Abstract
Eotaxin-3/CCL26 is an agonist for chemokine receptor 3 (CCR3) and a natural antagonist for CCR1, CCR2 and CCR5. CCL26 expression by non-haematopoietic cells has been well documented; however, no studies to date have demonstrated CCL26 expression by leucocytes. In this study, we investigated the ability of human monocytic cells to produce CCL26 in response to cytokines. We found that interleukin-4 (IL-4) increased the expression of CCL26 messenger RNA (mRNA) and protein in U937 cells, in human monocytes and in human monocyte-derived macrophages. Tumour necrosis factor-α (TNF-α) and interleukin-1β (IL-1β) alone did not induce CCL26 expression, yet these pro-inflammatory cytokines synergized with IL-4 to increase CCL26 protein expression. Signal transducer and activator of transcription 6 (STAT6) was not affected by costimulation with TNF-α, suggesting that the synergy between IL-4 and TNF-α occurs at a step downstream of STAT6 activation. Co-incubation of interferon-γ (IFN-γ) with IL-4 had no effect on CCL26 protein release. By contrast, pretreatment with IFN-γ decreased total STAT6 protein, blocked IL-4-mediated STAT6 phosphorylation and decreased IL-4-mediated CCL26 mRNA expression and protein release. These data show that IL-4 and pro-inflammatory cytokines such as TNF-α, IL-1β and IFN-γ regulate CCL26 synthesis in human monocytic cells, which may be important in regulating monocyte inflammatory responses.
Keywords: chemokine, eotaxin-3, interleukin-4, monocyte/macrophage, signalling
Introduction
The eotaxin subfamily of CC chemokines consists of eotaxin/CCL11, eotaxin-2/CCL24 and eotaxin-3/CCL26.1–3 Although they only share 34–39% protein homology, all eotaxins activate cells via CC chemokine receptor 3 (CCR3), which is expressed on several different cell types including eosinophils, basophils, dendritic cells, smooth muscle cells, epithelial cells and fibroblasts (reviewed in Ref. 4). CCL11 and CCL24 are expressed by haematopoietic and non-haematopoietic cells.5–7 By contrast, CCL26 expression has only been described in non-haematopoietic cells.3,6,8,9 Interleukin-4 (IL-4) is the principal stimulus for CCL26 expression,10 whereas CCL11 and CCL24 are upregulated by IL-4 and pro-inflammatory cytokines such as interleukin-1β (IL-1β) and tumour necrosis factor-α (TNF-α).11
CCL26 acts predominately as a CCR3 agonist,3 yet it also acts as an antagonist for CCR1, CCR2 and CCR5.12,13 This has led to the speculation that CCL26 may have a modulatory role in inflammation. CCR2, in particular, is a major pro-inflammatory chemokine receptor expressed by monocytes and macrophages, and CCL26 has been shown to block monocyte responses to monocyte chemotactic protein-1 (MCP-1), a major ligand for CCR2.12
The purpose of this study was to determine if monocytic cells could synthesize and express CCL26, because this could provide an autoregulatory mechanism during inflammation. We examined the ability of human peripheral blood monocytes, monocyte-derived macrophages (MDMs) and the monocytic cell line U937 to express CCL26 messenger RNA (mRNA) and protein. We showed that monocytic cells express CCL26 in response to IL-4 and that TNF-α, IL-1β and interferon-γ (IFN-γ) modulate IL-4-mediated CCL26 synthesis and expression.
Materials and methods
Reagents
Human recombinant TNF-α, IL-1β, IFN-γ, IL-4 and mouse non-immune immunoglobulin G1 (IgG1) were purchased from R&D Systems, Inc. (Minneapolis, MN). Lymphoprep was from BioLynx Inc. (Brockville, ON, Canada) Advanced RPMI-1640, penicillin–streptomycin–glutamine (PSG), TRIzol reagent, Superscript II and NeutrAvidin were from Invitrogen Life Technologies (Carlsbad, CA). Fetal bovine serum (FBS) was from Hyclone (Logan, UT). Hanks’ balanced salt solution (HBSS), 3,3′,5,5′ tetramethyl benzidine liquid substrate (TMB), Tween-20 and Triton X-100 were purchased from Sigma Chemicals (Oakville, Canada). Affinity purified goat anti-(human eotaxin-3) sera and biotinylated anti-(human eotaxin-3) Ig were purchased from PeproTech (Rocky Hill, NJ). Supersignal West Pico chemiluminescent reagent was from Pierce (Rockford, IL). TaqMAN PCR master mix for use in standard polymerase chain reaction (PCR) was from Qiagen (Mississauga, Canada). TaqMAN universal PCR master mix for use in real-time PCR and the 18S primer/probe kit were from Applied Biosystems (Warrington, UK). Rabbit anti-[human signal transducer and activation of transcription 6 (STAT6)], rabbit anti-(human phospho-STAT6) and rabbit anti-(human β-actin) Igs were purchased from New England Biolabs Ltd (Pickering, Canada). All other reagents were from VWR International (Edmonton, Canada).
Cell culture and isolation
Human promonocytic U937 cells were obtained from the American Type Culture Collection (Manassas, VA) and maintained as recommended. Whole blood was obtained from healthy volunteers, as approved by the Ethics Committee at the University of Calgary. Platelet-rich plasma was removed from heparinized whole blood following centrifugation at 250 g for 20 min. The remaining leucocytes and red blood cells were diluted in HBSS, carefully layered onto Lymphoprep and centrifuged at 400 g for 30 min. The mononuclear cells were harvested and washed with HBSS, and 8 × 106 cells/well were allowed to adhere onto six-well tissue culture plates for 2 hr at 37° in serum-free RPMI-1640. Non-adherent cells and contaminating platelets were carefully removed from the plate by multiple wash steps using HBSS. The purity of cells remaining on the plate after 2 hr of adhesion was > 90% monocytes, with contaminating cells being platelets and lymphocytes. The remaining adherent cells were cultured overnight in RPMI-1640 containing 5% FBS. For studies using monocytes, adherent cells were washed and incubated in serum-free RPMI-1640 in the presence or absence of cytokines for 24 hr. In control experiments, purified lymphocytes or platelets were stimulated with IL-4 for 24 hr and the expression of CCL26 was determined. Neither cell type showed an increase in CCL26 (data not shown). For MDM cultures, fresh RPMI-1640 containing 5% FBS and 5% human serum was added to the monocyte cultures after the overnight incubation. The cells were cultured for an additional 7 days to allow their differentiation into macrophages. Human serum, which contains monocyte colony-stimulating factor, was used to differentiate monocytes into macrophages as opposed to exogenous cytokines, as previously described by our group.14 Differentiation was determined morphologically, by flow cytometry, showing expression of CD14, but not CD83 (a dendritic cell marker), and by immunohistochemistry examining CD14 and CD83 (data not shown).
