Summary
Purpose
Medial temporal epilepsy (MTLE) is associated with extrahippocampal brain atrophy. The mechanisms underlying brain damage in MTLE are unknown. Seizures may lead to neuronal damage, but another possible explanation is deafferentation from loss of hippocampal connections. This study aimed to investigate the relationship between hippocampal deafferentation and brain atrophy in MTLE.
Methods
Three different MRI studies were performed involving 23 patients with unilateral MTLE (8 left and 15 right) and 34 healthy controls: (1) voxel-based morphometry (VBM), (2) diffusion tensor imaging (DTI) and (3) probabilistic tractography (PT). VBM was employed to define differences in regional gray matter volume (GMV) between controls and patients. Voxel-wise analyses of DTI evaluated differences in fractional anisotropy (FA), mean diffusivity (MD) and hippocampal PT. Z-scores were computed for regions-of-interest (ROI) GMV and perihippocampal FA and MD (to quantify hippocampal fiber integrity). The relationship between hippocampal deafferentation and regional GMV was investigated through the association between ROI Z scores and hippocampal fiber integrity.
Results
Patients with MTLE exhibited a significant reduction in GMV and FA in perihippocampal and limbic areas. There was a decrease in hippocampal PT in patients with MTLE in limbic areas. A significant relationship between loss of hippocampal connections and regional GMV atrophy was found involving the putamen, pallidum, middle and inferior temporal areas, amygdala and ceberellar hemisphere.
Discussion
There is a relationship between hippocampal disconnection and regional brain atrophy in MTLE. These results indicate that hippocampal deafferentation plays a contributory role in extrahippocampal brain damage in MTLE.
Keywords: Temporal lobe epilepsy, Atrophy, Neural network, Magnetic resonance imaging
Hippocampal sclerosis (HS) is the most common histological abnormality observed in patients with medial temporal lobe epilepsy (MTLE) (Margerison & Corsellis, 1966). It usually represents marked cell loss in the hippocampal regions CA1 and hilus, with less intense loss in the end folium (CA3/4) and relative sparing of CA2 (Blumcke et al., 2002). Magnetic resonance imaging (MRI) can identify in vivo signs that are reliably associated with the neuronal loss encountered in HS (Jack et al., 1990; Cendes et al., 1993). In particular, hippocampal atrophy or loss of hippocampal internal structure on T1-weighted images and increased hippocampal T2 signal are consistently associated with HS (Cendes et al., 1993). More refined MRI analyses have advanced the understanding of the pathological mechanisms underlying MTLE. Postprocessing computerized analyses of brain MRI demonstrate that patients with MTLE exhibit subtle brain abnormalities outside the hippocampus that are not fully appreciated by qualitative visual inspection of diagnostic images. Careful manual and automated morphometrical studies showed that patients with MTLE show significant extrahippocampal atrophy that involves the temporal lobe and extratemporal brain structures (Keller et al., 2002; Bernasconi et al., 2003; Bonilha et al., 2003; Bernasconi et al., 2004; Bonilha et al., 2004; McDonald et al., 2008). Notably, the distribution of brain atrophy in MTLE preferentially affects a network of regions that are functionally and anatomically connected to the hippocampus (Spencer, 2002; Bonilha et al., 2005; Keller & Roberts, 2008; Riederer et al., 2008).
Morphometrical MRI studies of patients with epilepsy have also demonstrated that extrahippocampal gray matter loss likely follows a progressive course (Bonilha et al., 2006). Cognitive deficits commonly exhibited by patients with MTLE, in particular memory impairment, are directly related to the degree of medial temporal and frontal lobe atrophy (Alessio et al., 2006; Bonilha et al., 2007; Focke et al., 2008). Even though hippocampal and extrahippocampal gray matter atrophy are directly related to seizure control and cognitive performance, the mechanisms underlying brain damage in patients with MTLE remain largely unknown. One theory postulates that seizure toxicity plays a major role in the distribution of damage (Spencer, 2002; Sutula et al., 2003; Riederer et al., 2008). However, this is still controversial and a recent study suggested that seizure control and gray matter atrophy may be unrelated (Liu et al., 2003), and it is unknown what the main reasons leading to damage are.
Another complementary theory suggests that loss of hippocampal connections can lead to remote deafferentation and thereby neuronal damage, particularly since structures directly connected to the hippocampus, such as the thalamus, typically exhibit a large degree of atrophy (Bonilha et al., 2005). This theory suggests that the impoverishment of hippocampal connections can lead to reduction in the complexity of remotely located neocortical circuitry. Interestingly, remote neuronal damage related to seizure spread (but not to deafferentation) may depend on the integrity of hippocampal connections, as this is the route for seizure travel. Therefore, it is possible that the two mechanisms, i.e., direct toxicity from seizure spread and deafferentation may contribute to extrahippocampal damage. The extent to which each mechanism contributes to the overall neuronal loss in patients with MTLE is largely unknown. It is also unclear whether both processes interact and result in the overall pattern of damage.
This study aimed to investigate the relationship between the loss of hippocampal fibers and extrahippocampal gray matter loss in patients with MTLE. In order to comprehensively evaluate to pattern of loss of connectivity from the hippocampus, this study employed a combination of automated voxel-based morphometrical analyses, diffusion tensor imaging (DTI) and hippocampal probabilistic tractography. DTI assesses the diffusion properties of water molecules and provides indirect information regarding white matter fibers. Traditional investigation of DTI data comprises the voxel by voxel assessment of mean diffusivity (MD) and fractional anisotropy (FA), which can therefore provide information regarding the location and extent of disconnection between brain structures in association with disease. DTI studies involving the assessment of FA and MD have been successfully used in patients with MTLE. For instance, Focke et al. (2008) recently confirmed that patients with MTLE exhibit reduction in FA and increase in MD, changes traditionally considered compatible with neuronal loss, in the ipsilateral temporal lobe of patients with MTLE. These results suggest loss of connections from the atrophied hippocampi. DTI data can also be employed in the probabilistic reconstruction of white matter tracts, which can be quantified and also analyzed on a voxel-by-voxel manner. This technique, termed probabilistic tractography, provides information about specific tracts as opposed voxel-wise MD and FA analyses, which provide information on the integrity of fibers within a region, not taking into account the direction of information travel. Probabilistic tractography has not been extensively investigated in MTLE.
This manuscript employed a cross-sectional study aiming to investigate the relationship between hippocampal connection loss and gray matter atrophy. We aimed to confirm that: (1) patients with MTLE would exhibit significant gray matter atrophy involving extrahippocampal structures, particularly affecting limbic regions, and (2) patients with MTLE would also exhibit loss in white matter integrity and reduction of fibers in the hippocampal and limbic pathways. Finally, we hypothesized that patients with MTLE would show a relationship between gray matter atrophy and loss of hippocampal connections, indicating a deafferentation mechanism underlying part of extrahippocampal atrophy.
