Abstract
FcαR (CD89) plays important roles in immunoglobulin A (IgA)-mediated immune responses. Soluble forms of FcαR (sFcαR) are found in the culture supernatants of FcαR-expressing cells, in human serum and in the serum of FcαR transgenic mice, and have been suggested to be produced through a proteolytic process. However, little is known about the mechanism involved in the proteolytic release of sFcαR. In this study, we investigated the shedding mechanism of FcαR and determined the nature of the proteinase involved in FcαR shedding. In chemical inhibitor assays, shedding of FcαR was dramatically inhibited by EDTA, EGTA and a broad-spectrum metalloproteinase inhibitor, GM6001, suggesting that a metalloproteinase was responsible for FcαR shedding. Overexpression of dominant-negative mutants of ADAM (a disintegrin and metalloproteinase) 10 and ADAM17 markedly inhibited the production of sFcαR. Finally, knockdown of both endogenous ADAM10 and endogenous ADAM17 inhibited FcαR shedding, demonstrating that ADAM10 and ADAM17 were involved in the shedding of FcαR. The characterization of ADAM10 and ADAM17 as sFcαR-releasing enzymes provides a novel insight into the molecular mechanism of sFcαR production and will help in further elucidation of the physiological and pathological roles of sFcαR.
Keywords: ADAM10, ADAM17, FcαR, IgA nephropathy, shedding
Introduction
FcαR (CD89), the Fc receptor for immunoglobulin A (IgA), is a heterogeneously glycosylated type I transmembrane protein expressed on myeloid cells, including neutrophils, monocytes/macrophages, eosinophils and subpopulations of dendritic cells.1–5 FcαR plays crucial roles in IgA-mediated immune responses, including endocytosis, phagocytosis, antibody-dependent cell cytotoxicity, superoxide generation and the release of cytokines and inflammatory mediators.5
In addition to being expressed at the cell surface as a transmembrane receptor, two types of soluble FcαR (sFcαR) proteins have been described. One is a slightly glycosylated 30 000 molecular weight (MW) protein, with a 25 000 MW backbone, that is found in the culture supernatants of stimulated U937 and THP-1 cells and in human serum. This type of sFcαR was reported to be covalently associated with polymeric IgA and to circulate in the serum of both normal individuals and IgA nephropathy (IgAN) patients.6,7 Another sFcαR is a heavily glycosylated 50 000–70 000 MW molecule with a 24 000 MW backbone. This type of sFcαR was only found in serum from IgAN patients, and has been proposed to be responsible for the decreased expression of FcαR on monocytes from IgAN patients.8–10 sFcαR has also been detected in the serum of FcαR transgenic mice, which developed symptoms of IgAN spontaneously. It has been demonstrated that FcαR is cleaved from monocytes/macrophages of FcαR transgenic mice and forms sFcαR–IgA complexes, which play a central role in the pathogenesis of IgAN in these mice.8,11
Proteolytic release of membrane proteins from the cell surface, also termed ectodomain shedding, has been observed for many membrane proteins, including cytokines, growth factors, adhesion molecules and their receptors.12–16 Ectodomain shedding results in (i) adaptation of the cell phenotype by reducing the amount of surface-expressed membrane protein and (ii) the release of soluble ectodomains of membrane proteins that are capable of acting on other cells. The ADAMs family of metalloproteases comprises membrane-anchored metalloproteases that belong to the zinc protease superfamily and which have been implicated in the ectodomain shedding of many membrane receptors.17 Tumour necrosis factor-α (TNF-α)-converting enzyme (TACE or ADAM17) is the first member of this protease family for which a role in ectodomain shedding of TNF-α has been found.18,19 Of more than 30 ADAMs that have been identified, ADAM10 has been found to be highly homologous to ADAM17. Recently, the closely related ADAM10 and ADAM17 have emerged as main sheddases for many membrane receptors.
Understanding how FcαR is shed from the cell membrane and identifying an enzyme that is responsible for the shedding will help us to understand the physiological and pathological functions of sFcαR. In this study, we report, for the first time, that the ADAMs family members ADAM10 and ADAM17 are both involved in the shedding of FcαR.
Material and methods
Antibody and reagents
The murine monoclonal anti-FcαR IgG MIP7c, 8a, 10c, 38c, 59c, 65c, 68b and 71a, rabbit anti-FcαR polyclonal IgG, fluorescein isothiocyanate (FITC)-labelled F(ab′)2 fragments of MIP8a, and horseradish peroxidase (HRP)-labelled MIP7c, 8a, 38c, 59c and 65c, were prepared as described previously.20,21 Goat polyclonal anti-ADAM10 (A10) and anti-ADAM17 (C15) IgG were from Santa Cruz Biotechnology (Santa Cruz, CA). Phorbol 12-myristate 13-acetate (PMA) and ionomycin were from Sigma-Aldrich (St Louis, MO). GM6001 was from Millipore (Billerica, MA). N-glycosidase F was from NEB (Ipswich, MA).
Enzyme-linked immunosorbent assay for sFcαR
sFcαR was detected using a previously described sandwich enzyme-linked immunosorbent assay (ELISA), with a few modifications.21 In brief, plates were coated with mixed monoclonal antibodies (mAbs) against the EC1 domain of FcαR (MIP10c, 68b, 71a), overnight at 4°, and blocked with 2% bovine serum albumin (BSA)/phosphate-buffered saline (PBS) for 1 hr at room temperature. sFcαR-containing culture supernatant was incubated for 2 hr at 37°. Bound sFcαR was detected using a mixture of HRP-labelled mAbs against the EC2 domain of FcαR (HRP-MIP7c, 8a, 38c, 59c and 65c) and developed using the peroxidase substrate system (ABTS; Roche, South San Francisco, CA). The absorbance was measured at 405 nm. Samples were analyzed in triplicate. The recombinant extracellular domain of FcαR, expressed by Chinese hamster ovary (CHO) cells, was purified and used to establish a standard curve to quantify sFcαR.20 The cut-off for this detection system was 0·05 ng/ml and the absorbance values were proportional to the concentration of sFcαR over the range of 0·05–100 ng/ml.
