Abstract
The e-NMR project is a European cooperation initiative that aims at providing the bio-NMR user community with a software platform integrating and streamlining the computational approaches necessary for the analysis of bio-NMR data. The e-NMR platform is based on a Grid computational infrastructure. A main focus of the current implementation of the e-NMR platform is on streamlining structure determination protocols. Indeed, to facilitate the use of NMR spectroscopy in the life sciences, the eNMR consortium has set out to provide protocolized services through easy-to-use web interfaces, while still retaining sufficient flexibility to handle specific requests by expert users. Various programs relevant for structural biology applications are already available through the e-NMR portal, including HADDOCK, XPLOR-NIH, CYANA and csRosetta. The implementation of these services, and in particular the distribution of calculations to the GRID infrastructure, has required the development of specific tools. However, the GRID infrastructure is maintained completely transparent to the users. With more than 150 registered users, eNMR is currently the second largest European Virtual Organization in the life sciences.
Keywords: CASD-NMR, CS-ROSETTA, CYANA, HADDOCK, NMR, XPLOR-NIH
Introduction
Nuclear Magnetic Resonance Spectroscopy (NMR) is one of two techniques that allow determining three-dimensional structures of biological macromolecules, and their complexes at atomic resolution [4, 25]. NMR is the only structural technique that can be applied in the solution state, making it exquisitely complementary to X-ray crystallography in high-throughput structural determination initiatives. In addition, NMR provides unique information on the dynamics of proteins and nucleic acids in solution, which cannot be obtained with other techniques [14]. At present, a major difficulty with the application of NMR to the investigation of biological systems (bio-NMR) is the lack of standard procedures for these experiments, leading to the implementation of “personalized” protocols in each NMR laboratory. Such protocols are often not formalized and the lack of complete information on the various steps and parameters used, e.g., for protein structure determination, complicates significantly the comparison of the results obtained by different research teams. This situation makes it very difficult for new NMR teams to start up and gain the necessary expertise. It also prevents biochemists and biologists with structural biology projects from readily finding guidance for the best use of NMR techniques.
The standard data analysis in NMR structure determination of proteins consists of a sequence of successive steps that include [4, 25]: (1) Fourier transformation of the time-domain data to obtain the frequency-domain spectra, (2) assigning the NMR signals to individual atoms in the polypeptide chain, (3) collecting structural restraints (e.g., interatomic distances, torsion angles about chemical bonds, residual dipolar couplings), (4) using structural restraints to determine atomic coordinates, (5) validating the obtained structure. Automating as many as possible of these steps, and particularly those from 2 to 4, would transform NMR structure determination of proteins into a highly reproducible activity rather than one that depends on subjective decisions and expertise of an individual [8]. In addition, automated methods would free users from spending excessive time sorting out many trivial issues, from coping with unfriendly software to iteratively correcting errors in the input data.
Presently, specialized computer programs are available for each of the steps involved, each with its own characteristics and often with its own data formats. Processing of NMR data thus currently requires the intervention of specialists, who understand the data and the data formats, as well as the programs and their installation requirements. Furthermore, NMR data processing requires considerable data storage and computational resources. These factors contribute to slowing down the timely adoption of state-of-the-art protocols by the whole NMR community.
Within this context, the e-NMR project (http://www.e-nmr.eu) was designed as an initiative to facilitate NMR data analysis, in order to make it easier and more objective. The project aims at providing users with integrated protocols accessible through user-friendly web interfaces. To do this, e-NMR adopted GRID technology for its underlying computational infrastructure and committed to spreading the use of GRIDs in structural biology and, more generally, in the life sciences. E-NMR has been funded by the European Commission under contract 213010. Currently, the e-NMR project involves an operational GRID, on which the various software programs have been and are being installed. The project focused initially on CPU-intensive programs, to be operated remotely as GRID enabled applications. A specific aspect of the project is the implementation of easy to use web interfaces while retaining sufficient flexibility for expert users. This constitutes a challenge in itself, as it requires the development of specific mechanisms to handle job traffic to and from the GRID. Here we describe the main achievements of the project of interest to potential users are described. The contributions of the e-NMR project toward the definition and adoption of state-of-the-art protocols, particularly in the field of automated structure determination, are also mentioned.