Methods
Subjects
We studied 23 consecutive patients who were diagnosed with MTLE according to the parameters defined by the ILAE (Commission, 1989). All patients underwent a comprehensive neurological evaluation comprised of a careful interview, neurological examination and neurophysiological monitoring. All patients also underwent diagnostic MRI, which revealed unilateral hippocampal atrophy in all cases. All patients had unilateral seizure onset documented by video-EEG corresponding to the side of hippocampal atrophy. None of the patients had bilateral onset of seizures. Five patients (three patients with left MTLE and two with right MTLE) were considered to have benign MTLE and were not surgical candidate since good control of seizures was achieved with medications. The mean age of the patient group was 38 ± 11 years, and 12 patients were women. Eight patients had right hippocampal atrophy and 15 left hippocampal atrophy based on clinical visual evaluation.
We also studied a control group of 34 healthy individuals without any significant past medical, neurological or psychiatric history. The mean age of controls was 33 ± 11 years. Seventeen controls were women and the control group was similar to the patient group in age (t(55) = −1.9, p = 0.1) and gender distribution (Yates' chi = 0.012, p = 0.91).
The Medical University of South Carolina IRB committee approved this study. All subjects signed an informed consent to participate in this study.
Imaging
All subjects underwent high-resolution MRI in a 3T scanner equipped with an eight-channel head coil. From each subject, two sequences were obtained. First, a T1-weighted image with 1 mm isotropic voxels was acquired in the sagittal plane with the following parameters: TR = 8.1 ms, TE = 3.7 ms, flip angle = 8°, FOV = 256 × 256 mm. Second, a 15 direction DTI image with 2 × 2 × 3 mm voxels was acquired in the axial plane (TR = 8,933 ms, TE = 82 ms, flip angle = 90°, FOV = 224 × 224 mm.
Image processing
1. Voxel based morphometry
VBM was performed on the T1 images. Images from the control subjects were used to create a normalized template and tissue priors (of gray and white matter). All images were then submitted to iterative spatial normalization to the stereotaxic space, bias field correction and tissue segmentation, based on ICBM a priori templates (affine and 16 iteration nonlinear transformations) (Ashburner & Friston, 2005). These steps were performed employing the software SPM5 and the toolbox VBM5 (Christian Gaser, http://dbm.neuro.uni-jena.de/vbm/). We employed modulated gray matter maps (therefore providing gray matter volumes). Finally, spatially normalized (to the stereotaxic space), bias corrected, segmented images were submitted to spatial normalization employing a 10 mm isotropic Gaussian Kernel. Only pre-processed gray matter images were employed in subsequent statistical analyses.
The mean gray matter volume was then extracted from all regions of interest (45 supratentorial regions on each hemisphere, cerebellum hemisphere and vermis) comprised in the Anatomical Automatic Labeling (AAL) brain Atlas, which contains three-dimensional atlas of regions of interest (http://www.cyceron.fr/web/aal__anatomical_automatic_labeling.html) (Tzourio-Mazoyer et al., 2002). This step was performed with the software MRIcron (Chris Rorden, http://www.sph.sc.edu/comd/rorden/mricron/). MANOVA with group as a fixed factor (controls vs. patients) and regions as dependent variables were used to assess regional differences between groups. For consistency, the data from patients was grouped into sides ipsilateral or contralateral to the hippocampal atrophy, and the data from controls was averaged between sides. A level of statistical significance was set at p < 0.05.
We also computed the standardized Z-score (i.e., the number of standard deviations away from the mean) of each AAL region for patients compared with controls. Once the scores were obtained, they were categorized as belonging to the hemisphere of origin of seizures (ipsilateral) or to the contralateral hemispheres. Z-scores were used to investigate the relationship between regional gray matter and hippocampal connectivity (as explained below in methods item 4).
2. Diffusion tensor imaging
axonal integrity was assessed through a voxel-wise analysis of the magnitude (MD) and the directionality (FA) of molecular displacement. FSL's (FMRIB's Software Library, http://www.fmrib.ox.ac.uk/fsl) Diffusion Toolkit (FDT) was used for pre-processing diffusion weighted images (DWIs) and construct DTI data. The tool dcm2nii (http://www.mricro.com/mricron/dcm2nii.html) was used to convert the DICOM images to NIfTI format and extract the diffusion gradient directions. Images underwent eddy current correction through affine transformation of each DWI to the base b = 0 T2-weighted image. This procedure removes spatial distortion in the DWIs due to application of diffusion gradients in various directions. After eddy current correction, diffusion images acquired of the same slice are in alignment and a pixel-wise calculation of the diffusion tensor may be performed. Variations in acquisition geometry were corrected and gradients were updated using the software DtoA (http://www.nottingham.ac.uk/∼njzwww/paul/software/dtoa.html). FDT was used to perform the pixel-wise calculation of the diffusion tensor (employing an explicit binary mask calculated with FSL's Brain Extraction Tool (BET) with fractional threshold of 0.3 to prevent erroneous DTI calculation in the noise background outside the head). As a result, MD and FA maps are created in the same space as the b = 0 image volume of the original DTI acquisition. The b = 0 image volume (which is effectively a T2-weighted spin-echo echo planar image), was linearly normalized to a T2 template in stereotaxic space using FLIRT (FMRIB's Linear Image Registration Tool – http://www.fmrib.ox.ac.uk/fsl/flirt/). The same normalization matrix was then applied to FA and MD maps obtained from the diffusion tensor reconstruction step.
FA and MD maps were smoothed to a Gaussian kernel of 8 mm to minimize individual variability, improve the normality of data distribution and reduce false positives. Smoothed FA and MD maps were then submitted to a two-sample t-test using the software NPM (Rorden et al., 2007), (http://www.sph.sc.edu/comd/rorden/npm/) to investigate voxel-wise differences between patients and controls. Results were corrected for multiple comparisons using a False Discovery Rate (FDR) corrected threshold of p < 0.05.
3. Probabilistic tractography
was performed on DTI data after the pixel-wise calculation of the diffusion tensor using FSL's DTIFit. BEDPOST (Bayesian Estimation of Diffusion Parameters Obtained using Sampling Techniques) was applied to the data. BEDPOST runs Markov Chain Monte Carlo sampling to build up distributions on diffusion parameters at each voxel. Probabilistic tractography was estimated using FSL's probtrack. Probabilistic tractography was performed using a seed mask in order to generate the probabilistic distribution of fibers from the voxels in the seed mask. The seed masks were located in the hippocampi, using the hippocampi masks obtained from the Anatomical Automatic Labeling dataset (http://www.cyceron.fr/freeware/), which were linearly transformed into each subject's native space, where probtrack was run. The resulting probabilistic tractography images from each subject were linearly transformed into standard stereotaxic MNI space and smoothed using an Isotropic Gaussian Kernel of 4 mm. Probablistic tractography wields a voxel-wise map of fiber density, which is a two-dimensional representation of fiber tractography. For illustration of how these fibers would appear tridimensionally, Fig. 1 exemplifies the average location of the hippocampal fibers from one representative normal subject. This conventional tractography was reconstructed using the software MedINRIA (http://www-sop.inria.fr/asclepios/software/MedINRIA/) from one control subject randomly chosen. The fibers are reconstructed from the stereotaxic hippocampal mask from AAL representing the hippocampus, normalized onto the subject's native space. Since probabilistic tractography is a 2D map of fiber density, it can be submitted to voxel-by-voxel comparisons, differently than the 3D conventional tractography.
