Abstract
Background
Immunoassays used for routine drug of abuse (DOA) and toxicology screening may be limited by cross-reacting compounds able to bind to the antibodies in a manner similar to the target molecule(s). To date, there has been little systematic investigation using computational tools to predict cross-reactive compounds.
Methods
Commonly used molecular similarity methods enabled calculation of structural similarity for a wide range of compounds (prescription and over-the-counter medications, illicit drugs, and clinically significant metabolites) to the target molecules of DOA/toxicology screening assays. We utilized different molecular descriptors (MDL public keys, functional class fingerprints, and pharmacophore fingerprints) and the Tanimoto similarity coefficient. These data were then compared with cross-reactivity data in the package inserts of immunoassays marketed for in vitro diagnostic use. Previously untested compounds that were predicted to have a high probability of cross-reactivity were tested.
Results
Molecular similarity calculated using MDL public keys and the Tanimoto similarity coefficient showed a strong and statistically significant separation between cross-reactive and non-cross-reactive compounds. This was validated experimentally by discovery of additional cross-reactive compounds based on computational predictions.
Conclusions
The computational methods employed are amenable towards rapid screening of databases of drugs, metabolites, and endogenous molecules, and may be useful for identifying cross-reactive molecules that would be otherwise unsuspected. These methods may also have value in focusing cross-reactivity testing on compounds with high similarity to the target molecule(s) and limiting testing of compounds with low similarity and very low probability of cross-reacting with the assay.
Keywords: toxicology, molecular conformations, molecular models, immunoassay, similarity, substance abuse detection
Introduction
Immunoassays are widely used for detection of drugs or drug metabolites (1). A common application is drug of abuse/toxicology (DOA/Tox) screening performed on urine or other body fluids (2). The use of immunoassays as ‘screening tests’ is distinguished from confirmation methods such as gas chromatography/mass spectrometry and liquid chromatography/tandem mass spectrometry, which can provide definitive identification of drugs and their metabolites. Positive DOA/Tox immunoassay screening results are often designated as “preliminary”, “presumptive”, or “unconfirmed” positives (2). One limitation of immunoassays is interference caused by compounds with structural similarity to the target molecule(s) of the assay (i.e., typically the hapten(s) against which the assay antibodies were generated) (3). Such cross-reactive molecules can be structurally related drugs, drug metabolites, or endogenous compounds. During the development of commercially marketed immunoassays, manufacturers typically test common drugs for cross-reactivity as well as endogenous compounds (4).
DOA/Tox screening assays may have broad specificity towards classes of drugs such as amphetamines, barbiturates, benzodiazepines, cannabinoids, and opiates. Other DOA/Tox screening assays are directed towards detection of a single target drug or metabolite (e.g., buprenorphine, benzoylecgonine). There are advantages to using broad specificity DOA/Tox screening assays. First, for some classes of drugs, the management of an overdose involves the same treatment regardless of which particular drug within the class is involved (e.g., flumazenil as an antidote for benzodiazepine overdose). Second, using a single assay to screen for multiple drugs within a class is less expensive than using separate assays for each individual drug.
While the package inserts or manufacturers' documents collectively contain extensive data on assay cross-reactivity, many of these compounds have been reported post-marketing over the last several decades. Examples of published reports of DOA/Tox assay cross-reactivity include fluoroquinolone antibiotic cross-reactivity with opiates assays (5, 6), quetiapine cross-reactivity with tricyclic antidepressant (TCA) assays (7-9), fentanyl cross-reactivity with LSD immunoassays (10, 11), sertraline and sertraline metabolite cross-reactivity with benzodiazepine assays (12, 13), and venlafaxine and O-desmethylvenlafaxine cross-reactivity with PCP assays (14).
To date there has been no comprehensive computational analysis aimed at predicting cross-reactivity of DOA/Tox screening assays. Our hypothesis was that a given compound is more likely to cross-react with an immunoassay if the compound shares a high level of structural similarity to the target molecule(s) of the assay.
We utilize an in silico method known as similarity analysis which determines the similarity between molecules independent of any in vitro data (15-17). Similarity can be assessed at the one-dimensional, two-dimensional (2D), and three-dimensional (3D) levels (17-20). Common 2D similarity approaches use fragment bit strings compared using the Tanimoto coefficient (0 being maximally dissimilar, 1 being maximally similar). 3D similarity methods essentially require the development of a pharmacophore pattern that represents the arrangement of the chemical features and distances between them that are important for biological activity (21).
In this study, we applied similarity analyses to a wide range of marketed immunoassays used for DOA/Tox screening. The overall goal was to develop computational methods that efficiently predict compounds likely to cross-react with immunoassays.
Materials and Methods
Standards and Reagents
Quetiapine fumarate and escitalopram oxalate were obtained from Sequoia Research Products (Pangbourne, United Kingdom). Citalopram hydrobromide was purchased from Molcan (Toronto, Canada). All other drugs were obtained from Sigma.