Western blotting for STAT6 in whole-cell lysates
Following stimulation, U937 cells were lysed with hot 2 × Laemelli buffer. Proteins were separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE), and Western blotting was performed using phospho-specific STAT6, total STAT6 or β-actin antibodies. Immunoblots were visualized using a Fluor-S MAX™ MultiImager and analysed using quantity one software (Bio-Rad Laboratories, Hercules, CA).
RNA extraction, reverse transcription–PCR and real-time PCR
Total RNA was extracted from cells, and first-strand complementary DNA (cDNA) was synthesized using Superscript II, as described in the manufacturer’s instructions. cDNA was amplified by PCR using either Taq polymerase or TaqMAN Universal master mix. Primer sequences for standard PCR amplification were as follows.
| CCL26 | forward primer: 5′-AGTCACAATTGTTTCGGAGTT-3′ |
| reverse primer: 5′-AGTCTCCACCTTGGAACTG-3′ | |
| β-actin | forward primer: 5′-CATGGATGATGATATCGCCG-3′ |
| reverse primer: 5′-ACAGCCTGGATAGCAACGTA-3′ |
Primer sequences for real-time PCR were as follows.
| CCL26 | forward primer: 5′-ACACGTGGGAGTGACATATCCA-3′ |
| reverse primer: 5′-GACTTTCTTGCCTCTTTTGGTAGTG-3′ | |
| probe: TACAGCCACAAGCCCCTTCCCTGG. |
A commercially purchased primer and probe were used for 18S ribosomal RNA (rRNA). The amount of CCL26 mRNA in each sample was calculated using the −delta delta Ct (−ddCt) method.
CCL26 detection by enzyme-linked immunosorbent assay
Following stimulation, supernatants were harvested and stored at −20°. A goat anti-(human CCL26) capture Ig and a biotinylated anti-(human CCL26) detection Ig were used in an enzyme-linked immunosorbent assay (ELISA) to measure CCL26 protein. The detection limit for the ELISA was 12·5 pg/ml.
Statistics
All experiments were performed at least three times. Data are presented as mean ± standard error of the mean (SEM). Statistical differences between groups were determined using either one-way or two-way analysis of variance (anova) with the appropriate post-test comparison. P-values of less than 0·05 were considered statistically significant.
Results
CCL26 mRNA is expressed in monocytic cells following IL-4 stimulation
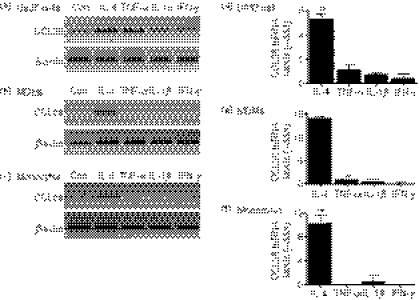
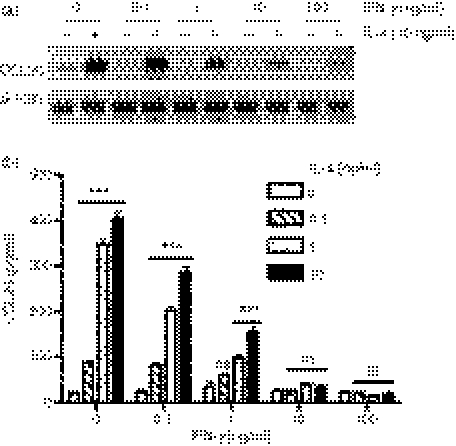
We investigated both constitutive and cytokine-induced expression of CCL26 mRNA in the monocytic cell line, U937, and in primary human MDMs and monocytes. Cells were stimulated for 24 hr in the presence or absence of 10 ng/ml of IL-4, TNF-α, IL-1β or IFN-γ. This concentration of the respective cytokines has been shown to increase the expression of CCL11 and/or CCL24 in other cell types.6,7,11 We previously showed that 10 ng/ml of IL-4 induced robust expression of CCL26 in human endothelial cells.15 RNA was harvested and CCL26 mRNA was detected by RT-PCR. With the exception of U937 cells, there was no constitutive expression of CCL26 by monocytic cells (Fig. 1a–c). Treatment with IL-4 led to increased expression of CCL26 mRNA in U937, MDMs and monocytes, whereas the other cytokines tested had little to no effect on CCL26 mRNA expression (Fig. 1a–c). Neither increasing the concentration of TNF-α, IL-1β or IFN-γ nor increasing the time to 48 hr resulted in CCL26 expression in U937 cells (data not shown). Treatment of other leucocyte subclasses, including neutrophils, lymphocytes or platelets, with IL-4 did not induce CCL26 expression (data not shown). We used real-time PCR and quantified these results by means of the −ddCt method, using the housekeeping gene 18S rRNA to normalize the data and using control cells as the calibrator (Fig. 1d–f). A value equal to the control will be 0. The results showed that treatment with IL-4 resulted in a significant increase in CCL26 over control values (U937 cells: 5·30 ± 0·43, n = 6, P < 0·01; MDMs: 13·83 ± 0·51, n = 3, P < 0·01; monocytes: 10·32 ± 1·43, n = 3, P < 0·01).
Figure 1.