Figure 1.
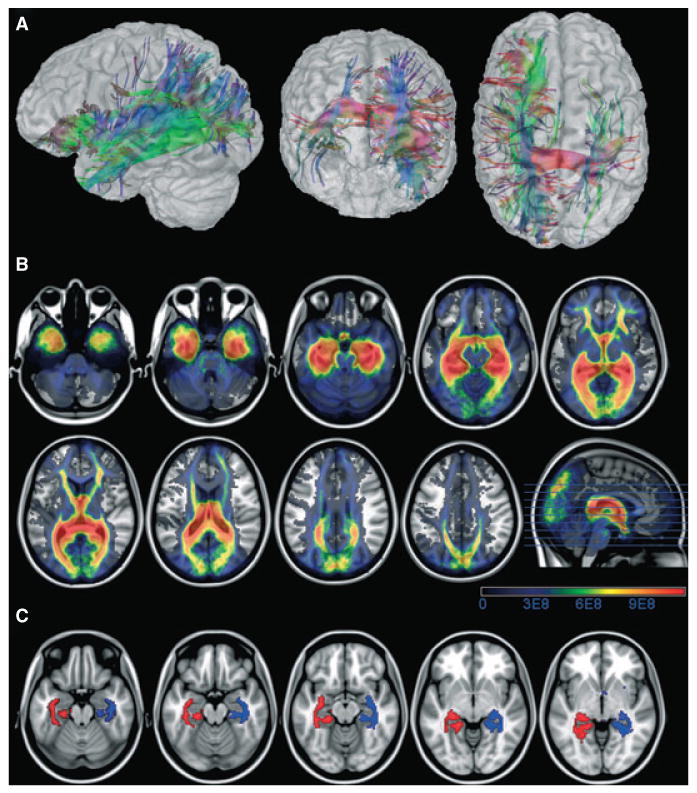
The upper panel shows the tridimensional representation of fibers from the hippocampus in one representative healthy subject (A), the middle panel (B) shows the location of the probabilistic tractography of fibers from the hippocampus on average amongst healthy subjects (the scale bars represent the number of fibers) and the lower panel (C) shows the perihippocampal masks with 95% of chance of encompassing all fibers traveling from the hippocampus.
Statistical analyses were applied to spatially normalized smoothed probabilistic tractography maps, which were submitted to a two-sample t-test to investigate voxel-wise differences between patients and controls. Results were corrected for multiple comparisons using a False Discovery Rate (FDR) corrected threshold of p < 0.05.
4. Estimation of hippocampal connectivity (relationship between VBM and DTI)
once normalized probabilistic tractography maps were obtained, we reconstructed an average image comprising patients and controls. From this image, a binary mask was obtained by applying an intensity filter in order to select only fibers within the highest 5th percentile of probability of being connected to the hippocampus, thereby focusing on the maximal intensity of fiber density around the hippocampus. The resulting mask corresponded to the location around the hippocampi where the majority of fibers from the hippocampi are located. This mask is shown in Fig. 1. This mask was used to construct an ROI (henceforth referred as perihippocampal ROI) from which the mean FA and mean MD was computed from all subjects (patients and controls). The mean FA and MD values from these ROIs were then standardized as Z-scores compared with the values from controls. Even though there is possibility of mild hippocampal asymmetry (and therefore of asymmetry of hippocampal fibers) in normal individuals, there was not a significant difference between left and right perihippocampal MD (t(62) = −1.4, p = 0.14) and FA (t(62) = −1.7, p = 0.09). Hence, all values were grouped and Z-scores from patients were based on the overall average from controls. For each individual with MTLE, the Z-score was categorized as belonging to the hippocampus ipsilateral or contralateral to the side of seizure onset.
The relationship between hippocampal connectivity and regional gray matter atrophy was assessed as follows: perihippocampal MD and FA Z-scores and regional gray matter Z-scores (i.e., Z-scores from AAL regions, as explained in methods item 1) were compared to evaluate synchronous changes. A simple correlation was performed evaluating the relationship in each subject between perihippocampal Z-scores and regional AAL Z-scores. The level of statistical significance was set at p < 0.05. We also performed a correction for multiple comparisons using a False Discovery Rate (FDR) corrected threshold of p < 0.05. Both the results from the corrected and uncorrected analyses were reported, in order to account for the high number of potential false negatives from corrected results (Rothman, 1990), and the high probability of false positives from the noncorrected results. Specifically, in order to maximize the number of true positives, whilst minimizing false negatives, we applied an FDR corrected threshold on the results from 15 regions of interest (ipsilateral to the hippocampal atrophy), which were chosen because they have been consistently demonstrated to be atrophied in MTLE, and are functionally or anatomically close to the hippocampus, thereby likely to suffer the effect of deafferentation. These regions were the amygdala, cingulum, caudate, cerebellum, inferior and middle and superior temporal cortex, insula, pallidum, parahippocampal, putamen, and thalamus (Bernasconi et al., 2004; Bonilha et al., 2004; Mueller et al., 2006; Keller & Roberts, 2008; Riederer et al., 2008). An evaluation of the corrected and uncorrected results was made in the context of the findings from the other experiments (numbers 1 to 3 above).
Results
Results are described item by item, in accordance with the experiments numbered in the Methods section.
1. Voxel based morphometry
Overall, patients with MTLE exhibited a significant reduction of gray matter volume diffusely distributed within the ipsilateral and contralateral cortex. These results are similar to previously published VBM studies (Keller & Roberts, 2008), and the region-by-region numbers are displayed in Table 1. Importantly, patients with MTLE exhibited widespread atrophy involving the hippocampus, parahippocampal gyrus, thalamus, lateral temporal regions and occipital, frontal and parietal regions.
