutgers.edu/) 53. The srfA operon binding sequence PDB was constructed based on the NarLC DNA PDB 35; the DNA in this solved structure was a bent/curved alternating B- to A-DNA conformation. Modeling of the ComAC:DNA interaction was completed using the program HADDOCK 54. Default HADDOCK parameters were used throughout the docking procedure. Experimental restraints known as ambiguous interaction restraints (AIR’s) were derived from NMR chemical shift perturbation experiments. The AIR’s are defined as both active and passive residues. Active residues are defined as those residues shown through the NMR protein:DNA titrations as having an effect upon binding DNA with a solvent accessibility of at least 40% as determined by NACCESS 33. Passive residues are defined as residues surrounding the active residues with at least 40% solvent accessibility.
The DNA sequence used along with its complementary strand was: GAATCTTTCGGCATCCCGCATGAAA, the known DNA binding sequence of ComA for the srfA operon 14. The underlined portions correspond to the response element boxes with dyad symmetry. A total of 1000 structures were generated in the static iteration, and the 200 lowest-energy structures further refined in semi-flexible iteration with the 20 lowest-energy structures undergoing water refinement. Each docking attempt was performed ten times, and the solution with the lowest HADDOCK score was retained. The HADDOCK score is a weighted sum of intermolecular electrostatic (EElec), van der Waals (EvdW), desolvation (Edesolv) and AIR (EAIR) energies and a buried surface area (BSA) term. The Cα r.m.s.d. values of the complexes were calculated by using ProFit 55. Values for base pair and base pair step parameters, as well as torsion angles for the sugar-phosphate backbone and sugar pucker were obtained by using the program 3DNA. A cluster analysis was performed on the final docking solutions by using a minimum cluster size of 4. The cut-off for clustering was manually determined for each docking run. The Cα r.m.s.d. matrix was calculated over the backbone atoms of the interface residues of the DNA after fitting on the interface residues of the protein.
Supplementary Material
MALDI-TOF spectrum from the ComAC subunit mixing experiment. The m/z of 9843.88 corresponds to the tagged-ComAC and the m/z of 8075.20 corresponds to the untagged-ComAC.
1H/15N-HSQC spectrum of ComAC. Inlay in upper left hand corner is an expansion of the dashed line box. Solid lines connecting two peaks designate side chain amide resonances.
1H/15N-HSQC ComAC:DNA titration. Molar ratios of consensus RE3 DNA:protein are 0:1 (black), 0.25:1 (red), 0.5:1 (green), 0.75:1 (blue), 1:1 (yellow), 2:1 (pink), and 4:1 (light blue).
Acknowledgments
This research was supported by NIH grants RO1 GM65156 (JC), RO1 GM55769 (JC), GM55594 (MP), and The V Foundation for Cancer Research (JC). The authors are grateful to Dr. Amanda Stewart (NCSU) for helpful comments. We would also like to thank Dr. Patrick McLaughlin and Erin Regel for their help with the ComAN and ComAN-D55N constructs.