CCL26 messenger RNA (mRNA) is expressed in monocytic cells following stimulation with interleukin (IL)-4. (a and d) U937 cells, (b and e) monocyte-derived macrophages (MDMs), or (c and f) monocytes were cultured for 24 hr in medium alone (Con) or in mediim containing 10 ng/ml of IL-4, tumour necrosis factor-α (TNF-α), IL-1β or interferon-γ (IFN-γ). RNA was isolated from the cells using TRIzol. (a–c) The expression of mRNA for CCL26 and β-actin was evaluated using the reverse transcription–polymerase chain reaction (RT-PCR), and the PCR products were visualized by gel electrophoresis in the presence of ethidium bromide. Data are representative of results obtained from at least three independent experiments. (d–f) The expression of mRNA for CCL26 was evaluated using real-time PCR and data were normalized using 18S ribosomal RNA (rRNA) expression. Values for cytokine-stimulated cells were compared with values for control cells and data were then expressed as the mean ± standard error of the mean (SEM) of −delta delta Ct (−ddCt) from at least three independent experiments. **P<0·01 indicates statistical significance compared with the control.
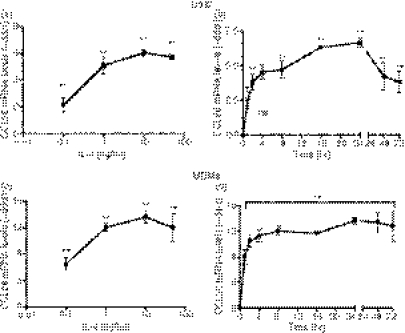
IL-4-induced CCL26 mRNA expression in monocytic cells is concentration-dependent and time-dependent
To further examine CCL26 gene expression in U937 cells and MDMs, cells were incubated with a range of concentrations of IL-4 for 24 hr. CCL26 mRNA levels were analyzed using real-time PCR. As shown in Fig. 2a,c, the increased levels of CCL26 mRNA correlated with increasing concentrations of IL-4, with a plateau at 10 ng/ml in both U937 cells and MDMs. To determine the kinetics of CCL26 gene expression, U937 cells and MDMs were stimulated with 10 ng/ml of IL-4 for 1–72 hr. IL-4 induced a very rapid (within 1 hr) and robust increase in CCL26 gene expression in both U937 cells (4·5 ± 0·5, n = 5, P < 0·01) (Fig. 2b) and MDMs (8·0 ± 1·2, n = 4, P < 0·01) (Fig. 2d). Expression in U937 cells began early at 1 hr, followed by a prolonged increase that continued to 24 hr. The levels of CCL26 then showed a decrease at 48 and 72 hr, although they were still elevated over control values (48 hr: 4·2 ± 1·0, n = 4, P < 0·01; 72 hr: 3·8 ± 0·78, n = 4, P < 0·01) (Fig. 2c). Unlike U937 cells, in MDMs early expression of CCL26 was sustained for as long as 72 hr following stimulation (Fig. 2d).
Figure 2.
Interleukin (IL)-4-induced CCL26 messenger RNA (mRNA) expression in monocytic cells is concentration-dependent and time-dependent. (a) U937 cells and (c) monocyte-derived macrophages (MDMs) were cultured for 24 hr in medium alone or in medium containing increasing amounts of IL-4 (0·1–50 ng/ml). (b) U937 cells and (d) MDMs were cultured in medium alone, or in medium containing 10 ng/ml of IL-4, for 1–72 hr. RNA was extracted from the cells using TRIzol. The expression of mRNA for CCL26 was evaluated by real-time polymerase chain reaction (PCR) and the data were normalized using 18S ribosomal RNA (rRNA) expression. Values for cytokine-stimulated cells were compared with those of control cells and data were then expressed as the mean ± standard error of the mean (SEM) of −delta delta Ct (−ddCt) from at least four independent experiments. **P<0·01 indicates statistical significance compared with the control; and ns is not significant.
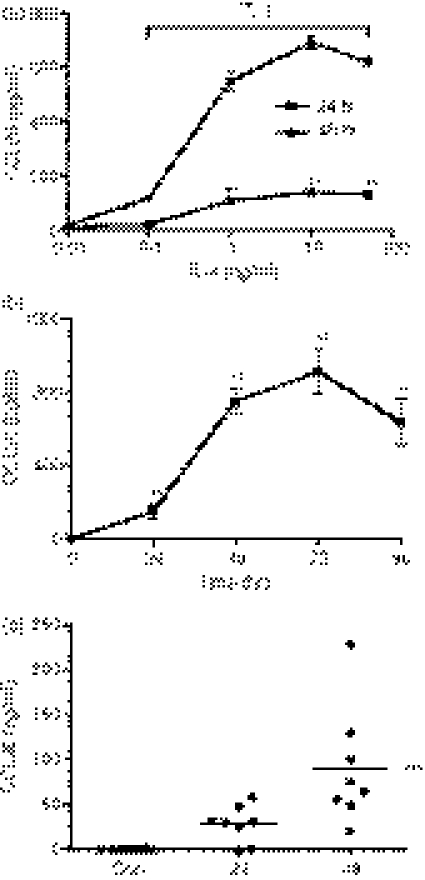
IL-4 induces CCL26 protein release from U937 cells and MDMs
To investigate whether U937 cells secrete CCL26, the cells were incubated with a range of concentrations of IL-4 for 24 and 48 hr. The supernatants were harvested and then assayed for CCL26 using an ELISA. No CCL26 was detected in the supernatants from U937 cells treated with medium alone, suggesting that U937 cells do not constitutively release CCL26 protein (Fig. 3a,b). IL-4 induced robust CCL26 release from U937 cells, with maximal levels detected using 10 ng/ml of IL-4 for 48 hr (692·83 ± 57·44 pg/ml, n = 6, P < 0·01 compared with the control). Similarly to U937 cells, no detectable levels of CCL26 were measured in supernatants from MDMs treated with medium alone (Fig. 3c). Stimulation with 10 ng/ml of IL-4 induced the release of CCL26 protein at 24 and 48 hr (control: 0·12 ± 0·12 pg/ml, n = 8; 24 hr: 28·00 ± 7·2 pg/ml n = 8, not significant; 48 hr: 90·25 ± 22·91 pg/mL n = 8, P < 0·001) (Fig. 3c). Consistent with mRNA data, no CCL26 protein was detected following stimulation of either U937 or MDMs with TNF-α, IL-1β or IFN-γ (data not shown). Notably, we found a high degree of donor-to-donor variation in the levels of CCL26 released from MDMs when the cells from eight different individuals were used. Owing to the variability in the levels of CCL26 released from MDMs, U937 cells were used in subsequent experiments.