Table 1. Voxel based morphometry results illustrating the location of gray matter volume reduction in patients with MTLE.
| MANOVA | ||||
|---|---|---|---|---|
| Region | Ipsilateral | Contralateral | ||
| F | p | F | p | |
| Amygdala | 11.623 | 0.0014 | 6.992 | 0.0115 |
| Angular gyrus | 10.581 | 0.0023 | 5.859 | 0.0199 |
| Calcarine | 0.195 | 0.6611 | 22.712 | 0.0000 |
| Caudate | 12.855 | 0.0009 | 1.383 | 0.2462 |
| Cerebelar hemisphere | 6.816 | 0.0125 | 12.367 | 0.0011 |
| Anterior cingulum | 9.265 | 0.0040 | 0.163 | 0.6885 |
| Middle cingulum | 7.238 | 0.0102 | 9.634 | 0.0034 |
| Posterior cingulum | 8.077 | 0.0069 | 10.002 | 0.0029 |
| Cuneus | 15.295 | 0.0003 | 9.725 | 0.0033 |
| Inferior opercular frontal | 5.354 | 0.0256 | 6.379 | 0.0154 |
| Inferior orbital frontal | 2.634 | 0.1121 | 15.770 | 0.0003 |
| Inferior frontal (triangularis) | 8.910 | 0.0047 | 4.380 | 0.0425 |
| Inferior frontal (orbitalis) | 10.113 | 0.0028 | 0.197 | 0.6592 |
| Middle frontal | 7.851 | 0.0076 | 2.827 | 0.1001 |
| Middle orbital frontal | 2.259 | 0.1403 | 7.662 | 0.0084 |
| Superior frontal | 8.766 | 0.0050 | 2.451 | 0.1249 |
| Medial superior frontal | 6.098 | 0.0177 | 4.256 | 0.0453 |
| Superior orbital frontal | 2.272 | 0.1392 | 10.360 | 0.0025 |
| Fusiform | 8.257 | 0.0063 | 6.178 | 0.0170 |
| Heschl gyrus | 0.134 | 0.7163 | 9.915 | 0.0030 |
| Hippocampus | 22.608 | 0.0000 | 0.890 | 0.3508 |
| Insula | 0.269 | 0.6065 | 14.012 | 0.0005 |
| Lingual gyrus | 13.436 | 0.0007 | 13.575 | 0.0007 |
| Inferior occipital | 14.087 | 0.0005 | 3.781 | 0.0586 |
| Middle occipital | 28.888 | 0.0000 | 4.580 | 0.0382 |
| Superior occipital | 13.300 | 0.0007 | 3.962 | 0.0531 |
| Olfactory gyrus | 6.250 | 0.0164 | 4.303 | 0.0442 |
| Pallidum | 15.701 | 0.0003 | 2.761 | 0.1040 |
| Paracentral lobule | 7.174 | 0.0105 | 14.619 | 0.0004 |
| Parahippocampal gyrus | 6.427 | 0.0150 | 8.073 | 0.0069 |
| Inferior parietal | 23.750 | 0.0000 | 8.058 | 0.0070 |
| Superior parietal | 16.583 | 0.0002 | 10.058 | 0.0028 |
| Postcentral | 13.876 | 0.0006 | 0.044 | 0.8353 |
| Precentral | 18.368 | 0.0001 | 1.405 | 0.2426 |
| Precuneus | 1.014 | 0.3197 | 22.026 | 0.0000 |
| Putamen | 0.019 | 0.8912 | 7.491 | 0.0091 |
| Rectus | 7.708 | 0.0082 | 4.844 | 0.0333 |
| Rolandic (opercularis) | 0.023 | 0.8796 | 6.212 | 0.0167 |
| Supplemetary motor area | 14.685 | 0.0004 | 19.389 | 0.0001 |
| Supramarginal gyrus | 13.016 | 0.0008 | 0.421 | 0.5198 |
| Inferior temporal | 0.316 | 0.5770 | 12.301 | 0.0011 |
| Middle temporal | 4.383 | 0.0424 | 4.683 | 0.0362 |
| Middle temporal pole | 10.484 | 0.0024 | 5.034 | 0.0302 |
| Superior temporal pole | 11.172 | 0.0018 | 10.168 | 0.0027 |
| Superior temporal | 6.007 | 0.0185 | 5.289 | 0.0265 |
| Thalamus | 2.325 | 0.1348 | 0.014 | 0.9076 |
| Vermis | 21.132 | 0.0000 | ||
Bold values signify statistically significant results (i.e., p < 0.05).
2. DTI
Patients with left and right MTLE showed a significant reduction in FA in the white matter underlying the medial temporal lobe, the insula, the frontotemporal stem and the orbitofrontal cortex. In both groups, the reduction of FA was more intense in the temporal lobe ipsilateral to the onset of seizures. The pattern of atrophy was similar in right and left MTLE groups and affected mostly the limbic rim of white matter surrounding the medial temporal lobe. These results are displayed in Fig. 2. Patients with right MTLE also displayed an increase in MD in the white matter within the temporal and frontal lobes. The difference in the MD results between left and right MTLE is likely related to the small sample size in both groups, with the right MTLE group being more affected than the left. Although this study included 57 subjects and robust effects were observed, the study may have been underpowered to observe differences between left and right MTLE.
Figure 2.
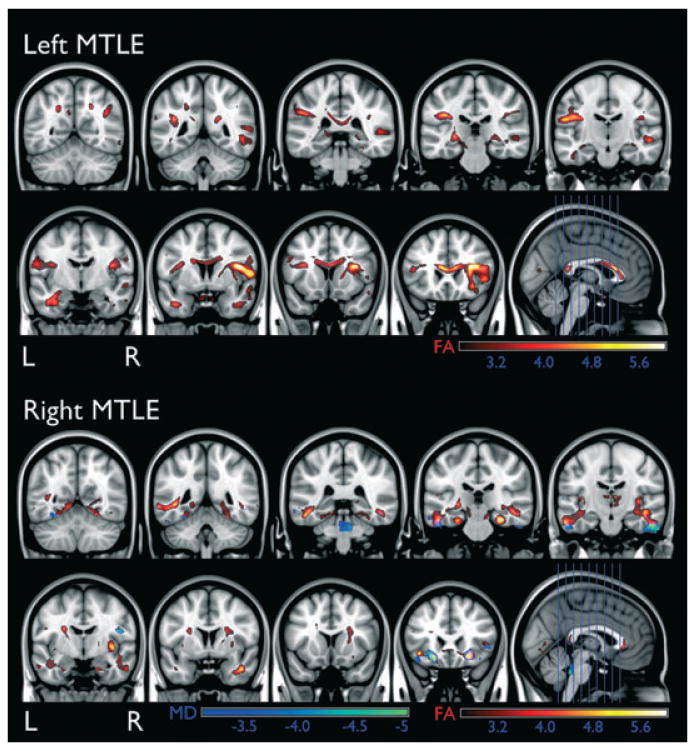
The location of FA reduction and MD increase in patients with left and right MTLE. The scale bars represent Z-scores.
3. Probabilistic tractography
Patients with left and right MTLE exhibited a significant decline in fiber density from the hippocampi in the regions highlighted in Fig. 3. Patients with left MTLE showed less fibers in the white matter located in the ipsilateral temporal pole, medial temporal region, orbitofrontal area, thalamus, temporooccipital and frontal regions, and contralateral thalamus and temporooccipital regions. Patients with right MTLE displayed less fibers in the white matter located at the cerebellum, ispilateral medial temporal lobe and occipital pole and contralateral medial temporal lobe and temporooccipital region. Interestingly, the results from probabilistic tractography are similar to the results from MD and FA analyses. Even though probabilistic tractography derives in part from FA measurements, it is also constructed from directionality. Furthermore, abnormal FA and MD do not necessarily correspond to fibers from or to the hippocampus, but rather represent the fibers in that particular white matter location, without further information on which bundle is affected. Therefore tractography complements the FA and MD analysis.