The e-NMR platform
The main goal of e-NMR is to offer complete protocols online for processing NMR data, including all the steps for structure determination described in the introduction, thereby creating a full platform for NMR-based structural biology that can be accessed via the web. To achieve this goal, we started by implementing individual programs as web based servers; the programs were prioritized on the basis of their popularity, CPU demands, as well as usefulness for the project itself. Currently, a variety of programs are operational and can be accessed by registered e-NMR users through the e-NMR portal at http://www.enmr.eu/webportal/ (Fig. 1). In the remainder, we will describe the five programs that have been available for the longest time. Four of these allow users to calculate the structure of proteins and/or protein complexes from NMR data: CS-ROSETTA [21, 22], CYANA [9, 10], HADDOCK [6, 7], XPLOR-NIH [19]. In addition the FormatConverter developed within the CCPN initiative [23] is available for data format conversion. The various programs and their use within the e-NMR portal are briefly described below.
Fig. 1.
The e-NMR portal
CS-ROSETTA
Chemical-Shift ROSETTA or CS-ROSETTA [21, 22] is a rather innovative program for protein structure determination. Unlike XPLOR-NIH and CYANA, it allows structure determination of proteins using only chemical shift information. This results in a very significant time saving as there is no need to obtain NOE-based distance restraints, which is one of the slowest and most tedious steps in the structure determination procedure. Another important advantage of CS-ROSETTA is that chemical shifts are among the most reliable parameters that can be obtained from NMR spectroscopy. The (current) major limitations of CS-ROSETTA are that it is computationally much more expensive than traditional structure calculation programs such as CYANA or XPLOR-NIH, and that it is limited in the size and complexity of systems whose structure determination can be successfully tackled. Because the most time consuming part of the calculations consists of a large number of independent calculations, the implementation of CS-ROSETTA in a GRID computational infrastructure is particularly advantageous.
Structure determination using CS-ROSETTA requires as only input a list of chemical shifts and a number of parameters to control the process. Backbone chemical shifts are initially validated and stored as the target shifts. They are first used to select a set of protein fragments from a structure database, e.g., the Protein Data Bank [2], based on the list of chemical shifts predicted by the SPARTA program [20]. Then the regular ROSETTA protocol [17] for Monte Carlo assembly and relaxation is used to reassemble the protein from the fragments. For the resulting models the chemical shifts are again predicted using SPARTA and the deviations between the predicted and target values are used as a pseudo-energy term in the scoring of the models, yielding a ranking based on both overall structural quality and agreement with the experimental data. The step computationally most expensive is the construction of a model using Monte Carlo assembly and relaxation. To obtain a reliable prediction, a set of 10,000–50,000 models has to be built, each starting from the same fragment library. Using different seeds for generation of random numbers ensures independence of the results from different runs. For the eNMR implementation of CS-ROSETTA only the Monte Carlo search is performed on the GRID. The computational cost involved in chemical shift based structure determination makes CS-ROSETTA a typical example of a program that is beyond the capacity of most local sites. Indeed, we estimate that a typical run that is on the low side of the above-mentioned range (10,000–20,000 models) would cost about 5 days of a dedicated cluster comprising 100 CPUs. The same calculation can be done on the eNMR GRID within about 1 day (depending on how busy the grid is). Furthermore, and perhaps more importantly, several different calculations can be run in parallel without experiencing a decrease in performance. For CS-ROSETTA therefore, the access to GRID resources through a web-portal, combining computational power and ease of use, provides a significant added value for the users.
CYANA
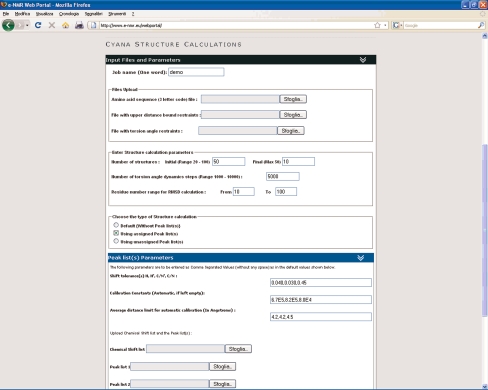
CYANA (Combined Assignment and dYnamics Algorithm for NMR Applications) [9, 10] is one of the most popular programs for calculating structures using “traditional” approaches, i.e., mainly based on distance restraints (typically supplemented with a variety of other restraints, such as torsional angle restraints). CYANA allows for iterative assignment of NOE peak lists, from which distance restrains are derived. This greatly reduces the amount of time needed to analyze and manually assign individual peaks in NOESY spectra. CYANA exploits molecular dynamics in torsion angle space coupled with a simulated annealing algorithm [9]. For NOE assignments, several structure calculation runs are performed in an iterative fashion. Structure calculations start with the generation of a random extended conformation of the macromolecule. Distance, orientation and other restraints derived from NMR data then drive the system to a folded state using simulated annealing. This procedure is repeated several times to obtain sufficient statistics regarding the goodness of fit of the structures determined against the experimental data. Since the different annealing runs are independent of each other, they can be easily distributed over multiple CPUs. After the annealing runs have finished, the best structures are usually selected for further refinement, e.g., including solvent in the calculations. These cannot be run at present within CYANA but can be performed on the e-NMR portal using the AMBER package for molecular dynamics [5].