Abbreviations
- NMR
nuclear magnetic resonance
- SPR
surface plasmon resonance
- CD
circular dichroism
- PDB
Protein Data Bank
- HPLC
high performance liquid chromatography
- HADDOCK
high ambiguity driven protein-protein docking
- MALDI-TOF MS
matrix assisted laser desorption/ionization-time of flight mass spectrometry
- PCA
principal components analysis
Footnotes
Protein Data Bank accession codes
The atomic coordinates for ComAC have been deposited in the Protein Data bank (accession number 2krf); the chemical shift list is available at the Biological Magnetic Resonance Data Bank (accession number rcsb101480).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Giammarinaro P, Sicard M, Gasc AM. Genetic and physiological studies of the CiaH-CiaR two-component signal-transducing system involved in cefotaxime resistance and competence of Streptococcus pneumoniae. Microbiology. 1999;145 ( Pt 8):1859–69. doi: 10.1099/13500872-145-8-1859. [DOI] [PubMed] [Google Scholar]
- 2.Gromkova RC, Mottalini TC, Dove MG. Genetic transformation in Haemophilus parainfluenzae clinical isolates. Curr Microbiol. 1998;37:123–6. doi: 10.1007/s002849900349. [DOI] [PubMed] [Google Scholar]
- 3.Orus P, Vinas M. Transfer of penicillin resistance between Neisseriae in microcosm. Microb Drug Resist. 2000;6:99–104. doi: 10.1089/107662900419393. [DOI] [PubMed] [Google Scholar]
- 4.Sjostrom JE, Lofdahl S, Philipson L. Transformation reveals a chromosomal locus of the gene(s) for methicillin resistance in Staphylococcus aureus. J Bacteriol. 1975;123:905–15. doi: 10.1128/jb.123.3.905-915.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kroll JS, Moxon ER, Loynds BM. Natural genetic transfer of a putative virulence-enhancing mutation to Haemophilus influenzae type a. J Infect Dis. 1994;169:676–9. doi: 10.1093/infdis/169.3.676. [DOI] [PubMed] [Google Scholar]
- 6.Dubnau D. Genetic competence in Bacillus subtilis. Microbiol Rev. 1991;55:395–424. doi: 10.1128/mr.55.3.395-424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakano MM, Zuber P. The primary role of comA in establishment of the competent state in Bacillus subtilis is to activate expression of srfA. J Bacteriol. 1991;173:7269–74. doi: 10.1128/jb.173.22.7269-7274.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hahn J, Kong L, Dubnau D. The regulation of competence transcription factor synthesis constitutes a critical control point in the regulation of competence in Bacillus subtilis. J Bacteriol. 1994;176:5753–61. doi: 10.1128/jb.176.18.5753-5761.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubnau D, Hahn J, Roggiani M, Piazza F, Weinrauch Y. Two-component regulators and genetic competence in Bacillus subtilis. Res Microbiol. 1994;145:403–11. doi: 10.1016/0923-2508(94)90088-4. [DOI] [PubMed] [Google Scholar]
- 10.Nakano MM, Xia LA, Zuber P. Transcription initiation region of the srfA operon, which is controlled by the comP-comA signal transduction system in Bacillus subtilis. J Bacteriol. 1991;173:5487–93. doi: 10.1128/jb.173.17.5487-5493.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magnuson R, Solomon J, Grossman AD. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell. 1994;77:207–16. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 12.Okada M, Sato I, Cho SJ, Iwata H, Nishio T, Dubnau D, Sakagami Y. Structure of the Bacillus subtilis quorum-sensing peptide pheromone ComX. Nat Chem Biol. 2005;1:23–4. doi: 10.1038/nchembio709. [DOI] [PubMed] [Google Scholar]
- 13.Nakano MM, Zuber P. Mutational analysis of the regulatory region of the srfA operon in Bacillus subtilis. J Bacteriol. 1993;175:3188–91. doi: 10.1128/jb.175.10.3188-3191.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roggiani M, Dubnau D. ComA, a phosphorylated response regulator protein of Bacillus subtilis, binds to the promoter region of srfA. J Bacteriol. 1993;175:3182–7. doi: 10.1128/jb.175.10.3182-3187.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solomon JM, Magnuson R, Srivastava A, Grossman AD. Convergent sensing pathways mediate response to two extracellular competence factors in Bacillus subtilis. Genes Dev. 1995;9:547–58. doi: 10.1101/gad.9.5.547. [DOI] [PubMed] [Google Scholar]