Figure 3.
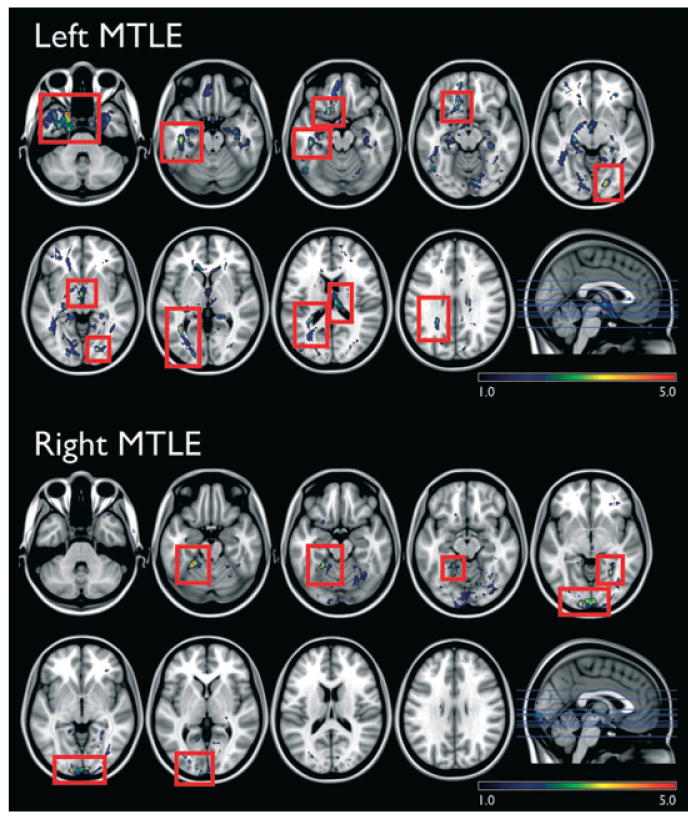
The location of significantly reduced hippocampal probabilistic tractography in patients compared with controls (red rectangles highlight the significant areas, above a critical threshold for repeated measures correction, Z = 3.1, left side and Z = 2.9, right side). The images are displayed in neurological convention, i.e., the left side represents the left side of the brain. The scale bars demonstrate the Z-scores.
4. Relationship between VBM and hippocampal fibers
Moderate to high correlations were observed in Z-scores between perihippocampal MD or FA and the ipsilateral amygdala, cerebellum, cingulum, cuneus, middle orbitofrontal, superior frontal, pallidum, putamen, supplementary motor area, inferior, middle and superior temporal regions. Furthermore, a significant Z-score relationship was observed between the perihippocampal FA or MD and the contralateral amygdala, cerebellum, cingulum, superior frontal, middle orbitofrontal, insula, occipital, pallidum, parahippocampal, putamen, rolandic, supplementary motor area, inferior temporal and thalamus (Table 3). The correlations between atrophy on the putamen, inferior and middle temporal areas, pallidum, amygdala, and cerebellar hemisphere decreased hippocampal connectivity (MD) remained significant after correction for multiple comparisons (Table 4). Compared with noncorrected results, the caudate, the middle and the anterior cingulate did not show a significant relationship when the results were corrected for multiple comparisons.
Table 3. A priori isolated regions of interest (based on literature review of frequently atrophied regions in MTLE), where the reduction of gray matter corresponded to standardized reduction in hippocampal fibers, as measured by the average MD. These results are corrected for multiple comparisons using FDR (the FDR corrected p-values marked with a star are statistically significant).
| Structure | |||
|---|---|---|---|
| Ipsilateral to the side of HA | MD | ||
| CC | p | FDR corrected | |
| Amygdala | 0.542857 | 0.018261 | 0.0133* |
| Anterior cingulum | 0.442857 | 0.049147 | 0.0333 |
| Caudate | 0.489286 | 0.03208 | 0.03 |
| Cerebelar hemisphere | 0.532143 | 0.020579 | 0.0200* |
| Inferior temporal | 0.571429 | 0.013032 | 0.0067* |
| Insula | 0.385714 | 0.077815 | 0.04 |
| Middle cingulum | 0.492857 | 0.030976 | 0.0267 |
| Middle temporal | 0.528571 | 0.021399 | 0.0233* |
| Middle temporal pole | 0.535714 | 0.019783 | 0.0167* |
| Pallidum | 0.546429 | 0.017533 | 0.0100* |
| Parahippocampal | 0.382143 | 0.079912 | 0.0467 |
| Posterior cingulum | 0.385714 | 0.077815 | 0.0433 |
| Putamen | 0.592857 | 0.009923 | 0.0033* |
| Superior temporal | 0.3 | 0.138658 | 0.05 |
| Thalamus | 0.435714 | 0.052247 | 0.0367 |
Interestingly, there was not a correlation between the ipsilateral hippocampal volume and the perihippocampal fibers, suggesting that there may be a critical threshold of volume loss until which there is a correspondence of fiber loss, but beyond this point the level of atrophy surpasses the degree of fiber loss.
Discussion
This study aimed to investigate the presence and extent of gray matter loss and white matter disconnection in patients with MTLE. The results obtained confirmed the presence of extrahippocampal and extratemporal gray matter atrophy in patients with MTLE. The results also indicated the presence of significant white matter abnormalities in limbic structures in patients with MTLE, in particular regarding hippocampal fiber loss and disconnection. Interestingly, there was a significant relationship between hippocampal fiber abnormalities and regional gray matter atrophy in MTLE patients. While this last finding does not prove causality between fiber loss and atrophy, it shows that, amongst the areas of extrahippocampal atrophy in MTLE, some areas are damaged independently from hippocampal fiber loss, whilst other are closely related.
Extrahippocampal gray matter atrophy in patients with MTLE was first observed via manual morphometry studies employing high-resolution MRI and anatomical protocols for segmentation of the medial temporal lobe (Andermann, 2003; Bernasconi et al., 2003; Bonilha et al., 2003). Those studies demonstrated that medial temporal structures such as the entorhinal and perirhinal cortices exhibit significant atrophy in patients with MTLE. The atrophy is usually more intense in the entorhinal cortex, which is anatomically closer to the hippocampus. Later studies, employing automatic whole-brain voxel-wise morphometry (VBM), confirmed the findings from manual morphometry studies but also atrophy extending beyond the temporal lobe, affecting cortical and subcortical structures, mostly within the limbic system (Bernasconi et al., 2004; Bonilha et al., 2004; Mueller et al., 2006; Keller & Roberts, 2008; Riederer et al., 2008).