Plasmids
The complementary DNA (cDNA) of FcαR was cloned from human peripheral white blood cells using the reverse transcription–polymerase chain reaction (RT-PCR) and then inserted into the BamHI and XhoI sites of the pcDNA 3.1 vector (Invitrogen, Carlsbad, CA). Flag-tagged mADAM8ΔMP (amino acids 228–397 deleted), mADAM10ΔMP (amino acids 56–457 deleted), mADAM17ΔMP (amino acids 189–378 deleted) and mADAM10 E385A in the pQCXIP vector (Clontech, Mountain View, CA) were provided by Zena Werb.22 To knock down endogenous ADAM10 and ADAM17 in U937 cells, a lentiviral short hairpin RNA (shRNA) vector, pLVTHM, was used.23 This vector expresses green fluorescent protein (GFP) under an independent promoter, and therefore transduced cells can be easily identified. The target sequences against human ADAM10 (GACATTTCAACCTACGAAT) and ADAM17 (CACATGTAGAAACACTACT) were from previous publications.24,25
Cell culture and transfection
U937 cells were grown in RPMI-1640 containing 10% fetal bovine serum. HEK293T cells, CHO cells and rat basophilic leukaemia (RBL) cells were grown in Dulbecco’s modified Eagle’s minimal essential medium (DMEM) containing 10% fetal bovine serum and antibiotics, at 37° in a humidified atmosphere of 5% CO2. CHO cells were transfected with plasmids encoding the FcαR using Lipofectamine™ 2000 (Invitrogen). Stably transfected CHO cells were selected by growth in 800 μg/ml of G418 and evaluated for receptor expression by flow cytometry. RBL cells stably expressing FcαR were established by lentiviral transduction. FcαR was inserted in a lentiviral vector (pWPXL) between BamHI and MluI. Virus produced in 293T cells was used to infect RBL cells. Stable RBL cells expressing FcαR were examined by flow cytometry.
Lentiviral transduction
Stable cell lines expressing shRNA against human ADAM10 and ADAM17 were produced by lentiviral transduction using the packaging vector psPAX2 and the envelope vector pMD2.G. pLVTHM-scramble shRNA, pLVTHM-shADAM10 and pLVTHM-shADAM17 were co-transfected with psPAX2 and pMD2.G into 293T cells using Lipofectamine™ 2000. Then, U937 cells were infected with the supernatants from packing cells supplemented with 8 μg/ml of Polybrene. After three infections, GFP-positive cells were sorted by flow cytometry. The expression of ADAM10 and ADAM17 in GFP-positive U937 cells was determined using quantitative real-time PCR.
Measurement of FcαR shedding
A total of 2 × 105 U937 cells were seeded in 96-well plates in 0·3 ml of medium. PMA (10 ng/ml) and ionomycin (5 μm) were used to stimulate FcαR shedding. EDTA, EGTA, leupeptin, pepstatin and GM6001 were used at the indicated concentrations. Cells were cultured for 24 hr. The culture medium was harvested by centrifugation (400 g for 5 min and then 16 500 g for 15 min at 4°), after which the sFcαR concentration was determined using ELISA analysis. For FcαR shedding from stable transfectants of CHO and RBL cells, 2 × 105 cells were plated in 24-well plates with 0·45 ml of medium (or with medium containing the indicated protease inhibitors) and cultured for 24 hr. For FcαR shedding from 293T cells, 2 × 105 293T cells were seeded into a 24-well plate and allowed to grow overnight. Cells were co-transfected with FcαR and ADAM8ΔMP, ADAM10ΔMP, ADAM10E385A or ADAM17ΔMP. The medium was replaced with culture medium 24 hr after transfection. The medium was harvested between 24 and 48 hr post-transfection and the concentration of sFcαR was determined using an ELISA.
Western blot
A total of 1 × 107 U937 cells were stimulated with 10 ng/ml of PMA for 24 hr and the supernatants were collected by centrifugation. Cells were lysed with RIPA buffer [20 mM Tris (pH 7·5), 150 mM NaCl, 1% Nonidet P-40, 0·5% sodium deoxycholate, 1 mm EDTA, 0·1% SDS] supplemented with 0·5 mm phenylmethylsulphonyl fluoride (PMSF), 1 μg/ml of aprotinin, 1 μg/ml of leupeptin and 1 μg/ml of pepstatin, and the lysates were clarified by centrifugation (16 500 g, 15 min, 4°). sFcαR in the supernatants and FcαR in the cell lysates were purified with MIP8a-coupled beads by rotation at 4° overnight. Purified protein was treated with N-glycosidase F and separated by electrophoresis on a 10% sodium dodecyl sulphate–polyacrylamide gel under reducing conditions, then transferred onto a nitrocellulose membrane. FcαR was detected by rabbit anti-FcαR polyclonal IgG.
Quantitative real-time PCR
Total RNA was extracted with Trizol reagent, used according to the manufacturer’s instructions (TransGen BioTec, Beijing, China). Total RNA (1 μg) from each sample was used for first-strand cDNA synthesis with EasyScript First-Strand cDNA Synthesis SuperMix (TransGen, Beijing, China). Real-time PCR was performed on a Rotor-Gene 3000 cycler to determine the expression of ADAM10, ADAM17 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) using the SYBR Premix Ex Taq™ II kit (Takara, Shiga, Japan). The following experimental protocol for PCR reaction (40 cycles) was performed: denaturation for 5 min at 95°, followed by 40 amplification cycles at 95° for 10 seconds, 60° for 15 seconds and 72° for 15 seconds. The specific primers for real-time PCR were: ADAM10 forward, 5′-AATGGATTGTGGCTCATTGGTGGG-3′ and reverse, 5′-TGGAAGTGGTTTAGGAGGAGGCAA-3′; ADAM17 forward, 5′-AGTGCAGTGACAGGAACAGTCCTT-3′ and reverse, 5′-GGACACGCCTTTGCAAGTAGCATT-3′; and GAPDH forward, 5′-GCTCACTGGCATGGCCTTCCG-3′ and reverse, 5′-GTGGGCCATGAGGTCCACCAC-3′. The expression level for each gene was expressed as a ratio relative to GAPDH.