The design of the web portal for CYANA (Fig. 2) was taken from that of HADDOCK (see next section). Foldable menus are used to hide optional sets of parameters; a default standard structure calculation protocol is offered at first glance. In its current implementation, CYANA can be run in three different modes: using a set of upper distance restraints provided by the user, providing a list of assigned peaks from which CYANA generates the distance restraints, or providing a list of unassigned peaks. In the latter case CYANA will perform a fully automated peak assignment procedure.
Fig. 2.
The CYANA web server
To use CYANA on the e-NMR platform, a valid CYANA license is required in addition to the registration with the e-NMR Virtual Organization.
HADDOCK
HADDOCK [6, 7] stands for High Ambiguity Driven DOCKing; it is a program to calculate the structures of complexes from individual components. The distinguishing feature of HADDOCK with respect to other approaches for biomolecular docking is the use of external information to guide the calculation. Such information can be empirical, theoretical or both, and describes the residues or atoms involved in the binding interface. This information is converted into ambiguous restraints that drive the docking. HADDOCK is particularly useful in predicting complexes from known experimental structures of the partners using NMR data, such as chemical shift perturbations and residual dipolar couplings. These experimental parameters are relatively straightforward to obtain even in the case of large macromolecular assemblies, thus making HADDOCK a program of wide use in structural biology. HADDOCK has proven its value within the CAPRI experiments (Critical Assessment of PRediction of Interactions) [6, 13].
The docking process starts with random placement of the individual components with a given separation and random orientations. Subsequently, a large number of initial complex structures, typically in the order of thousands, are generated by rigid-body docking driven by the ambiguous restraints. From these a number of structures, typically several hundred, are selected for further refinement. These structures are first subjected to a further cycle of simulated annealing, introducing flexibility to allow optimization of contacts. Then, a final cycle of refinement follows, in which the complex is solvated. The results are then scored, analyzed and returned to the user.
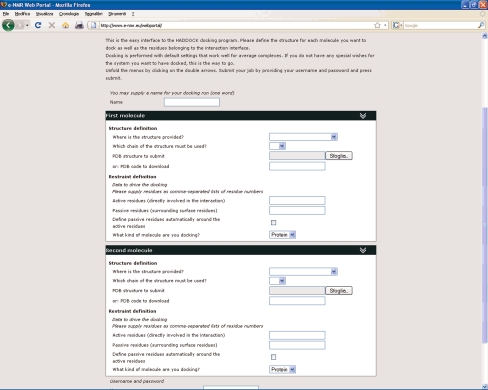
Users can have almost full control of the many parameters involved in the docking process. To offer the full functionality of HADDOCK through a web portal (Fig. 3) required putting forth a complicated form, contrasting with the objective of having a simple interface. To tune the balance between user friendliness and functionality, the portal was split in various interfaces, corresponding to different levels of control and user experience:
The Easy Interface requires no more than providing the two components of a complex and the residues of each that are involved in the interaction (Fig. 3).
The Expert Interface allows the user to provide restraints to include in the docking process and to specify certain aspects of the sampling and analysis. In addition, using this interface the user can set protonation states of histidine residues, and define regions of the interacting molecules to be kept flexible during the docking. This allows a certain degree of conformational change to take place during docking.
The Guru Interface offers almost full control of parameters, allowing e.g., specification of symmetry and relaxation anisotropy restraints and residual dipolar couplings as well as of parameters pertaining to the energy, the scoring and the analysis of results.
For complete control a File Upload Interface is available, where a HADDOCK run parameter file can be provided. This is particularly useful for those who have their own standard protocol or who want to replicate a previous run with minor modifications.
Other interfaces that have been added recently (or that are still in the process of being implemented) include the prediction interface, to be used with predicted rather than experimentally determined interfaces; the multi-body interface; the refinement interface.
Fig. 3.