The etiology of extrahippocampal atrophy in patients with MTLE is debatable. There is a relationship between time and severity of epilepsy and the degree of extrahippocampal atrophy, suggesting that brain atrophy is a dynamic process that progresses with time (Bonilha et al., 2006). Electroencephalogram studies have demonstrated that seizure spread in patients with MTLE follows a route that matches the anatomical pattern of atrophy, encompassing limbic structures (Wennberg et al., 2002). In a review paper, Dr. Spencer suggested that patients with MTLE exhibit a network of atrophy, involving predominantly medial temporal and limbic structures (Spencer, 2002). This pattern has been regularly reproduced by further automatic MRI studies. A prominent hypothesis to explain this phenomenon is the excitotoxic injury from seizures since the location of atrophy matches the route of seizure spread. Specifically, neuronal damage due to seizure excitotoxicity may lead to extrahippocampal gray matter loss and detectable volume atrophy in patients with MTLE.
While this hypothesis might explain a considerable component of the extratemporal atrophy in patients with MTLE, another possible explanation for atrophy is that the loss of hippocampal cells in HS can lead to loss of input from the hippocampus into limbic structures and thereby neuronal loss from deafferentation. If there is a direct relationship between hippocampal fiber loss and limbic gray matter atrophy, it is possible that deafferentation plays a role in extrahippocampal volume loss. Interestingly, if the efferent routes from the hippocampus are reduced, seizure spread would decrease to directly connected areas, thereby minimizing the excitotoxic effects of seizures. Hence, if atrophy evolves in the same rate as hippocampal disconnection, deafferentation is a plausible mechanism. Here, we investigated this relationship, combining measures of gray matter volume (VBM), whole brain connectivity (DTI) and hippocampal connectivity (probabilistic tractography) in patients with MTLE. From this data, this study was able to assess the relationship between hippocampal fiber loss (as standardized Z-scores) and regional gray matter atrophy. There was a significant relationship between gray matter loss and perihippocampal fiber loss in diffuse brain regions, notably in temporal areas, basal nuclei and the cerebellum. If the results from the noncorrected analyses are also taken into account, there is a possibility that deafferentation plays a role in the atrophy of the orbitofrontal cortex and cingulum as well. Since noncorrected results may incur into type I error, these may be interpreted with caution and this is a limitation of this study. Further studies may elucidate if frontal or cingulated atrophy may be related to hippocampal disconnection.
The results from this study suggest that extrahippocampal atrophy in MTLE may be, to some extent, related to hippocampal deafferentation. Conversely, these results also suggest that a significant portion of extrahippocampal atrophy in MTLE is independent from deafferentation and may be a direct consequence of the effect of seizures.
We speculate that the dissociation between seizure effects versus deafferentation as a cause for regional brain atrophy could be further examined in patients with hippocampal atrophy but without epilepsy, such as first degree relatives of patients with familial MTLE (Kobayashi et al., 2003) or hypoxic hippocampal damage. Moreover, the effects of longstanding seizures can also be examined in patients with refractory primary generalized epilepsy.
In conclusion, we suggest that patients with MTLE exhibit white matter fiber disconnections that involve predominantly limbic structures. We also suggest that MTLE is associated with hippocampal deafferentation and we speculate that deafferentation from hippocampal fiber loss is partially responsible for gray matter atrophy involving the lateral temporal areas, basal nuclei and cerebellar areas.
Table 2. Areas where standardized reduction of gray matter corresponded to standardized reduction in hippocampal fibers, as measured by the average FA and MD in the perihippocampal ROI.
| Structure | Structure | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ipsilateral to the side of HA | FA | MD | Contralateral to the side of HA | FA | MD | ||||
| CC | p | CC | p | CC | p | CC | p | ||
| Amygdala | −0.09643 | 0.366226 | 0.542857 | 0.018261 | Amygdala | −0.12857 | 0.323958 | 0.628571 | 0.00604 |
| Angular gyrus | −0.38929 | 0.075756 | 0.310714 | 0.129834 | Angular gyrus | −0.28571 | 0.150968 | 0.332143 | 0.113236 |
| Calcarine | −0.36071 | 0.093277 | 0.153571 | 0.292382 | Calcarine | −0.36786 | 0.088671 | 0.371429 | 0.086425 |
| Caudate | −0.03571 | 0.449723 | 0.489286 | 0.03208 | Caudate | −0.175 | 0.266374 | 0.396429 | 0.071747 |
| Cerebellar hemisphere | −0.08929 | 0.375836 | 0.532143 | 0.020579 | Cerebellar hemisphere | −0.25357 | 0.180908 | 0.471429 | 0.038035 |
| Anterior cingulum | −0.21429 | 0.22157 | 0.442857 | 0.049147 | Anterior cingulum | −0.38929 | 0.075756 | 0.328571 | 0.115905 |
| Middle cingulum | −0.31786 | 0.124145 | 0.492857 | 0.030976 | Middle cingulum* | −0.29286 | 0.144736 | 0.478571 | 0.035565 |