Figure 3.
Interleukin (IL)-4 induces CCL26 protein release from U937 cells and monocyte-derived macrophages (MDMs). U937 cells were cultured (a) for 24 or 48 hr in the absence or presence of 0·1–50 ng/ml of IL-4 or (b) for 24–72 hr in the presence of 10 ng/ml of IL-4. (c) MDMs were cultured for 24 or 48 hr in the absence or presence of 10 ng/ml of IL-4. Supernatants were collected and the levels of CCL26 protein were measured using enzyme-linked immunosorbent assay (ELISA). (a and b) Data are expressed as mean ± standard error of the mean (SEM) of at least three independent experiments. (c) Data are expressed as the mean of eight independent experiments. **P<0·01 and ***P < 0·001 indicates statistical significance compared with the control; † indicates statistical significance as compared to cells stimulated with the same concentration of IL-4 for 24 hr; and ns is not significant. Con, control.
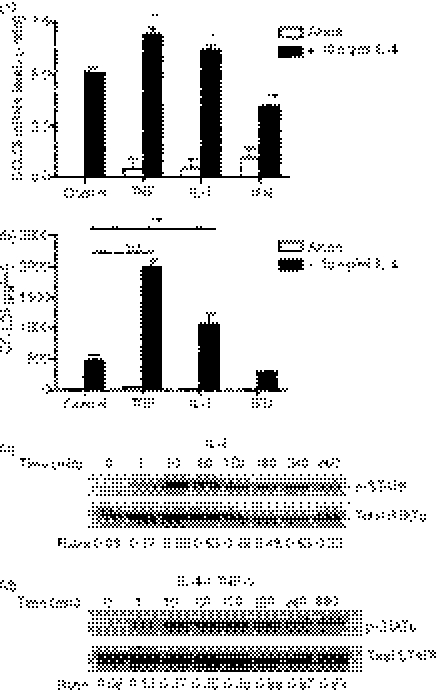
TNF-α and IL-1β synergize with IL-4 to induce enhanced expression of CCL26 protein
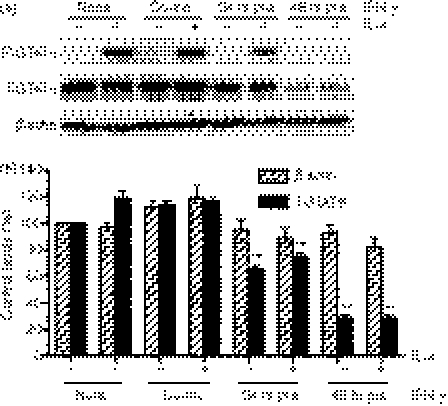
TNF-α or IL-1β synergize with IL-4 in A549 airway epithelial cells to enhance CCL26 expression and release.8 To investigate whether pro-inflammatory cytokines could synergize with IL-4 to enhance CCL26 mRNA and protein release in U937 cells, cells were treated with IL-4, either alone or with TNF-α, IL-1β or IFN-γ, for 48 hr. U937 cells treated with IL-4, together with TNF-α or IL-1β, demonstrated a slight, but significant, increase in CCL26 mRNA expression when compared with the CCL26 mRNA levels obtained from U937 cells treated with IL-4 alone (Fig. 4a). CCL26 protein release was substantially enhanced in supernatants harvested from U937 cells stimulated with a combination of IL-4/TNF-α and IL-4/IL-1β when compared to U937 cells stimulated with IL-4 alone (IL-4 alone: 474 ± 89 pg/ml, n = 5; IL-4 + TNF-α: 2004 ± 99·27 pg/ml, n = 5, P < 0·001 compared to stimulation with IL-4 alone; IL-4 + IL-1β: 1069 ± 172 pg/ml, n = 5, P < 0·01 compared to stimulation with IL-4 alone) (Fig. 4b). The levels of CCL26 protein detected were greater than the sum of the release induced by the cytokines on their own, clearly demonstrating synergy between IL-4 and either TNF-α or IL-1β. Costimulation with IFN-γ led to a significant increase in CCL26 mRNA, but had no effect on IL-4-mediated CCL26 protein release (Fig. 4).
Figure 4.
Tumour necrosis factor-α (TNF-α) and interleukin (IL)-1β synergize with IL-4 to induce enhanced CCL26 expression from U937 cells. U937 cells were cultured for 48 hr in medium alone (Control) or in medium containing 10 ng/ml of IL-4, TNF-α, IL-1β or IFN-γ, alone, or in combination with IL-4. (a) RNA was extracted from the cells using TRIzol. The expression of CCL26 mRNA was evaluated by real-time polymerase chain reaction (PCR) and data were normalized using 18S ribosomal RNA (rRNA) expression. Values for cytokine-stimulated cells were compared with those for control cells and data were then expressed as the mean ± standard error of the mean (SEM) of −delta delta Ct (−ddCt) from at least three independent experiments. (b) Supernatants were collected and CCL26 protein was measured using enzyme-linked immunosorbent assay (ELISA). Data are expressed as mean ± standard error of the mean (SEM) of at least three independent experiments. *P < 0·05; **P < 0·01 and ***P < 0·001 compared to stimulation with IL-4 alone. U937 cells were stimulated with (c) IL-4 or (d) IL-4 and TNF-α for 1–360 min. Cell lysates were harvested in 2 × Laemelli sample buffer and analysed by Western blotting for phosphorylated signal transducer and activator of transcription 6 (p-STAT6) and Total STAT6. The blots shown are representative of the results obtained in more than three independent experiments. Densitometry was performed, the ratio of the signal in P-STAT6 to total STAT6 was determined and the value is shown.