The easy interface to the HADDOCK web server
The Expert and Guru interface offer control of the docking process at the expense of making the forms to be filled in more complex. Thus, to facilitate the user and keep the forms manageable, foldable menus were introduced that group related parameters under a single header so that the users need only unfold groups of options that should be changed from their default values. In the Easy and Expert interfaces part of the variables is not accessible and thus effectively locked to the default values. All the interfaces, besides the File Upload interface, access to the same backend CGI script to handle the request. After issuing a request, the user is given a link to a site where the progress can be followed. After the run is finished, the results can be viewed online and selected complexes or the complete run can be downloaded.
XPLOR-NIH
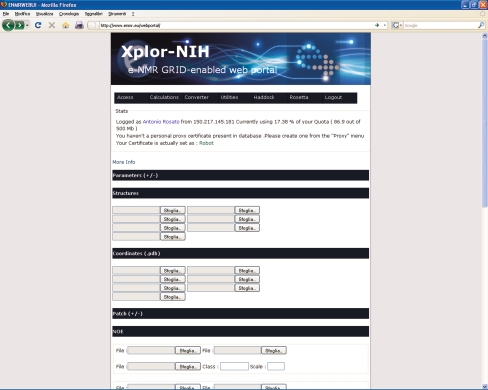
XPLOR-NIH [19] is another very popular program for structure calculations using traditional restraints. It is a quite versatile program, and can be operated through a command line interface or with scripts in its own specific language. The portal uses a design aimed at direct user interaction during the process (Fig. 4). A feature that is currently uniquely implemented for XPLOR-NIH is direct log in of the users with their GRID certificate loaded in the web browser (i.e., no password or user names are required to registered e-NMR users). After the user is identified, he/she obtains access to an environment where projects can be started, stored and managed. Structure calculations with XPLOR-NIH, similarly to CYANA, are run using molecular dynamics in the torsion angle space coupled to a simulated annealing protocol. To run a calculation, the user must provide files containing the description of the structures and topologies of the molecules (which can be proteins, nucleic acids or other molecular types), as well as the different restraint lists to be used to drive the simulated annealing. Note that in CYANA, the structures and topologies of proteins and nucleic acids containing respectively only standard amino acids and bases are instead automatically generated from the sequence.
Fig. 4.
The XPLOR-NIH web server
A specific feature of the XPLOR-NIH server is that it permits the use of some non-standard restraints, namely pseudocontact shifts [1]. Pseudocontact shifts can be measured in proteins containing paramagnetic metal ions, which can be either natively present in the system or introduced through a specific metal-binding tag added to the protein [11, 24]. The usefulness of pseudocontact shifts, especially when employed together with residual dipolar couplings, to investigate the structure and dynamics of proteins has been extensively documented in the literature [3, 16].
When the structure calculations have finished, the user can view and download the results. In addition, it is possible to select a number of structures for further refinement using the AMBER package [5] for molecular dynamics simulations.
The CCPN FormatConverter
The fifth portal allows users to exploit the Format Converter which was developed by the CCPN initiative [23] without having to install the entire CCPN data model. It allows relatively easy conversion of data into different formats in a consistent manner, a task which is very often a major hurdle in the course of a structure determination project, due to the lack of standards in the field of biological NMR that resulted in the majority of the various software tools along the structure determination workflow having their own data formats. The correct conversion of data formats is thus a crucial need in order to be able to pass data from one program to another (e.g., using the output of spectra processing software as input to data analysis software). Very similar considerations apply also to the e-NMR platform itself, as it ultimately aims at linking together various software tools in order to build a full structure determination pipeline. Indeed, establishing interoperability of the programs requires automated conversion of output from one step to match the input of a next step, which can be tackled through the use of the Format Converter.
The current implementation of the Format Converter in the e-NMR platform allows e-NMR users to shuffle their data among the various programs that they employ either locally or within the platform. File conversions are performed locally, as they are not computationally intensive.
The status of the e-NMR GRID
Currently, the computational infrastructure supporting the e-NMR portal is distributed over three sites, which together provide a body of 252 dedicated CPUs (661 kSI2K), and 2.87 Tb of storage. Resources are shared with seven other sites, giving access to a further 4185 CPU (7794 kSI2K) and 35.25 Tb of storage. Since the start of the project, a total of 420 k jobs have been run on the GRID, corresponding to almost 130 years of normalized CPU time. Of these jobs, the majority was run since May 2009. In addition to the software tools available through the web portal of the project, which can be run from the web browser, there are a few other that can be run on the e-NMR GRID from shell (see www.enmr.eu for details); for some of these, a web interface is being developed.