| Posterior cingulum | −0.33929 | 0.108014 | 0.385714 | 0.077815 | Posterior cingulum | −0.36786 | 0.088671 | 0.371429 | 0.086425 |
| Cuneus | −0.48929 | 0.03208 | 0.085714 | 0.380667 | Cuneus | −0.36786 | 0.088671 | 0.167857 | 0.274928 |
| Inferior opercular frontal | −0.42857 | 0.05548 | 0.339286 | 0.108014 | Inferior opercular frontal | −0.375 | 0.084216 | 0.342857 | 0.105462 |
| Inferior orbital frontal | −0.38214 | 0.079912 | 0.425 | 0.057148 | Inferior orbital frontal | −0.25 | 0.184423 | 0.407143 | 0.066006 |
| Inferior frontal (triangularis) | −0.27143 | 0.163895 | 0.367857 | 0.088671 | Inferior frontal (triangularis) | −0.28929 | 0.147833 | 0.382143 | 0.079912 |
| Inferior frontal (orbitalis) | −0.33571 | 0.110606 | 0.282143 | 0.154142 | Inferior frontal (orbitalis) | −0.40357 | 0.067884 | 0.257143 | 0.17743 |
| Middle frontal | −0.26429 | 0.170587 | 0.425 | 0.057148 | Middle frontal | −0.26429 | 0.170587 | 0.385714 | 0.077815 |
| Middle orbital frontal* | −0.19643 | 0.241449 | 0.617857 | 0.007052 | Middle orbital frontal | −0.31071 | 0.129834 | 0.45 | 0.046179 |
| Superior frontal | −0.28571 | 0.150968 | 0.382143 | 0.079912 | Superior frontal | −0.15 | 0.296815 | 0.507143 | 0.026832 |
| Medial superior frontal | −0.21429 | 0.22157 | 0.528571 | 0.021399 | Medial superior frontal | −0.275 | 0.160606 | 0.453571 | 0.044743 |
| Superior orbital frontal* | −0.08929 | 0.375836 | 0.496429 | 0.029899 | Superior orbital frontal | 0.003571 | 0.494961 | 0.460714 | 0.041967 |
| Fusiform | −0.30714 | 0.132736 | 0.382143 | 0.079912 | Fusiform | −0.26429 | 0.170587 | 0.335714 | 0.110606 |
| Heschl gyrus | −0.11429 | 0.342533 | 0.067857 | 0.405054 | Heschl gyrus | −0.22143 | 0.213857 | 0.139286 | 0.310273 |
| Hippocampus | −0.04286 | 0.43973 | 0.2 | 0.237407 | Hippocampus | −0.225 | 0.210052 | 0.439286 | 0.05068 |
| Insula | −0.25714 | 0.17743 | 0.385714 | 0.077815 | Insula | −0.22143 | 0.213857 | 0.446429 | 0.047647 |
| Lingual gyrus | −0.18571 | 0.25377 | 0.207143 | 0.229421 | Lingual gyrus | −0.30357 | 0.135678 | 0.275 | 0.160606 |
| Inferior occipital | −0.3 | 0.138658 | 0.210714 | 0.225479 | Inferior occipital | −0.36786 | 0.088671 | 0.271429 | 0.163895 |
| Middle occipital | −0.43571 | 0.052247 | 0.014286 | 0.47985 | Middle occipital | −0.475 | 0.036785 | 0.232143 | 0.202549 |
| Superior occipital | −0.31071 | 0.129834 | 0.185714 | 0.25377 | Superior occipital | −0.20357 | 0.233398 | 0.296429 | 0.141678 |
| Olfactory gyrus | −0.075 | 0.395255 | 0.496429 | 0.029899 | Olfactory gyrus | −0.225 | 0.210052 | 0.496429 | 0.029899 |
| Pallidum | −0.08214 | 0.385514 | 0.546429 | 0.017533 | Pallidum* | −0.02857 | 0.459745 | 0.721429 | 0.001199 |
| Paracentralobule | −0.42143 | 0.05885 | 0.382143 | 0.079912 | Paracentralobule | −0.30714 | 0.132736 | 0.25 | 0.184423 |
| Parahippocampal | −0.11786 | 0.337856 | 0.382143 | 0.079912 | Parahippocampal | −0.225 | 0.210052 | 0.485714 | 0.033213 |
| Inferior parietal | −0.29286 | 0.144736 | 0.146429 | 0.301275 | Inferior parietal | −0.43214 | 0.053847 | 0.032143 | 0.454731 |
| Superior parietal | −0.16071 | 0.283598 | −0.27143 | 0.163895 | Superior parietal | −0.475 | 0.036785 | −0.23929 | 0.19519 |
| Postcentral | −0.34286 | 0.105462 | 0.378571 | 0.082045 | Postcentral | −0.33214 | 0.113236 | 0.289286 | 0.147833 |
| Precentral | −0.26786 | 0.167222 | 0.414286 | 0.062358 | Precentral | −0.36071 | 0.093277 | 0.346429 | 0.102948 |
| Precuneus | −0.40714 | 0.066006 | 0.196429 | 0.241449 | Precuneus | −0.39286 | 0.073733 | 0.267857 | 0.167222 |
| Putamen* | 0.175 | 0.266374 | 0.592857 | 0.009923 | Putamen | 0.021429 | 0.469789 | 0.6 | 0.009025 |
| Rectus | −0.275 | 0.160606 | 0.375 | 0.084216 | Rectus | −0.32143 | 0.12136 | 0.282143 | 0.154142 |
| Rolandic (opercularis) | −0.40714 | 0.066006 | 0.267857 | 0.167222 | Rolandic (opercularis) | −0.23571 | 0.198851 | 0.517857 | 0.024002 |
| Supplemetary motor area | −0.37143 | 0.086425 | 0.442857 | 0.049147 | Supplemetary motor area | −0.28571 | 0.150968 | 0.475 | 0.036785 |
| Supramarginal gyrus | −0.35357 | 0.098036 | 0.425 | 0.057148 | Supramarginal gyrus | −0.34643 | 0.102948 | 0.185714 | 0.25377 |
| Inferior temporal* | −0.20714 | 0.229421 | 0.571429 | 0.013032 | Inferior temporal | −0.35 | 0.100473 | 0.45 | 0.046179 |
| Middle temporal | −0.25 | 0.184423 | 0.528571 | 0.021399 | Middle temporal | −0.27857 | 0.157355 | 0.371429 | 0.086425 |
| Middle temporal pole | −0.17143 | 0.270636 | 0.535714 | 0.019783 | Middle temporal pole | −0.24643 | 0.187975 | 0.335714 | 0.110606 |
| Superior temporal pole | −0.31786 | 0.124145 | 0.321429 | 0.12136 | Superior temporal pole | −0.25 | 0.184423 | 0.296429 | 0.141678 |
| Superior temporal | −0.46071 | 0.041967 | 0.3 | 0.138658 | Superior temporal | −0.28214 | 0.154142 | 0.314286 | 0.12697 |
| Thalamus | −0.26071 | 0.17399 | 0.435714 | 0.052247 | Thalamus* | −0.49286 | 0.030976 | 0.428571 | 0.05548 |
| Vermis | −0.32143 | 0.12136 | 0.396429 | 0.071747 | – | – | – | – | – |
Significant correlations are highlighted in bold. A star indicates a region that was not significantly atrophied in patients compared with controls.
Acknowledgments
We confirm that we have read the Journal's position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Footnotes
Disclosure: The authors report no financial or non-financial conflicts of interest associated with this study.