Levels of total and phosphorylated STAT6 in IL-4-stimulated U937 cells
Several investigators have suggested that the synergistic effects observed following cytokine costimulation occur as a result of cytokine-enhanced expression of the IL-4 receptor α subunit (IL-4Rα). We investigated the change in expression of IL-4Rα mRNA under these conditions using real-time PCR and were unable to detect any significant alteration in the expression of this receptor subunit under any of the conditions tested (data not shown). We next examined STAT6 phosphorylation to determine if there were changes in the extent or kinetics of activation. U937 cells were stimulated with IL-4 or TNF-α, alone or in combination, for 1–360 min. Whole-cell lysates were immediately harvested and assayed, by Western blotting, for phosphorylated and total STAT6 expression. As expected, IL-4 induced a time-dependent phosphorylation of STAT6 (Fig. 4c). A similar pattern of STAT6 phosphorylation was seen following stimulation of U937 cells with the combination of IL-4 and TNF-α (Fig. 4d), suggesting that the phosphorylation of STAT6 was neither prolonged nor enhanced by combined cytokine treatment. The levels of total STAT6 varied slightly, and thus densitometry was performed and the ratio of P-STAT6 to total STAT6 was determined. This showed that TNF did not alter the extent or the kinetics of STAT6 phosphorylation induced by IL-4 (Fig. 4c,d).
Pretreatment of U937 cells with IFN-γ decreases IL-4-induced CCL26 gene expression and CCL26 release
Yamamoto et al.16 showed that a 48-hr pretreatment of bronchial epithelial cells with IFN-γ enhanced CCL26 gene expression and protein production induced by IL-4. To determine whether this was also observed in monocytic cells, U937 cells were pretreated with IFN-γ for 48 hr and then stimulated with IL-4. Surprisingly, this resulted in a substantial decrease in expression of CCL26 mRNA (Fig. 5a), suggesting that monocytic cells regulate CCL26 differently than epithelial cells. We next measured the levels of CCL26 protein release and found that pretreatment with IFN-γ led to a reduction in IL-4-induced CCL26 release (10 ng/ml of IL-4 alone: 404 ± 32 pg/ml, n = 5; IL-4 + 10 ng/ml of IFN-γ: 36 ± 7 pg/ml, n = 5; P < 0·001) (Fig. 5b). The influence of IFN-γ pretreatment was concentration-dependent, with maximal reductions seen following pretreatment of the U937 cells with 10 and 100 ng/ml of IFN-γ (Fig. 5b).
Figure 5.
Treatment with interferon-γ (IFN-γ) prevents subsequent interleukin (IL)-4-induced CCL26 messenger RNA (mRNA) and protein expression in U937 cells. U937 cells were cultured for 48 hr in medium alone (0) or in medium containing 0·1–100 ng/ml of IFN-γ. The U937 cells were then washed and incubated with fresh serum-free medium, either alone or containing IL-4 (1 or 10 ng/ml), for 24 hr. (a) RNA was isolated from the cells using TRIzol. The expression of mRNAs for CCL26 and β-actin was evaluated using the reverse transcription–polymerase chain reaction (RT-PCR). PCR products were visualized by gel electrophoresis in the presence of ethidium bromide. Data are representative of the results obtained from at least three independent experiments. (b) Supernatants were collected and CCL26 protein was measured using enzyme-linked immunosorbent assay (ELISA). Data are expressed as mean ± standard error of the mean (SEM) of at least three independent experiments. ***P < 0·001 compared to the Control (0) with no IL-4; ns, not significant.
Pretreatment with IFN-γ decreases total STAT6 expression levels in U937 cells
To determine whether the IFN-γ pretreatment affected IL-4-induced STAT6 phosphorylation in monocytic cells, U937 cells were cultured in the presence of medium alone, or in medium containing IFN-γ, for 24 and 48 hr. The cells were then stimulated with IL-4, either alone or with IFN-γ, for 10 min. Whole-cell lysates were immediately harvested and Western blotted for phosphorylated STAT6, total STAT6 and β-actin. As expected, IL-4 alone induced robust phosphorylation of STAT6 (Fig. 6a). Pretreatment of U937 cells with IFN-γ for 48 hr before stimulation with IL-4 blocked phosphorylation of STAT6 (Fig. 6a). A 24-hour pretreatment with IFN-γ also decreased IL-4-induced STAT6 phosphorylation, but to a lesser extent (Fig. 6a). When total STAT6 levels were examined, we found that total STAT6 was decreased following IFN-γ pretreatments (Fig. 6a). This decline in total STAT6 was not caused by global changes in protein levels, because β-actin expression was not significantly affected by IFN-γ pretreatment (Fig. 6a). Densitometry revealed a significant decrease in total STAT6 protein levels following 24 and 48 hr of treatment with IFN-γ (Fig. 6b). The decrease in total STAT6 mirrored the decrease we observed in phosphorylated STAT6, suggesting that the reduction in phosphorylated STAT6 was, in part, related to a decrease in total STAT6 protein. These data suggest that pretreatment with IFN-γ decreases STAT6 protein levels, thus inhibiting IL-4-induced CCL26 expression in U937 cells.
Figure 6.
Treatment with interferon-γ (IFN-γ) decreases total signal transducer and activator of transcription 6 (STAT6) protein and subsequent interleukin (IL)-4-induced STAT6 phosphorylation in U937 cells. (a) U937 cells were cultured for 24 or 48 hr in medium alone (none) or in medium containing 100 ng/ml of IFN-γ. U937 cells were then washed and incubated with fresh serum-free medium alone or with medium containing IL-4 (10 ng/ml) or IL-4 and IFN-γ for 10 min (co-inc). Cell lysates were harvested in 2 × Laemelli sample buffer and analysed by Western blotting for phosphorylated STAT6 (P-STAT6), total STAT6 (T-STAT6) and β-actin. (a) The blots shown are representative of the results obtained in three independent experiments. (b) Densitometry was used to measure the levels of total STAT6 and β-actin, and the values were expressed as a percentage of the density levels of each respective protein present in U937 cells cultured for 48 hr in the presence of medium alone (none) ± standard error of the mean (SEM) from four independent experiments. **P < 0·01 compared to cells cultured for 24 hr in the presence of medium alone.