How to become an e-NMR user
Registering with e-NMR is less straightforward than for many other web-based services of use in the life sciences as it involves obtaining a so-called personal certificate that warrants access to the GRID infrastructure. Requesting and obtaining a certificate is indeed a pre-requisite to be able to apply for e-NMR membership. The exact procedure for obtaining a certificate can vary from country to country. Generally, it involves identifying oneself to a Registration Authority (RA). In a country, there can be multiple RA’s, which are often located within universities or research institutions. The RA will transmit the request to the national Certification Authority, which will then process it and release the certificate. At this point, the user will have to download the certificate and install it in his/her web browser. This will allow him/her to access the website of the e-NMR Virtual Organization (VO) and apply for membership (the VO site will automatically detect and check the validity of the certificate). One of the e-NMR VO administrators will approve your request (typically within a few hours). Detailed instructions and links to the various National Authorities are available at http://www.enmr.eu/eNMR-registration. Note that the above procedure applies in Europe; Users from the US can request a user certificate directly to the Structural Biology Grid (SBGrid, http://sbgrid.org/). Other international users can visit the website of the International Grid Trust Federation (IGTF, http://www.igtf.net/). Currently (February 2010), there are more than 150 users registered with e-NMR, of which 15 from outside Europe (10 from USA, two from South Korea, one from Canada, one from New Zealand, one from South Africa).
Other initiatives within the e-NMR project
In addition to developing the e-NMR platform, the e-NMR project is committed to the dissemination of the use of GRID computing within the life sciences. To this end, it organizes events to spread the news about relevant achievements as well as to train (potential) users, which are advertised on the project web site.
In addition, the e-NMR consortium is involved in assessing the performance of software tools that are being considered for inclusion in the portal. In particular, the comparative assessment of the different tools available to tackle a defined problem allows the consortium to define the state-of-the-art in that particular respect. Such an assessment has been carried out in May 2009 focusing on automated protein structure calculations. To guarantee that the comparison is done using each tool at its best, the program developers are directly involved into the experiment. In the first assessment, an ensemble of ten experimental datasets for various protein systems of known structure and two data sets for protein structures not yet released in the public domain (“blind” data sets) were collected by the organizers. The blind data sets were provided by the Northeast Structural Genomics consortium (NESG, www.nesg.org). Seven research teams involved in the development or innovative use of software tools for automated protein structure calculations based on NOE plus chemical shifts or chemical shifts alone were then given access to this input data. The resulting automatically generated structures were then analyzed and validated through various tools by independent researchers. The teams met in Florence, Italy on May 4–6, 2009 to discuss the outcome of this evaluation. The concept closely resembles that of other well-known community-wide “competitions”, such as, within the realm of structural biology, CASP, the Critical Assessment of Techniques for Protein Structure Prediction [15], and CAPRI, the Critical Assessment of Prediction of Interactions [12]. A noteworthy difference is, however, that here only experimental data were used. The participants in this exercise reckoned its value also to drive and provide hints for further development in the field. It was thus agreed that the initiative would continue in the form of an ongoing, community-wide experiment: CASD-NMR (Critical Assessment of automated Structure Determination from NMR data). CASD-NMR is open for any laboratory to participate and/or to submit new targets. The CASD-NMR manifesto has appeared in Nature Methods [18].
Conclusion
The e-NMR project is providing a platform for the analysis of biological NMR data that is open to all users under the requisite that they possess a valid GRID certificate and register with the e-NMR Virtual Organization (expect for specific programs that additionally require ownership of a valid license). The e-NMR platform can be accessed free of charge through the e-NMR web portal. Calculations submitted via the portal are efficiently run on a GRID-based computational infrastructure. The latter enables the use of applications that are computationally very demanding by distributing the tasks over hundreds of nodes.
The e-NMR consortium is also active in disseminating the use of GRID computing in the life sciences and in identifying the state-of-the-art for specific NMR-related applications.
Further information, regarding the state of the project, the available services, and how to join the e-NMR virtual organization, can be found on the project web page at http://www.e-nmr.eu/.