References
- Alessio A, Bonilha L, Rorden C, Kobayashi E, Min LL, Damasceno BP, Cendes F. Memory and language impairments and their relationships to hippocampal and perirhinal cortex damage in patients with medial temporal lobe epilepsy. Epilepsy Behav. 2006;8:593–600. doi: 10.1016/j.yebeh.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Andermann F. Why study mesial temporal atrophy in patients with intractable temporal lobe epilepsy? J Neurol Neurosurg Psychiatry. 2003;74:1606–1607. doi: 10.1136/jnnp.74.12.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Bernasconi N, Bernasconi A, Caramanos Z, Antel SB, Andermann F, Arnold DL. Mesial temporal damage in temporal lobe epilepsy: a volumetric MRI study of the hippocampus, amygdala and parahippocampal region. Brain. 2003;126:462–469. doi: 10.1093/brain/awg034. [DOI] [PubMed] [Google Scholar]
- Bernasconi N, Duchesne S, Janke A, Lerch J, Collins DL, Bernasconi A. Whole-brain voxel-based statistical analysis of gray matter and white matter in temporal lobe epilepsy. Neuroimage. 2004;23:717–723. doi: 10.1016/j.neuroimage.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Blumcke I, Thom M, Wiestler OD. Ammon's horn sclerosis: a maldevelopmental disorder associated with temporal lobe epilepsy. Brain Pathol. 2002;12:199–211. doi: 10.1111/j.1750-3639.2002.tb00436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilha L, Kobayashi E, Rorden C, Cendes F, Li LM. Medial temporal lobe atrophy in patients with refractory temporal lobe epilepsy. J Neurol Neurosurg Psychiatry. 2003;74:1627–1630. doi: 10.1136/jnnp.74.12.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilha L, Rorden C, Castellano G, Pereira F, Rio PA, Cendes F, Li LM. Voxel-based morphometry reveals gray matter network atrophy in refractory medial temporal lobe epilepsy. Arch Neurol. 2004;61:1379–1384. doi: 10.1001/archneur.61.9.1379. [DOI] [PubMed] [Google Scholar]
- Bonilha L, Rorden C, Castellano G, Cendes F, Li LM. Voxel-based morphometry of the thalamus in patients with refractory medial temporal lobe epilepsy. Neuroimage. 2005;25:1016–1021. doi: 10.1016/j.neuroimage.2004.11.050. [DOI] [PubMed] [Google Scholar]
- Bonilha L, Rorden C, Appenzeller S, Coan AC, Cendes F, Li LM. Gray matter atrophy associated with duration of temporal lobe epilepsy. Neuroimage. 2006;32:1070–1079. doi: 10.1016/j.neuroimage.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Bonilha L, Alessio A, Rorden C, Baylis G, Damasceno BP, Min LL, Cendes F. Extrahippocampal gray matter atrophy and memory impairment in patients with medial temporal lobe epilepsy. Hum Brain Mapp. 2007;28:1376–1390. doi: 10.1002/hbm.20373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cendes F, Andermann F, Gloor P, Evans A, Jones-Gotman M, Watson C, Melanson D, Olivier A, Peters T, Lopes-Cendes I, Leroux G. MRI volumetric measurement of amygdala and hippocampus in temporal lobe epilepsy. Neurology. 1993;43:719–725. doi: 10.1212/wnl.43.4.719. [DOI] [PubMed] [Google Scholar]
- Commission on Classification and Terminology of the International League Against Epilepsy. Proposal for revised classification of epilepsies and epileptic syndromes. Epilepsia. 1989;30:389–399. doi: 10.1111/j.1528-1157.1989.tb05316.x. [DOI] [PubMed] [Google Scholar]
- Focke NK, Yogarajah M, Bonelli SB, Bartlett PA, Symms MR, Duncan JS. Voxel-based diffusion tensor imaging in patients with mesial temporal lobe epilepsy and hippocampal sclerosis. Neuroimage. 2008;40:728–737. doi: 10.1016/j.neuroimage.2007.12.031. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr, Sharbrough FW, Twomey CK, Cascino GD, Hirschorn KA, Marsh WR, Zinsmeister AR, Scheithauer B. Temporal lobe seizures: lateralization with MR volume measurements of the hippocampal formation. Radiology. 1990;175:423–429. doi: 10.1148/radiology.175.2.2183282. [DOI] [PubMed] [Google Scholar]
- Keller SS, Mackay CE, Barrick TR, Wieshmann UC, Howard MA, Roberts N. Voxel-based morphometric comparison of hippocampal and extrahippocampal abnormalities in patients with left and right hippocampal atrophy. Neuroimage. 2002;16:23–31. doi: 10.1006/nimg.2001.1072. [DOI] [PubMed] [Google Scholar]
- Keller SS, Roberts N. Voxel-based morphometry of temporal lobe epilepsy: an introduction and review of the literature. Epilepsia. 2008;49:741–757. doi: 10.1111/j.1528-1167.2007.01485.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi E, D'Agostino MD, Lopes-Cendes I, Berkovic SF, Li ML, Andermann E, Andermann F, Cendes F. Hippocampal atrophy and T2-weighted signal changes in familial mesial temporal lobe epilepsy. Neurology. 2003;60:405–409. doi: 10.1212/wnl.60.3.405. [DOI] [PubMed] [Google Scholar]
- Liu RS, Lemieux L, Bell GS, Hammers A, Sisodiya SM, Bartlett PA, Shorvon SD, Sander JW, Duncan JS. Progressive neocortical damage in epilepsy. Ann Neurol. 2003;53:312–324. doi: 10.1002/ana.10463. [DOI] [PubMed] [Google Scholar]
- Margerison JH, Corsellis JA. Epilepsy and the temporal lobes. A clinical, electroencephalographic and neuropathological study of the brain in epilepsy, with particular reference to the temporal lobes. Brain. 1966;89:499–530. doi: 10.1093/brain/89.3.499. [DOI] [PubMed] [Google Scholar]
- McDonald CR, Hagler DJ, Jr, Ahmadi ME, Tecoma E, Iragui V, Gharapetian L, Dale AM, Halgren E. Regional neocortical thinning in mesial temporal lobe epilepsy. Epilepsia. 2008;49:794–803. doi: 10.1111/j.1528-1167.2008.01539.x. [DOI] [PubMed] [Google Scholar]
- Mueller SG, Laxer KD, Cashdollar N, Buckley S, Paul C, Weiner MW. Voxel-based optimized morphometry (VBM) of gray and white matter in temporal lobe epilepsy (TLE) with and without mesial temporal sclerosis. Epilepsia. 2006;47:900–907. doi: 10.1111/j.1528-1167.2006.00512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer F, Lanzenberger R, Kaya M, Prayer D, Serles W, Baumgartner C. Network atrophy in temporal lobe epilepsy: a voxel-based morphometry study. Neurology. 2008;71:419–425. doi: 10.1212/01.wnl.0000324264.96100.e0. [DOI] [PubMed] [Google Scholar]
- Rorden C, Bonilha L, Nichols TE. Rank-order versus mean based statistics for neuroimaging. Neuroimage. 2007;35:1531–1537. doi: 10.1016/j.neuroimage.2006.12.043. [DOI] [PubMed] [Google Scholar]
- Rothman KJ. No adjustments are needed for multiple comparisons. Epidemiology. 1990;1:43–46. [PubMed] [Google Scholar]
- Spencer SS. Neural networks in human epilepsy: evidence of and implications for treatment. Epilepsia. 2002;43:219–227. doi: 10.1046/j.1528-1157.2002.26901.x. [DOI] [PubMed] [Google Scholar]
- Sutula TP, Hagen J, Pitkanen A. Do epileptic seizures damage the brain? Curr Opin Neurol. 2003;16:189–195. doi: 10.1097/01.wco.0000063770.15877.bc. [DOI] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Wennberg R, Arruda F, Quesney LF, Olivier A. Preeminence of extrahippocampal structures in the generation of mesial temporal seizures: evidence from human depth electrode recordings. Epilepsia. 2002;43:716–726. doi: 10.1046/j.1528-1157.2002.31101.x. [DOI] [PubMed] [Google Scholar]