Statistical analysis
Data are expressed as the mean ± standard error of the mean (SEM). Statistical significance was calculated using the two-tailed unpaired t-test. Differences were considered significant when P<0·05.
Results
PMA-stimulated shedding of FcαR from U937 cells
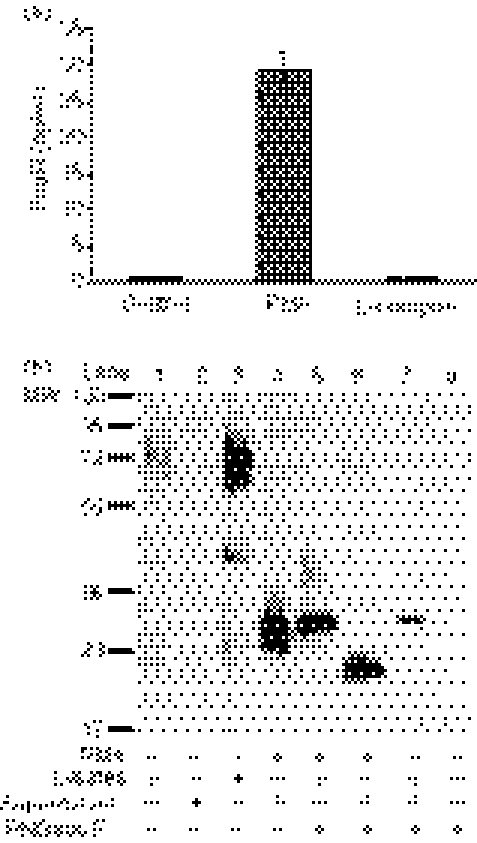
U937 is a monocytic cell line that naturally expresses FcαR, and the results of a previous study showed that sFcαR could be detected in the supernatants of U937 cells after stimulation.6 Therefore, we first examined FcαR shedding from U937 cells after stimulation with the protein kinase C activator PMA and the Ca2+ ionophore ionomycin, two common chemical stimulators of receptor shedding.26,27 As shown in Fig. 1(a), PMA, but not ionomycin, induced FcαR shedding from U937 cells.
Figure 1.
Phorbol 12-myristate 13-acetate (PMA)-stimulated shedding of FcαR from U937 cells. (a) Detection of soluble FcαR (sFcαR) in the supernatant of U937 cells. A total of 2 × 105 U937 cells were cultured in the presence of PMA (10 ng/ml) or ionomycin (5 μm) for 24 hr, after which the concentration of sFcαR in the supernatants was determined using enzyme-linked immunosorbent assay (ELISA) analysis. The control was the equivalent concentration of solvent (dimethylsulphoxide) for PMA and ionomycin added to the culture medium. Data are the mean ± standard error of the mean (SEM) from three independent experiments. (b) Biochemical analysis of sFcαR from U937 cells. A total of 1 × 107 U937 cells were stimulated with 10 ng/ml of PMA for 24 hr. FcαR in the cell lysate and sFcαR in the supernatant were purified using MIP8a-coupled beads and treated with N-glycosidase F. Proteins were separated by electrophoresis on 10% sodium dodecyl sulphate–polyacrylamide, transferred onto nitrocellulose membranes and detected using rabbit anti-FcαR polyclonal IgG. The result is representative of five independent experiments. PGNase F, N-glycosidase F.
To further characterize sFcαR shedding from PMA-stimulated U937 cells, sFcαR was affinity purified by MIP8a-coupled beads and analyzed by Western blotting. As shown in Fig. 1(b), U937 cells expressed a low level of FcαR with a MW of 55 000–75 000 (lane 1), and no sFcαR could be detected in the absence of stimulation with PMA (lane 2). PMA treatment greatly enhanced FcαR expression in U937 cells (lane 3), and sFcαR shed from PMA-treated U937 cells was a heterogeneous glycosylated protein with a MW ranging from 28 000 to 36 000 (lane 4). Treatment of sFcαR with N-glycosidase F revealed a backbone of 25 000 MW (lane 6), whereas full-length FcαR had a backbone of 32 000 MW after treatment with N-glycosidase F (lanes 5 and 7).