Discussion
CCL26 may play an important role in several human diseases including eosinophilic oesophagitis, atopic dermatitis and asthma.17–20 Furthermore, single nucleotide polymorphism (SNP) analysis has revealed that polymorphisms in CCL26 are associated with increased susceptibility to these diseases as well as to rhinitis and rheumatoid arthritis.19–23 Also, low CCL26 levels in the peripheral blood have been shown be an independent indicator of future mortality and morbidity in patients with established coronary artery disease.24 These chronic diseases are often associated with monocyte and/or macrophage activation; thus, understanding the mechanisms that regulate CCL26 expression and function in monocytic cells may provide new insights into these conditions.
The results of this study showed that human peripheral blood monocytes, MDMs and U937 cells are capable of expressing CCL26 mRNA and protein following stimulation with the T helper 2 (Th2) cytokine, IL-4. The studies that originally characterized CCL26 stated that CCL26 mRNA was not detected in peripheral blood leucocytes.3,25 Our data are consistent with these studies, as CCL26 mRNA was only detected in primary human monocytic cells following stimulation with IL-4. CCL26 mRNA expression was rapidly upregulated in U937 cells, monocytes and MDMs following stimulation with IL-4. This time course is consistent with the reported kinetics of IL-4-induced CCL26 mRNA expression in other cell types, such as lung and intestinal epithelial cells,26,27 where mRNA is detected early and is sustained for at least 48 hr. U937 cells, monocytes and MDMs also expressed significant amounts of CCL26 protein. Our findings are further supported by a recent study examining the effects of hypoxia on immature dentritic cells. In this study, peripheral blood monocytes were treated with IL-4 and granulocyte–macrophage colony-stimulating factor (GM-CSF) for 72 hr to induce an immature dentritic cell phenotype. Under these conditions, CCL26 mRNA and protein levels were elevated to levels similar to this study.28
Pro-inflammatory cytokines, such as TNF-α, IL-1β and IFN-γ, are released in the early stages of allergic inflammation. The CCL11 promoter contains binding sites for many transcription factors, including specificity protein-1 (SP1), nuclear factor-κB (NF-κB), activator protein 1 (AP-1), CCAAT/enhancer binding protein and STAT6.11,29 As a result, CCL11 expression can be regulated by TNF-α via NF-κB and by IL-4 via STAT6.30 In contrast, the CCL26 promoter only contains a single STAT6-binding motif located upstream of the transcription initiation site;10 hence, as shown in Figs 1 and 2, neither TNF-α nor IL-1β alone were able to induce significant gene expression or protein synthesis of CCL26 in monocytic cells. Furthermore, TNF-α did not alter IL-4-mediated STAT6 activation. Despite this, TNF-α and IL-1β synergized with IL-4 to increase CCL26 protein expression in U937 cells (Fig. 4b). This occurred with only a modest increase in CCL26 mRNA, suggesting that the synergistic effect could have occurred following transcription. There is precedent for this in the eotaxin family, as shown by data in human epithelial cells where TNF-α and IL-4 regulate CCL11 expression both at the level of transcription as well as during translation by increasing mRNA stability.31 The time course for CCL26 expression also suggests that CCL26 may be regulated at the level of translation. Peak mRNA transcription occurred as early as 1 hr following stimulation, yet protein levels did not reach maximal levels until 48 hr. Future studies will explore the role of translational regulation in CCL26 expression in monocytic cells.
Modulation of CCL26 expression by IFN-γ was very different from that observed with TNF-α and IL-1β. IFN-γ had no effect on CCL26 expression when introduced simultaneously with IL-4, but had a profound effect on both mRNA and protein levels if cells were pre-exposed to IFN-γ before stimulation with IL-4. This is in part because of decreased expression and phosphorylation of STAT6. Previous studies of the effect of IFN-γ on IL-4/STAT6 signal transduction in human monocytes suggested that there are several possible mechanisms by which IFN-γ could inhibit the IL-4-activated STAT6 pathway, such as the downregulation of IL-4R receptors on the cell surface, inhibition of Janus kinase (JAK), induction of phosphatases and the degradation of STAT proteins.32–34 Our data show that pretreatment with IFN-γ for 48 hr decreased the expression of CCL26 mRNA, and had an even more pronounced effect on protein expression. This correlates with the results of Heller et al.,35 who showed that IL-4-induced CCL26 protein production in epithelial cells is fivefold more sensitive to IFN-γ pretreatment than mRNA expression. The decrease in CCL26 protein and gene expression in U937 cells pretreated for 48 hr with IFN-γ before IL-4 stimulation (Fig. 5) correlated with a reduction in both phosphorylated and total STAT6 protein (Fig. 6). This differs in part from the mechanism used by IFN-γ in epithelial cells where IFN-γ decreased phosphorylated STAT6, but did not affect total cytoplasmic STAT6 levels.35 These results suggest that there is a complex communication between the Th2 cytokine, IL-4, and pro-inflammatory cytokines such as TNF-α, IL-1β and IFN-γ in the regulation of CCL26 synthesis in monocytic cells.
Acknowledgments
We thank Evelyn Lailey and Claudia Silva for their excellent technical assistance; the Canadian Foundation for Innovation for providing key infrastructure. Dr K. D. Patel and Dr C. Power are both Canada Research Chairs. KDP is an Alberta Heritage Foundation for Medical Research Scientist and CP is an AHFMR Senior Scholar. Dr V.E.L. Stubbs is supported by fellowships from the Alzheimer Society of Canada, the CIHR Institute of Aging and the CIHR Strategic Training Program. This research was supported by grants from the Heart and Stroke Foundation and the CIHR.