- 16.Nakano MM, Magnuson R, Myers A, Curry J, Grossman AD, Zuber P. srfA is an operon required for surfactin production, competence development, and efficient sporulation in Bacillus subtilis. J Bacteriol. 1991;173:1770–8. doi: 10.1128/jb.173.5.1770-1778.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grossman AD. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu Rev Genet. 1995;29:477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]
- 18.Turgay K, Hahn J, Burghoorn J, Dubnau D. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 1998;17:6730–8. doi: 10.1093/emboj/17.22.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guillen N, Weinrauch Y, Dubnau DA. Cloning and characterization of the regulatory Bacillus subtilis competence genes comA and comB. J Bacteriol. 1989;171:5354–61. doi: 10.1128/jb.171.10.5354-5361.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weinrauch Y, Guillen N, Dubnau DA. Sequence and transcription mapping of Bacillus subtilis competence genes comB and comA, one of which is related to a family of bacterial regulatory determinants. J Bacteriol. 1989;171:5362–75. doi: 10.1128/jb.171.10.5362-5375.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hahn J, Dubnau D. Growth stage signal transduction and the requirements for srfA induction in development of competence. J Bacteriol. 1991;173:7275–82. doi: 10.1128/jb.173.22.7275-7282.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Msadek T, Kunst F, Klier A, Rapoport G. DegS-DegU and ComP-ComA modulator-effector pairs control expression of the Bacillus subtilis pleiotropic regulatory gene degQ. J Bacteriol. 1991;173:2366–77. doi: 10.1128/jb.173.7.2366-2377.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mueller JP, Bukusoglu G, Sonenshein AL. Transcriptional regulation of Bacillus subtilis glucose starvation-inducible genes: control of gsiA by the ComP-ComA signal transduction system. J Bacteriol. 1992;174:4361–73. doi: 10.1128/jb.174.13.4361-4373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Solomon JM, Lazazzera BA, Grossman AD. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev. 1996;10:2014–24. doi: 10.1101/gad.10.16.2014. [DOI] [PubMed] [Google Scholar]
- 25.Jiang M, Grau R, Perego M. Differential processing of propeptide inhibitors of Rap phosphatases in Bacillus subtilis. J Bacteriol. 2000;182:303–10. doi: 10.1128/jb.182.2.303-310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Core L, Perego M. TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis. Mol Microbiol. 2003;49:1509–22. doi: 10.1046/j.1365-2958.2003.03659.x. [DOI] [PubMed] [Google Scholar]
- 27.Bongiorni C, Ishikawa S, Stephenson S, Ogasawara N, Perego M. Synergistic regulation of competence development in Bacillus subtilis by two Rap-Phr systems. J Bacteriol. 2005;187:4353–61. doi: 10.1128/JB.187.13.4353-4361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auchtung JM, Lee CA, Grossman AD. Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides. J Bacteriol. 2006;188:5273–85. doi: 10.1128/JB.00300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Comella N, Grossman AD. Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis. Mol Microbiol. 2005;57:1159–74. doi: 10.1111/j.1365-2958.2005.04749.x. [DOI] [PubMed] [Google Scholar]
- 30.Griffith KL, Grossman AD. A degenerate tripartite DNA-binding site required for activation of ComA-dependent quorum response gene expression in Bacillus subtilis. J Mol Biol. 2008;381:261–75. doi: 10.1016/j.jmb.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavanagh J, Fairbrother WJ, Palmer A, Rance M, Skelton NJ. Protein NMR Spectroscopy: Principles and practice. 2. Elsevier Academic Press; 2007. [Google Scholar]
- 32.Stuart A, Borzilleri KA, Withka JM, Palmer AG., III Compensating for Variations in 1H-13C Scalar Coupling Constants in Isotope-Filtered NMR experiments. J Am Chem Soc. 1999;121:5346–5347. [Google Scholar]
- 33.Hubbard SJ, Thornton JM. NACCESS. University College London, Department of Biochemistry and Molecular Biology; 1993. [Google Scholar]
- 34.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–86. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 35.Maris AE, Sawaya MR, Kaczor-Grzeskowiak M, Jarvis MR, Bearson SM, Kopka ML, Schroder I, Gunsalus RP, Dickerson RE. Dimerization allows DNA target site recognition by the NarL response regulator. Nat Struct Biol. 2002;9:771–8. doi: 10.1038/nsb845. [DOI] [PubMed] [Google Scholar]