FcαR shedding is inhibited by metalloproteinase inhibitors
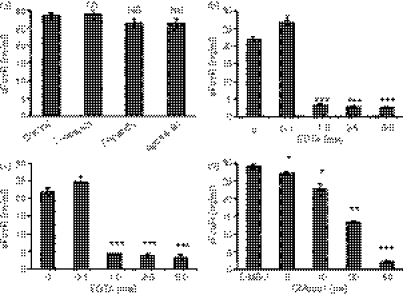
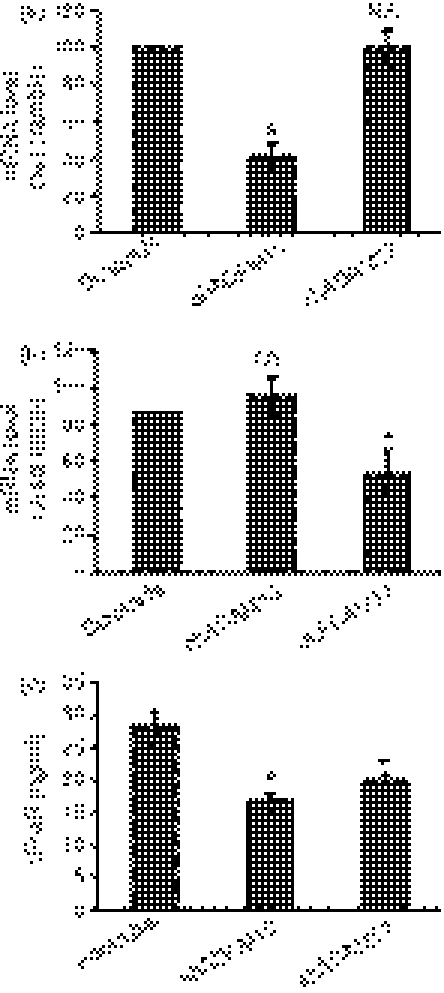
To investigate the mechanism of FcαR shedding, we analyzed the ability of inhibitors of different classes of proteases to inhibit FcαR shedding from U937 cells. Leupeptin is an inhibitor of serine and cysteine proteinases, pepstatin inhibits aspartate proteinase and aprotinin inhibits serine proteinase. EDTA and EGTA are chelators of bivalent metal ions, which can inhibit the activities of metalloproteinase. GM6001 is a broad-spectrum metalloproteinase inhibitor, which inhibits both matrix metalloproteinase (MMPs) and some members of the ADAM family of proteases.28 As shown in Fig. 2(a), inhibitors of serine, cysteine and aspartate proteases had no effect on FcαR shedding. By contrast, EDTA, EGTA and GM6001 inhibited the shedding of FcαR in a dose-dependent manner (Fig. 2), suggesting the involvement of one or more metalloproteinases in FcαR shedding. Treatment of U937 cells with these inhibitors had no effect on cell viability (data not shown) and therefore the reduction in the levels of released sFcαR was caused by the inhibition of sheddase(s), not toxicity. We also observed that FcαR shedding increased slightly when U937 cells were treated with a low concentration (0·1 mm) of EDTA or EGTA; this phenomenon has also been observed in the shedding of other receptors.29,30
Figure 2.
FcαR shedding is inhibited by metalloproteinase inhibitors. (a) A total of 2 × 105 U937 cells were cultured in the presence of phorbol 12-myristate 13-acetate (PMA) (10 ng/ml) and leupeptin (2 μg/ml), pepstatin (2 μg/ml) or aprotinin (2 μg/ml) for 24 hr, and then the concentration of soluble FcαR (sFcαR) in the supernatants was determined using enzyme-linked immunosorbent assay (ELISA) analyses. (b), (c) and (d) A total of 2 × 105 U937 cells were cultured in the presence of PMA (10 ng/ml) and EDTA (b), EGTA (c) or GM6001(d), at the indicated concentrations, for 24 hr, and then the concentration of sFcαR in the supernatants was determined using ELISA. Data are the mean ± standard error of the mean (SEM) from three independent experiments. *P <0·05, **P <0·01, ***P <0·001. NS, not significant.
FcαR shedding from FcαR-transfected CHO and RBL cells
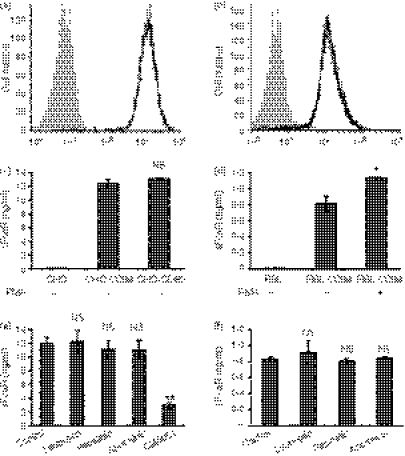
In addition to cells naturally expressing FcαR, we also investigated the shedding of FcαR in two stable transfectants of FcαR, namely CHO cells and RBL cells. The CHO cell line is widely used to study cell-based shedding mechanisms of various membrane receptors, and RBL is a common cell line used to investigate the function of FcαR. The expression of FcαR on stably transfected CHO and RBL cells was examined by flow cytometry analysis (Fig. 3a,b). The FcαR shedding assay showed that both CHO cells and RBL cells were able to shed FcαR spontaneously, and that treatment with PMA could increase the shedding of FcαR from RBL cells, but not from CHO cells (Fig. 3b,c).
Figure 3.
FcαR shedding from stable transfectants of Chinese Hamster ovary (CHO) cells and rat basophilic leukaemia (RBL) cells. (a) and (b) Flow cytometry analysis of FcαR stably expressed on CHO (a) and RBL (b) cells. CHO or RBL cells (filled histogram) and CHO-CD89 or RBL-CD89 cells (black line) were incubated with fluorescein isothiocyanate (FITC)-conjugated MIP8a-F(ab′)2 for 60 min on ice, then washed and analysed using flow cytometry. (c) and (d) FcαR shedding from CHO and RBL cells stably expressing FcαR. A total of 2 × 105 cells were cultured with or without phorbol 12-myristate 13-acetate (PMA) (10 ng/ml) for 24 hr, and the concentration of soluble FcαR (sFcαR) in the supernatants was determined using enzyme-linked immunosorbent assay (ELISA). (e) and (f) CHO-CD89 cells or RBL-CD89 cells were cultured in the presence of leupeptin (2 μg/ml), pepstatin (2 μg/ml), aprotinin (2 μg/ml) or GM6001 (20 μm) for 24 hr. The concentration of sFcαR in the supernatants was determined using enzyme-linked immunosorbent assay (ELISA) analyses. Data are the mean ± standard error of the mean (SEM) of three independent experiments. *P <0·01, **P <0·001. NS, not significant.