Glossary
Abbreviations:
- CCR3
CC-chemokine receptor 3
- cDNA
complementary DNA
- ELISA
enzyme-linked immunosorbent assay
- FBS
fetal bovine serum
- HBSS
Hanks’ balanced salt solution
- IFN-γ
interferon-γ
- IL-1β
interleukin-1β
- IL-4
interleukin-4
- MDMs
monocyte-derived macrophages
- mRNA
messenger RNA
- NF-κB
nuclear factor-κB
- PCR
polymerase chain reaction
- rRNA
ribosomal RNA
- STAT6
signal transducer and activation of transcription 6
- Th2
T helper 2
- TNF-α
tumour necrosis factor-α
Disclosures
None.
References
- 1.Forssmann U, Uguccioni M, Loetscher P, Dahinden CA, Langen H, Thelen M, Baggiolini M. Eotaxin-2, a novel CC chemokine that is selective for the chemokine receptor CCR3, and acts like eotaxin on human eosinophil and basophil leukocytes. J Exp Med. 1997;185:2171–6. doi: 10.1084/jem.185.12.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffiths-Johnson DA, Collins PD, Rossi AG, Jose PJ, Williams TJ. The chemokine, eotaxin, activates guinea-pig eosinophils in vitro and causes their accumulation into the lung in vivo. Biochem Biophys Res Commun. 1993;197:1167–72. doi: 10.1006/bbrc.1993.2599. [DOI] [PubMed] [Google Scholar]
- 3.Shinkai A, Yoshisue H, Koike M, et al. A novel human CC chemokine, eotaxin-3, which is expressed in IL-4- stimulated vascular endothelial cells, exhibits potent activity toward eosinophils. J Immunol. 1999;163:1602–10. [PubMed] [Google Scholar]
- 4.De Lucca GV. Recent developments in CCR3 antagonists. Curr Opin Drug Discov Devel. 2006;9:516–24. [PubMed] [Google Scholar]
- 5.Ponath PD, Qin S, Ringler DJ, et al. Cloning of the human eosinophil chemoattractant, eotaxin. Expression, receptor binding, and functional properties suggest a mechanism for the selective recruitment of eosinophils. J Clin Invest. 1996;97:604–12. doi: 10.1172/JCI118456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dulkys Y, Schramm G, Kimmig D, Knoss S, Weyergraf A, Kapp A, Elsner J. Detection of mRNA for eotaxin-2 and eotaxin-3 in human dermal fibroblasts and their distinct activation profile on human eosinophils. J Invest Dermatol. 2001;116:498–505. doi: 10.1046/j.1523-1747.2001.01299.x. [DOI] [PubMed] [Google Scholar]
- 7.Lezcano-Meza D, Davila-Davila B, Vega-Miranda A, Negrete-Garcia MC, Teran LM. Interleukin (IL)-4 and to a lesser extent either IL-13 or interferon-gamma regulate the production of eotaxin-2/CCL24 in nasal polyps. Allergy. 2003;58:1011–7. doi: 10.1034/j.1398-9995.2003.00174.x. [DOI] [PubMed] [Google Scholar]
- 8.Heiman AS, Abonyo BO, Darling-Reed SF, Alexander MS. Cytokine-stimulated human lung alveolar epithelial cells release eotaxin-2 (CCL24) and eotaxin-3 (CCL26) J Interferon Cytokine Res. 2005;25:82–91. doi: 10.1089/jir.2005.25.82. [DOI] [PubMed] [Google Scholar]
- 9.Igawa K, Satoh T, Hirashima M, Yokozeki H. Regulatory mechanisms of galectin-9 and eotaxin-3 synthesis in epidermal keratinocytes: possible involvement of galectin-9 in dermal eosinophilia of Th1-polarized skin inflammation. Allergy. 2006;61:1385–91. doi: 10.1111/j.1398-9995.2006.01130.x. [DOI] [PubMed] [Google Scholar]
- 10.Hoeck J, Woisetschlager M. Activation of eotaxin-3/ccl26 gene expression in human dermal fibroblasts is mediated by stat6. J Immunol. 2001;167:3216–22. doi: 10.4049/jimmunol.167.6.3216. [DOI] [PubMed] [Google Scholar]
- 11.Matsukura S, Stellato C, Plitt JR, Bickel C, Miura K, Georas SN, Casolaro V, Schleimer RP. Activation of eotaxin gene transcription by NF-kappa B and STAT6 in human airway epithelial cells. J Immunol. 1999;163:6876–83. [PubMed] [Google Scholar]
- 12.Ogilvie P, Paoletti S, Clark-Lewis I, Uguccioni M. Eotaxin-3 is a natural antagonist for CCR2 and exerts a repulsive effect on human monocytes. Blood. 2003;102:789–94. doi: 10.1182/blood-2002-09-2773. Epub 2003 Apr 10. [DOI] [PubMed] [Google Scholar]
- 13.Petkovic V, Moghini C, Paoletti S, Uguccioni M, Gerber B. Eotaxin-3/CCL26 is a natural antagonist for CC chemokine receptors 1 and 5. A human chemokine with a regulatory role. J Biol Chem. 2004;279:23357–63. doi: 10.1074/jbc.M309283200. [DOI] [PubMed] [Google Scholar]
- 14.Jones G, Zhu Y, Silva C, Tsutsui S, Pardo CA, Keppler OT, McArthur JC, Power C. Peripheral nerve-derived HIV-1 is predominantly CCR5-dependent and causes neuronal degeneration and neuroinflammation. Virol. 2005;334:178–93. doi: 10.1016/j.virol.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 15.Cuvelier SL, Patel KD. Shear-dependent eosinophil transmigration on interleukin 4-stimulated endothelial cells: a role for endothelium-associated eotaxin-3. J Exp Med. 2001;194:1699–709. doi: 10.1084/jem.194.12.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto S, Kobayashi I, Tsuji K, et al. Upregulation of interleukin-4 receptor by interferon-gamma: enhanced interleukin-4-induced eotaxin-3 production in airway epithelium. Am J Respir Cell Mol Biol. 2004;31:456–62. doi: 10.1165/rcmb.2004-0128OC. [DOI] [PubMed] [Google Scholar]