Acknowledgments
The e-NMR project is funded by the European Commission (Contract no. 213010).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviation
- NMR
Nuclear magnetic resonance
References
- 1.Banci L, Bertini I, Cavallaro G, Giachetti A, Luchinat C, Parigi G. Paramagnetism-based restraints for Xplor-NIH. J Biomol NMR. 2004;28:249–261. doi: 10.1023/B:JNMR.0000013703.30623.f7. [DOI] [PubMed] [Google Scholar]
- 2.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertini I, Luchinat C, Parigi G, Pierattelli R. Perspectives in NMR of paramagnetic proteins. Dalton Trans. 2008;2008:3782–3790. doi: 10.1039/b719526e. [DOI] [PubMed] [Google Scholar]
- 4.Billeter M, Wagner G, Wüthrich K. Solution NMR structure determination of proteins revisited. J Biomol NMR. 2008;42:155–158. doi: 10.1007/s10858-008-9277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, Crowley M, Brozell S, TsuiV, Gohlke H, Mongan J, HornakV, Cui G, Beroza P, Schafmeister CE, Caldwell JW, Ross WS, Kollman PA (2008) AMBER 10. University of California, San Francisco, CA
- 6.de Vries SJ, van Dijk AD, Krzeminski M, van Dijk M, Thureau A, Hsu V, Wassenaar T, Bonvin AM. HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- 7.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 8.Guntert P. Automated structure determination from NMR spectra. Eur Biophys J. 2009;38:129–143. doi: 10.1007/s00249-008-0367-z. [DOI] [PubMed] [Google Scholar]
- 9.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 10.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319:209–227. doi: 10.1016/S0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 11.Ikegami T, Verdier L, Sakhaii P, Grimme S, Pescatore P, Saxena K, Fiebig KM, Griesinger C. Novel techniques for weak alignment of proteins in solution using chemical tags coordinating lanthanide ions. J Biomol NMR. 2004;29:339–349. doi: 10.1023/B:JNMR.0000032611.72827.de. [DOI] [PubMed] [Google Scholar]
- 12.Janin J, Henrick K, Moult J, Eyck LT, Sternberg MJ, Vajda S, Vakser I, Wodak SJ. CAPRI: a critical assessment of PRedicted interactions. Proteins. 2003;52:2–9. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- 13.Lensink MF, Mendez R, Wodak SJ. Docking and scoring protein complexes: CAPRI 3rd Edition. Proteins. 2007;69:704–718. doi: 10.1002/prot.21804. [DOI] [PubMed] [Google Scholar]
- 14.Mittermaier A, Kay LE. New tools provide new insights in NMR studies of protein dynamics. Science. 2006;312:224–228. doi: 10.1126/science.1124964. [DOI] [PubMed] [Google Scholar]
- 15.Moult J, Pedersen JT, Judson R, Fidelis K. A large-scale experiment to assess protein structure prediction methods. Proteins. 1995;23:ii–iv. doi: 10.1002/prot.340230303. [DOI] [PubMed] [Google Scholar]
- 16.Pintacuda G, John M, Su XC, Otting G. NMR structure determination of protein-ligand complexes by lanthanide labeling. Acc Chem Res. 2007;40:206–212. doi: 10.1021/ar050087z. [DOI] [PubMed] [Google Scholar]
- 17.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 18.Rosato A, Bagaria A, Baker D, Bardiaux B, Cavalli A, Doreleijers JF, Giachetti A, Guerry P, Guntert P, Herrmann T, Huang YJ, Jonker HR, Mao B, Malliavin TE, Montelione GT, Nilges M, Raman S, van der SG, Vranken WF, Vuister GW, Bonvin AM. CASD-NMR: critical assessment of automated structure determination by NMR. Nat Methods. 2009;6:625–626. doi: 10.1038/nmeth0909-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwieters CD, Kuszewski J, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/S1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 20.Shen Y, Bax A. Protein backbone chemical shifts predicted from searching a database for torsion angle and sequence homology. J Biomol NMR. 2007;38:289–302. doi: 10.1007/s10858-007-9166-6. [DOI] [PubMed] [Google Scholar]
- 21.Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, Ignatchenko A, Arrowsmith CH, Szyperski T, Montelione GT, Baker D, Bax A. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci USA. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Y, Vernon R, Baker D, Bax A. De novo protein structure generation from incomplete chemical shift assignments. J Biomol NMR. 2009;43:63–78. doi: 10.1007/s10858-008-9288-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectrooscopy development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 24.Wöhnert J, Franz KJ, Nitz M, Imperiali B, Schwalbe H. Protein alignment by a coexpressed lanthanide-binding tag for the measurement of residual dipolar couplings. J Am Chem Soc. 2003;125:13338–13339. doi: 10.1021/ja036022d. [DOI] [PubMed] [Google Scholar]
- 25.Wüthrich K. NMR of proteins and nucleic acids. New York: Wiley; 1986. [Google Scholar]