Next, protease inhibitors were used to characterize the shedding of FcαR from CHO cells and RBL cells. Because EDTA and EGTA detached CHO cells and RBL cells from the culture plate, and RBL cells were very sensitive to even a very low concentration of dimethylsulphoxide (DMSO; a solvent for GM6001), these inhibitors were not used. As shown in Fig. 3(e,f), inhibitors of serine, cysteine and aspartate proteases had no effect on FcαR shedding from CHO cells and RBL cells, whereas GM6001 dramatically decreased FcαR shedding from CHO cells, indicating that the sheddase(s) responsible for FcαR shedding from CHO cells and RBL cells was/were some member(s) of the metalloproteinases, but not member(s) of other classes of proteases.
ADAM10 and ADAM17 are involved in FcαR shedding from 293T cells
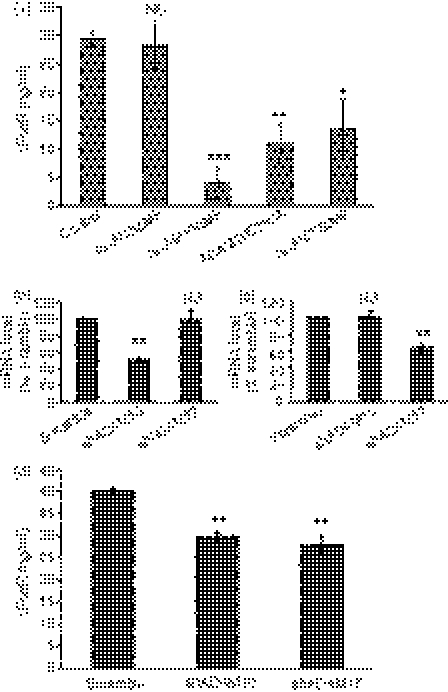
The ADAM family of proteases, especially ADAM10 and ADAM17, are responsible for various types of membrane protein shedding.28 Therefore, we examined whether these sheddases were involved in the shedding of FcαR. Mutant versions of ADAM proteins that lack metalloproteinase domains have been shown to function as dominant-negative mutants that suppress shedding of proteinase substrates.31–33 We examined the effect of dominant-negative mutants of ADAM8 (ADAM8ΔMP), ADAM10 (ADAM10ΔMP and ADAM10E385A) and ADAM17 (ADAM17ΔMP) on the shedding of FcαR in 293T cells via co-transfection experiments. Plasmids coding for FcαR were co-transfected with these dominant-negative mutants of ADAMs, then the concentration of sFcαR in the supernatants of the transfected cells was determined using an ELISA. As shown in Fig. 4(a), FcαR shedding was dramatically decreased when FcαR was co-transfected with ADAM10ΔMP, ADAM10E385A and ADAM17ΔMP, but not with ADAM8ΔMP, suggesting that both ADAM10 and ADAM17 were involved in FcαR shedding, whereas ADAM8 had nothing to do with FcαR shedding.
Figure 4.
ADAM10 and ADAM17 are responsible for FcαR shedding from 293T cells. (a) 293T cells were co-transfected with plasmids coding for FcαR and ADAM8ΔMP, ADAM10ΔMP, ADAM10E385A or ADAM17ΔMP. Supernatants were collected between 24 and 48 hr after transfection for detection of soluble FcαR (sFcαR) using enzyme-linked immunosorbent assay (ELISA). (b) and (c) Quantitive real-time polymerase chain reaction (PCR) analysis of the messenger RNA (mRNA) level in 293T cells transduced with short hairpin RNA (shRNA) against human ADAM10 and ADAM17. (d) FcαR was transfected into ADAM10 and ADAM17 knockdown 293T cells, and supernatants were collected between 24 and 48 hr after transfection for detection of sFcαR using ELISA. Data are the mean ± standard error of the mean (SEM) of three independent experiments. *P <0·05, **P <0·01, ***P <0·001. NS, not significant.
To explore, in further detail, the role of ADAM10 and ADAM17 in FcαR shedding, we established stable 293T cells expressing shRNA against human ADAM10 and ADAM17. The degree of expression of ADAM10 and ADAM17 in stable 293T cells was examined using quantitative real-time PCR. As shown in Fig. 4(b,c), the messenger RNA (mRNA) level of ADAM10 was decreased to about 50% of that of the control without affecting ADAM17 expression, and the mRNA level of ADAM17 was decreased to about 60% of that of the control without affecting ADAM10 expression. Then, FcαR was transfected into these ADAM10 and ADAM17 knockdown cells and the concentration of sFcαR in the supernatants was determined using an ELISA. Figure 4(d) showed that, compared with the scramble shRNA control, FcαR shedding from both ADAM10 and ADAM17 knockdown 293T cells was significantly decreased. These data demonstrated that both ADAM10 and ADAM17 were involved in the shedding of FcαR in 293T cells.
ADAM10 and ADAM17are responsible for FcαR shedding from U937 cells
To confirm whether ADAM10 and ADAM17 are responsible for FcαR shedding in cells that naturally express this receptor, ADAM10 and ADAM17 were knocked down in U937 cells by lentiviral transduction. Transduced (GFP positive) cells were sorted by fluorescence-activated cell sorting (FACS) (the purity was > 95%) and the degree of expression of ADAM10 and ADAM17 was examined using quantitive real-time PCR. Specificity of the knockdown was confirmed using scramble control shRNA and cross-detection of ADAM10 in ADAM17 shRNA cells, and vice versa. As shown in Fig. 5(a,b), the degree of expression of ADAM10 and of ADAM17 decreased in shADAM10 and shADAM17 U937 cells, respectively, without affecting the expression levels of each other. Then, FcαR shedding from ADAM10 and ADAM17 knockdown U937 cells were examined using ELISA. Figure 5(c) showed that shedding of sFcαR decreased both in ADAM10 and ADAM17 knockdown U937 cells, demonstrating that FcαR expressed on U937 cells was cleaved by ADAM10 and ADAM17.
Figure 5.