- 17.DeBrosse CW, Rothenberg ME. Allergy and eosinophil-associated gastrointestinal disorders (EGID) Curr Opin Immunol. 2008;20:703–8. doi: 10.1016/j.coi.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berkman N, Ohnona S, Chung FK, Breuer R. Eotaxin-3 but not eotaxin gene expression is upregulated in asthmatics 24 hours after allergen challenge. Am J Respir Cell Mol Biol. 2001;24:682–7. doi: 10.1165/ajrcmb.24.6.4301. [DOI] [PubMed] [Google Scholar]
- 19.Blanchard C, Wang N, Stringer KF, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–47. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kagami S, Kakinuma T, Saeki H, et al. Significant elevation of serum levels of eotaxin-3/CCL26, but not of eotaxin-2/CCL24, in patients with atopic dermatitis: serum eotaxin-3/CCL26 levels reflect the disease activity of atopic dermatitis. Clin Exp Immunol. 2003;134:309–13. doi: 10.1046/j.1365-2249.2003.02273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chae SC, Lee YC, Park YR, et al. Analysis of the polymorphisms in eotaxin gene family and their association with asthma, IgE, and eosinophil. Biochem Biophys Res Commun. 2004;320:131–7. doi: 10.1016/j.bbrc.2004.05.136. [DOI] [PubMed] [Google Scholar]
- 22.Chae SC, Park YR, Oh GJ, Lee JH, Chung HT. The suggestive association of eotaxin-2 and eotaxin-3 gene polymorphisms in Korean population with allergic rhinitis. Immunogenetics. 2005;56:760–4. doi: 10.1007/s00251-004-0746-2. [DOI] [PubMed] [Google Scholar]
- 23.Chae SC, Park YR, Shim SC, Lee IK, Chung HT. Eotaxin-3 gene polymorphisms are associated with rheumatoid arthritis in a Korean population. Hum Immunol. 2005;66:314–20. doi: 10.1016/j.humimm.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Falcone C, Minoretti P, D’Angelo A, et al. Markers of eosinophilic inflammation and risk prediction in patients with coronary artery disease. Eur J Clin Invest. 2006;36:211–7. doi: 10.1111/j.1365-2362.2006.01624.x. [DOI] [PubMed] [Google Scholar]
- 25.Kitaura M, Suzuki N, Imai T, et al. Molecular cloning of a novel human CC chemokine (Eotaxin-3) that is a functional ligand of CC chemokine receptor 3. J Biol Chem. 1999;274:27975–80. doi: 10.1074/jbc.274.39.27975. [DOI] [PubMed] [Google Scholar]
- 26.Banwell ME, Tolley NS, Williams TJ, Mitchell TJ. Regulation of human eotaxin-3/ccl26 expression: modulation by cytokines and glucocorticoids. Cytokine. 2002;17:317–23. doi: 10.1006/cyto.2002.1021. [DOI] [PubMed] [Google Scholar]
- 27.Blanchard C, Durual S, Estienne M, Emami S, Vasseur S, Cuber JC. Eotaxin-3/CCL26 gene expression in intestinal epithelial cells is up-regulated by interleukin-4 and interleukin-13 via the signal transducer and activator of transcription 6. Int J Biochem Cell Biol. 2005;37:2559–73. doi: 10.1016/j.biocel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 28.Ricciardi A, Elia AR, Cappello P, et al. Transcriptome of hypoxic immature dendritic cells: modulation of chemokine/receptor expression. Mol Cancer Res. 2008;6:175–85. doi: 10.1158/1541-7786.MCR-07-0391. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Zepeda EA, Rothenberg ME, Weremowicz S, Sarafi MN, Morton CC, Luster AD. Genomic organization, complete sequence, and chromosomal location of the gene for human eotaxin (SCYA11), an eosinophil-specific CC chemokine. Genomics. 1997;41:471–6. doi: 10.1006/geno.1997.4656. [DOI] [PubMed] [Google Scholar]
- 30.Hoeck J, Woisetschlager M. STAT6 mediates eotaxin-1 expression in IL-4 or TNF-alpha-induced fibroblasts. J Immunol. 2001;166:4507–15. doi: 10.4049/jimmunol.166.7.4507. [DOI] [PubMed] [Google Scholar]
- 31.Atasoy U, Curry SL, Lopez de Silanes I, Shyu AB, Casolaro V, Gorospe M, Stellato C. Regulation of eotaxin gene expression by TNF-alpha and IL-4 through mRNA stabilization: involvement of the RNA-binding protein HuR. J Immunol. 2003;171:4369–78. doi: 10.4049/jimmunol.171.8.4369. [DOI] [PubMed] [Google Scholar]
- 32.Bonder CS, Davies KV, Hosszu EK, Finlay-Jones JJ, Hart PH. IFN-gamma downregulates interleukin-4 functional activity on monocytes by multiple mechanisms. J Interferon Cytokine Res. 2002;22:287–93. doi: 10.1089/107999002753675703. [DOI] [PubMed] [Google Scholar]
- 33.Dickensheets HL, Venkataraman C, Schindler U, Donnelly RP. Interferons inhibit activation of STAT6 by interleukin 4 in human monocytes by inducing SOCS-1 gene expression. Proc Natl Acad Sci U S A. 1999;96:10800–5. doi: 10.1073/pnas.96.19.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.So EY, Park HH, Lee CE. IFN-gamma and IFN-alpha posttranscriptionally down-regulate the IL-4-induced IL-4 receptor gene expression. J Immunol. 2000;165:5472–9. doi: 10.4049/jimmunol.165.10.5472. [DOI] [PubMed] [Google Scholar]
- 35.Heller NM, Matsukura S, Georas SN, Boothby MR, Rothman PB, Stellato C, Schleimer RP. Interferon-{gamma} Inhibits STAT6 signal transduction and gene expression in human airway epithelial cells. Am J Respir Cell Mol Biol. 2004;31:573–82. doi: 10.1165/rcmb.2004-0195OC. [DOI] [PubMed] [Google Scholar]