- 36.Wisedchaisri G, Wu M, Rice AE, Roberts DM, Sherman DR, Hol WG. Structures of Mycobacterium tuberculosis DosR and DosR-DNA complex involved in gene activation during adaptation to hypoxic latency. J Mol Biol. 2005;354:630–41. doi: 10.1016/j.jmb.2005.09.048. [DOI] [PubMed] [Google Scholar]
- 37.Ducros VM, Lewis RJ, Verma CS, Dodson EJ, Leonard G, Turkenburg JP, Murshudov GN, Wilkinson AJ, Brannigan JA. Crystal structure of GerE, the ultimate transcriptional regulator of spore formation in Bacillus subtilis. J Mol Biol. 2001;306:759–71. doi: 10.1006/jmbi.2001.4443. [DOI] [PubMed] [Google Scholar]
- 38.Pristovsek P, Sengupta K, Lohr F, Schafer B, von Trebra MW, Ruterjans H, Bernhard F. Structural analysis of the DNA-binding domain of the Erwinia amylovora RcsB protein and its interaction with the RcsAB box. J Biol Chem. 2003;278:17752–9. doi: 10.1074/jbc.M301328200. [DOI] [PubMed] [Google Scholar]
- 39.Kurashima-Ito K, Kasai Y, Hosono K, Tamura K, Oue S, Isogai M, Ito Y, Nakamura H, Shiro Y. Solution structure of the C-terminal transcriptional activator domain of FixJ from Sinorhizobium meliloti and its recognition of the fixK promoter. Biochemistry. 2005;44:14835–44. doi: 10.1021/bi0509043. [DOI] [PubMed] [Google Scholar]
- 40.Vannini A, Volpari C, Gargioli C, Muraglia E, Cortese R, De Francesco R, Neddermann P, Marco SD. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 2002;21:4393–401. doi: 10.1093/emboj/cdf459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Theobald DL, Wuttke DS. Accurate structural correlations from maximum likelihood superpositions. PLoS Comput Biol. 2008;4:e43. doi: 10.1371/journal.pcbi.0040043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Logan TM, Olejniczak ET, Xu RX, Fesik SW. Side chain and backbone assignments in isotopically labeled proteins from two heteronuclear triple resonance experiments. FEBS Lett. 1992;314:413–8. doi: 10.1016/0014-5793(92)81517-p. [DOI] [PubMed] [Google Scholar]
- 43.Grzesiek S, Bax A. Measurement of amide proton exchange rates and NOEs with water in 13C/15N-enriched calcineurin B. J Biomol NMR. 1993;3:627–38. doi: 10.1007/BF00198368. [DOI] [PubMed] [Google Scholar]
- 44.Ikura M, Kay LE, Bax A. A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin. Biochemistry. 1990;29:4659–67. doi: 10.1021/bi00471a022. [DOI] [PubMed] [Google Scholar]
- 45.Logan TM, Olejniczak ET, Xu RX, Fesik SW. A general method for assigning NMR spectra of denatured proteins using 3D HC(CO)NH-TOCSY triple resonance experiments. J Biomol NMR. 1993;3:225–31. doi: 10.1007/BF00178264. [DOI] [PubMed] [Google Scholar]
- 46.Montelione GT, Emerson SD, Lyons BA. A general approach for determining scalar coupling constants in polypeptides and proteins. Biopolymers. 1992;32:327–34. doi: 10.1002/bip.360320406. [DOI] [PubMed] [Google Scholar]
- 47.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 48.Brunger AT, Adams PD, Rice LM. Recent developments for the efficient crystallographic refinement of macromolecular structures. Curr Opin Struct Biol. 1998;8:606–11. doi: 10.1016/s0959-440x(98)80152-8. [DOI] [PubMed] [Google Scholar]
- 49.Junius FK, O’Donoghue SI, Nilges M, Weiss AS, King GF. High resolution NMR solution structure of the leucine zipper domain of the c-Jun homodimer. J Biol Chem. 1996;271:13663–7. doi: 10.1074/jbc.271.23.13663. [DOI] [PubMed] [Google Scholar]
- 50.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–52. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 51.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 52.DeLano WL. The PyMOL molecular graphics system. DeLano Scientific; PaloAlto, CA: 2002. [Google Scholar]
- 53.Zheng G, Lu XJ, Olson WK. Web 3DNA--a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res. 2009;37:W240–6. doi: 10.1093/nar/gkp358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–7. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 55.Martin ACR. ProFit. www.bioing,org.uk/software/profit.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MALDI-TOF spectrum from the ComAC subunit mixing experiment. The m/z of 9843.88 corresponds to the tagged-ComAC and the m/z of 8075.20 corresponds to the untagged-ComAC.
1H/15N-HSQC spectrum of ComAC. Inlay in upper left hand corner is an expansion of the dashed line box. Solid lines connecting two peaks designate side chain amide resonances.
1H/15N-HSQC ComAC:DNA titration. Molar ratios of consensus RE3 DNA:protein are 0:1 (black), 0.25:1 (red), 0.5:1 (green), 0.75:1 (blue), 1:1 (yellow), 2:1 (pink), and 4:1 (light blue).