Depletion of endogenous ADAM10 and ADAM17 by short hairpin RNA (shRNA) reduces FcαR shedding from U937 cells. (a) and (b) Quantitive real-time PCR analysis of the messenger RNA (mRNA) level in U937 cells transduced with shRNA against human ADAM10 and ADAM17. These data are the mean ± standard error of the mean (SEM) of three independent experiments. (c) ADAM10 or ADAM17 knockdown U937 cells, or scramble shRNA U937 cells, were stimulated with 10 ng/ml of phorbol 12-myristate 13-acetate (PMA) for 24 hr, and the supernatants were collected to detect soluble FcαR (sFcαR) via enzyme-linked immunosorbent assay (ELISA). Data are the mean ± standard error of the mean (SEM) of five independent experiments. *P <0·01. NS, not significant.
Discussion
Proteolytic shedding of the ectodomain of membrane receptors is an evolutionarily conserved post-translational modification by which transmembrane molecules are converted into soluble forms. Cleavage and release of membrane proteins is a critical regulatory step in many physiological and pathological processes that allows the cell to adapt to the surface phenotype and generate soluble mediators to act on other cells. sFcαR has been found in the culture supernatant of FcαR-expressing cells, in human serum and in the serum of FcαR transgenic mice, but the identities of the proteinase activities responsible for generating sFcαR remain unknown. The present work is the first report demonstrating that two sheddases, ADAM10 and ADAM17, are both involved in the shedding of the FcαR.
Biochemical analysis of sFcαR shed from U937 cells revealed a 28 000–36 000 MW protein with a 25 000 MW backbone, which is in agreement with a previous report.6 sFcαR found in the serum of IgAN patients was reported to be a 50 000–70 000 MW protein with a 24 000 MW backbone.8 At present, it is not known why the heavily glycosylated sFcαR (50 000–70 000 MW with a 24 000 MW backbone) was only found in serum from IgAN patients, whereas the slightly glycosylated sFcαR (28 000–36 000 MW with a 25 000 MW backbone) can be found both in serum from all individuals and in supernatants of stimulated U937 cells.6–8 It is possible that the slight difference (1000 MW) in the core protein of two sFcαRs was the result of different methods used to calculate the molecular weight and suggests that the difference in these two sFcαRs is mainly caused by different levels of glycosylation.
Receptor shedding can be either constitutive or inductive. Both constitutive and induced shedding of FcαR have been reported previously.6,8 In our study, no detectable sFcαR was found in the supernatant of unstimulated U937 cells, and PMA was found to stimulate FcαR shedding from U937 cells. This result is consistent with a previous report.6 By contrast, FcαR shedding was also observed in this report when U937 cells were treated with ionomycin.6 However, in our study, ionomycin (1–10 μm) was unable to induce FcαR shedding from U937cells. This discrepancy might be a result of different conditions of cells or experimental procedures. We also observed spontaneous shedding of FcαR from transfected 293T cells, CHO cells and RBL cells. Spontaneous shedding of FcαR from transfected RBL cells was also reported previously; the same study also showed that sFcαR could be detected in the culture supernatants of monocytes isolated from IgAN patients but not from normal individuals.8 It remains unknown why FcαR can undergo constitutive shedding from transfected cells and monocytes of IgAN patients but not from cultured U937 cells and monocytes of normal individuals. One possibility is that there might be some unidentified factors in IgAN patients (either monocytes or factors in the serum of IgAN patients) that induce FcαR shedding. Collectively, these data suggest that FcαR shedding from different cells are regulated differentially, indicating that the activities of ADAM10 and/or ADAM17 in different cells might be controlled by different signals, or that the availability of the substrate (FcαR) to the ADAMs may be a regulated event that varies in different cells or under different situations. This cellular behaviour has also been observed with many other ADAM substrates, such as CD44, EGFR and Klotho.26,27,30,34
As an Fc receptor for IgA, FcαR is usually classified as a member of the Fc receptor family. However, the FcαR gene is localized in chromosome 19, whereas all other Fc receptor genes are localized in chromosome 1. Structurally and evolutionarily, FcαR belongs to another family of receptors, the so-called leucocyte receptor cluster.35 Interestingly, shedding of another member of this family, GPVI (a collagen receptor expressed on platelets), is also mediated by ADAM10 and/or ADAM17.36–39 Therefore, shedding of these two closely related receptors are mediated by the same sheddases, suggesting that shedding of other member of this family might also be attributed to these sheddases.
Our results from chemical inhibitor assays, overexpression of dominant-negative mutants of ADAMs and depletion of endogenous ADAM10 and ADAM17 via shRNA, all support both ADAM10 and ADAM17 as cellular sheddases of the FcαR. However, targeted inhibition of endogenous ADAM10 and ADAM17 to inhibit FcαR shedding did not reduce the shedding of FcαR as effectively as that by chemical inhibitors and dominant-negative mutants of ADAM10 and ADAM17. This is mainly because the depletion of ADAM10 and ADAM17 in U937 cells was not complete (about 50% at the mRNA level). Therefore, from our present results, the relative importance of ADAM10 and ADAM17 in FcαR shedding cannot be discriminated. In addition, we cannot rule out the possibility that other non-characterized metalloproteinase(s) might also be involved in the shedding of FcαR, considering that U937 cells express almost every proteolytically active member of the ADAM family.
In this study, we have shown that ADAM10 and ADAM17 are functional sheddases of the FcαR. Although a pathogenic role of sFcαR in human IgAN has not been determined, sFcαR has been demonstrated to play a central role in initiating IgAN in FcαR transgenic mice, which developed IgAN spontaneously.8,10,11 Because ADAMs are evolutionarily conserved sheddases, and the shedding of another Fc receptor (FcεRII, CD23) in both human and mice is mediated by the same sheddase (ADAM10), we speculate that the two sheddases of the FcαR – ADAM10 and ADAM17 – are also involved in the shedding of the FcαR in FcαR transgenic mice.22,40 Therefore, it is tempting and would be interesting to investigate whether inhibiting FcαR shedding can prevent the onset, or alleviate the symptoms, of IgAN in these mice, which will help us to determine the role of sFcαR in the pathogeneses of IgAN.
Acknowledgments
This study was supported by a grant (no. 30872287) from the National Natural Science Foundation of China. We thank Dr Didier Trono for providing the plasmids pWPXL and pLVTHM.
Disclosures
None of the authors of this paper have conflicts of interest to disclose.
References
- 1.Monteiro RC, Cooper MD, Kubagawa H. Molecular heterogeneity of Fcα receptors detected by receptor-specific monoclonal antibodies. J Immunol. 1992;148:1764–70. [PubMed] [Google Scholar]
- 2.Geissmann F, Launay P, Pasquier B, Lepelletier Y, Leborgne M, Lehuen A, Brousse N, Monteiro RC. A subset of human dendritic cells expresses IgA Fc receptor (CD89), which mediates internalization and activation upon cross-linking by IgA complexes. J Immunol. 2001;166:346–52. doi: 10.4049/jimmunol.166.1.346. [DOI] [PubMed] [Google Scholar]
- 3.Hamre R, Farstad IN, Brandtzaeg P, Morton HC. Expression and modulation of the human immunoglobulin A Fc receptor (CD89) and the FcR α chain on myeloid cells in blood and tissue. Scand J Immunol. 2003;57:506–16. doi: 10.1046/j.1365-3083.2003.01220.x. [DOI] [PubMed] [Google Scholar]
- 4.van Egmond M, van Garderen E, van Spriel AB, et al. FcαRI-positive liver Kupffer cells: reappraisal of the function of immunoglobulin A in immunity. Nat Med. 2000;6:680–5. doi: 10.1038/76261. [DOI] [PubMed] [Google Scholar]
- 5.Monteiro RC, van de Winkel JG. IgA Fc receptors. Annu Rev Immunol. 2003;21:177–204. doi: 10.1146/annurev.immunol.21.120601.141011. [DOI] [PubMed] [Google Scholar]
- 6.van Zandbergen G, Westerhuis R, Mohamad NK, van De Winkel JG, Daha MR, van Kooten C. Crosslinking of the human Fc receptor for IgA (FcαRI/CD89) triggers FcR γ-chain-dependent shedding of soluble CD89. J Immunol. 1999;163:5806–12. [PubMed] [Google Scholar]
- 7.van der Boog PJ, van Zandbergen G, de Fijter JW, Klar-Mohamad N, van Seggelen A, Brandtzaeg P, Daha MR, van Kooten C. FcαRI/CD89 circulates in human serum covalently linked to IgA in a polymeric state. J Immunol. 2002;168:1252–8. doi: 10.4049/jimmunol.168.3.1252. [DOI] [PubMed] [Google Scholar]
- 8.Launay P, Grossetête B, Arcos-Fajardo M, et al. Fcα receptor (CD89) mediates the development of immunoglobulin A (IgA) nephropathy (Berger’s disease): evidence for pathogenic soluble receptor-IgA complexes in patients and CD89 transgenic mice. J Exp Med. 2000;191:1999–2009. doi: 10.1084/jem.191.11.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grossetête B, Launay P, Lehuen A, Jungers P, Bach JF, Monteiro RC. Down-regulation of Fcα receptors on blood cells of IgA nephropathy patients: evicence for a negative regulatory role of serum IgA. Kidney Int. 1998;53:1321–35. doi: 10.1046/j.1523-1755.1998.00885.x. [DOI] [PubMed] [Google Scholar]
- 10.Monteiro RC, Moura IC, Launay P, Tsuge T, Haddad E, Benhamou M, Cooper MD, Arcos-Fajardo M. Pathogenic significance of IgA receptor interactions in IgA nephropathy. Trends Mol Med. 2002;8:464–8. doi: 10.1016/s1471-4914(02)02405-x. [DOI] [PubMed] [Google Scholar]
- 11.Kanamaru Y, Arcos-Fajardo M, Moura IC, et al. Fcα receptor I activation induces leukocyte recruitment and promotes aggravation of glomerulonephritis through the FcRγ adaptor. Eur J Immunol. 2007;37:1116–28. doi: 10.1002/eji.200636826. [DOI] [PubMed] [Google Scholar]
- 12.Schlöndorff J, Blobel CP. Metalloprotease-disintegrins: modular proteins capable of promoting cell-cell interactions and triggering signals by protein-ectodomain shedding. J Cell Sci. 1999;112:3603–17. doi: 10.1242/jcs.112.21.3603. [DOI] [PubMed] [Google Scholar]
- 13.Müllberg J, Althoff K, Jostock T, Rose-John S. The importance of shedding of membrane proteins for cytokine biology. Eur Cytokine Netw. 2000;11:27–38. [PubMed] [Google Scholar]
- 14.Herren B. ADAM-mediated shedding and adhesion: a vascular perspective. News Physiol Sci. 2002;17:73–6. doi: 10.1152/nips.01373.2001. [DOI] [PubMed] [Google Scholar]
- 15.Black RA. Tumor necrosis factor-alpha converting enzyme. Int J Biochem Cell Biol. 2002;34:1–5. doi: 10.1016/s1357-2725(01)00097-8. [DOI] [PubMed] [Google Scholar]
- 16.Garton KJ, Gough PJ, Raines EW. Emerging roles for ectodomain shedding in the regulation of inflammatory responses. J Leukoc Biol. 2006;79:1105–16. doi: 10.1189/jlb.0106038. [DOI] [PubMed] [Google Scholar]
- 17.Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- 18.Black RA, Rauch CT, Kozlosky CJ, et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385:729–33. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 19.Moss ML, Jin SL, Milla ME, et al. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-alpha. Nature. 1997;385:733–6. doi: 10.1038/385733a0. [DOI] [PubMed] [Google Scholar]
- 20.Zhang W, Bi B, Oldroyd RG, Lachmann PJ. Neutrophil lactoferrin release induced by IgA immune complexes differed from that induced cross-linking of fcalpha receptors (FcαR) with a monoclonal antibody, MIP8a. Clin Exp Immunol. 2000;121:106–11. doi: 10.1046/j.1365-2249.2000.01254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin N, Peng M, Xing Y, Zhang W. Intracellular pools of FcαR (CD89) in human neutrophils are localized in tertiary granules and secretory vesicles, and two FcαR isoforms are found in tertiary granules. J Leukoc Biol. 2007;82:551–8. doi: 10.1189/jlb.0207112. [DOI] [PubMed] [Google Scholar]
- 22.Lemieux GA, Blumenkron F, Yeung N, et al. The low affinity IgE receptor (CD23) is cleaved by the metalloproteinase ADAM10. J Biol Chem. 2007;282:14836–44. doi: 10.1074/jbc.M608414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–61. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reiss K, Maretzky T, Ludwig A, Tousseyn T, de Strooper B, Hartmann D, Saftig P. ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and beta-catenin nuclear signalling. EMBO J. 2005;24:742–52. doi: 10.1038/sj.emboj.7600548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maretzky T, Schulte M, Ludwig A, et al. L1 is sequentially processed by two differently activated metalloproteases and presenilin/gamma-secretase and regulates neural cell adhesion, cell migration, and neurite outgrowth. Mol Cell Biol. 2005;25:9040–53. doi: 10.1128/MCB.25.20.9040-9053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagano O, Murakami D, Hartmann D, et al. Cell-matrix interaction via CD44 is independently regulated by different metalloproteinases activated in response to extracellular Ca2+ influx and PKC activation. J Cell Biol. 2004;165:893–902. doi: 10.1083/jcb.200310024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horiuchi K, Le Gall S, Schulte M, et al. Substrate selectivity of epidermal growth factor-receptor ligand sheddases and their regulation by phorbol esters and calcium influx. Mol Biol Cell. 2007;18:176–88. doi: 10.1091/mbc.E06-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Overall CM, Blobel CP. In search of partners: linking extracellular proteases to substrates. Nat Rev Mol Cell Biol. 2007;8:245–57. doi: 10.1038/nrm2120. [DOI] [PubMed] [Google Scholar]
- 29.Imura A, Iwano A, Tohyama O, Tsuji Y, Nozaki K, Hashimoto N, Fujimori T, Nabeshima Y. Secreted Klotho protein in sera and CSF: implication for post-translational cleavage in release of Klotho protein from cell membrane. FEBS Lett. 2004;565:143–7. doi: 10.1016/j.febslet.2004.03.090. [DOI] [PubMed] [Google Scholar]
- 30.Chen CD, Podvin S, Gillespie E, Leeman SE, Abraham CR. Insulin stimulates the cleavage and release of the extracellular domain of Klotho by ADAM10 and ADAM17. Proc Natl Acad Sci USA. 2007;104:19796–801. doi: 10.1073/pnas.0709805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solomon KA, Pesti N, Wu G, Newton RC. Cutting edge: a dominant negative form of TNF-alpha converting enzyme inhibits proTNF and TNFRII secretion. J Immunol. 1999;163:4105–8. [PubMed] [Google Scholar]
- 32.Pan D, Rubin GM. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell. 1997;90:271–80. doi: 10.1016/s0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- 33.Schäfer B, Marg B, Gschwind A, Ullrich A. Distinct ADAM metalloproteinases regulate G protein-coupled receptor-induced cell proliferation and survival. J Biol Chem. 2004;279:47929–38. doi: 10.1074/jbc.M400129200. [DOI] [PubMed] [Google Scholar]
- 34.Sahin U, Weskamp G, Kelly K, et al. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J Cell Biol. 2004;164:769–79. doi: 10.1083/jcb.200307137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin AM, Kulski JK, Witt C, Pontarotti P, Christiansen FT. Leukocyte Ig-like receptor complex (LRC) in mice and men. Trends Immunol. 2002;23:81–8. doi: 10.1016/s1471-4906(01)02155-x. [DOI] [PubMed] [Google Scholar]
- 36.Bergmeier W, Rabie T, Strehl A, Piffath CL, Prostredna M, Wagner DD, Nieswandt B. GPVI down-regulation in murine platelets through metalloproteinase-dependent shedding. Thromb Haemost. 2004;91:951–8. doi: 10.1160/TH03-12-0795. [DOI] [PubMed] [Google Scholar]
- 37.Stephens G, Yan Y, Jandrot-Perrus M, Villeval JL, Clemetson KJ, Phillips DR. Platelet activation induces metalloproteinase-dependent GP VI cleavage to down-regulate platelet reactivity to collagen. Blood. 2005;105:186–91. doi: 10.1182/blood-2004-07-2842. [DOI] [PubMed] [Google Scholar]
- 38.Gardiner EE, Karunakaran D, Shen Y, Arthur JF, Andrews RK, Berndt MC. Controlled shedding of platelet glycoprotein (GP)VI and GPIb-IX-V by ADAM family metalloproteinases. J Thromb Haemost. 2007;5:1530–7. doi: 10.1111/j.1538-7836.2007.02590.x. [DOI] [PubMed] [Google Scholar]
- 39.Bergmeier W, Piffath CL, Cheng G, Dole VS, Zhang Y, von Andrian UH, Wagner DD. Tumor necrosis factor-alpha-converting enzyme (ADAM17) mediates GPIbalpha shedding from platelets in vitro and in vivo. Circ Res. 2004;95:677–83. doi: 10.1161/01.RES.0000143899.73453.11. [DOI] [PubMed] [Google Scholar]
- 40.Weskamp G, Ford JW, Sturgill J, et al. ADAM10 is a principal ‘sheddase’ of the low-affinity immunoglobulin E receptor CD23. Nat Immunol. 2006;7:1293–8. doi: 10.1038/ni1399. [DOI] [PubMed] [Google Scholar]