Classification of Immunoassay Cross-Reactivity Data
Cross-reactivity data was obtained from manufacturers' package inserts or supplemental manufacturers' documents (Data Supplement 1). Often this information is presented as the concentration of the tested compound that produces the same response in the assay as the cutoff concentration of the target compound, or occasionally as percent cross-reactivity. Additional data for cannabinoid (22), opiate (5, 6), and TCA assays (23, 24) were obtained from the peer-reviewed literature.
To classify cross-reactivity of compounds for the various assays, we broadly divided compounds into “Strong True Positives”, “Weak True Positives”, “Strong False Positives”, “Weak False Positives”, “True Negatives”, and “False Negatives” (Table 1). For any compound, meeting the criteria for strong or weak cross-reactivity in any one assay was sufficient for classification in that category. Within-class compounds were formally defined as drugs and their metabolites (if present) whose detection allows for interpretation that one or more members of the target drug class are present in the sample. For example, for benzodiazepines, within-class compounds include alprazolam, chlordiazepoxide, clonazepam, diazepam, and their metabolites but not the therapeutically similar but non-benzodiazepine drugs eszopiclone and zolpidem.
Table 1.
Criteria for classifying cross-reactivity of compounds.
| Classification | Definition |
|---|---|
| DOA/Tox assays | |
| Target compound | For a given DOA/Tox assay, the drug or drug metabolite used as the antigenic (hapten) target (e.g., morphine for opiates assay) |
| Strong true positives | Within-class compounds causing cross-reactivity equal to the positive cutoff of the assay at concentrations of less than 10,000 ng/mL |
| Weak true positives | Within-class compounds causing cross-reactivity equal to the positive cutoff of the assay at concentrations of 10,000 to 100,000 ng/mL |
| Strong false positives | Out-of-class compounds causing cross-reactivity equal to the positive cutoff of the assay at concentrations of less than 10,000 ng/mL |
| Weak false positives | Out-of-class compounds causing cross-reactivity equal to the positive cutoff of the assay at concentrations of 10,000 to 100,000 ng/mL |
| True negatives | Out-of-class compounds causing no cross-reactivity or cross-reactivity equal to the positive cutoff of the assay only at concentrations of greater than 100,000 ng/mL |
| False negatives | Within-class compounds causing no cross-reactivity or cross-reactivity equal to the positive cutoff of the assay only at concentrations of greater than 100,000 ng/mL |
Similarity Searching in Discovery Studio 2.0
Similarity searching used the ‘find similar molecules by fingerprints’ protocol in Discovery Studio 2.0 (Accelrys, San Diego, CA). The MDL public keys and long range functional class fingerprint description 6 keys (referred to as ‘FCFP_6’) (25) were used separately with the Tanimoto similarity coefficient and an input query molecule (17). It should be noted that these similarity algorithms do not recognize differences between stereoisomers and racemic mixtures (e.g., citalopram and escitalopram).
Pharmacophore Fingergrints
Three-point and four-point pharmacophore-based fingerprints were calculated from the 3D conformation using the Molecular Operating Environment (Chemical Computing Group, Montreal, Canada). Each atom in a molecule was given one of eight atom types computed from three atomic properties (“in π system”, “is donor”, “is acceptor”). All quadruplets of atoms were coded as features using the inter-atomic distance, atom types, and chirality.
Databases
The main database searched was created using the database of Food and Drug Administration (FDA)-approved drugs derived from the Clinician's Pocket Drug Reference (26-28) (‘SCUT 2008 database’). The database was supplemented with drugs of abuse and drug metabolites (n = 110) important in clinical toxicology testing. The total database of 786 compounds was referred to as the ‘Expanded SCUT database’.
Cross-Reactivity Testting
Tests for cross-reactivity determined the lowest concentration of a compound added to drug-free urine that caused a reaction result that equaled or exceeded the cutoff concentration for the target compound of the assay (4). Cross-reactivity testing was performed following manufacturers' instructions using two different assay systems: (1) Emit® II plus assays (amphetamines, barbiturate, benzodiazepine, cannabinoids, cocaine metabolite, methadone, opiate, phencyclidine, propoxyphene) and Emit® tox™ serum (tricyclic antidepressants) run on Siemens (Dade-Behring) Viva-E analyzers; and (2) Biosite Triage® Tox screen.
Results
Classification of Cross-Reactivity Data
We compiled cross-reactivity data for 84 marketed versions of 18 DOA/Tox screening assays (amphetamines, barbiturates, benzodiazepines, benzoylecgonine, buprenorphine, cannabinoids, cotinine, 6-AM, LSD, MDMA, methadone, EDDP, methaqualone, opiates, oxycodone, phencyclidine, propoxyphene, and TCAs) using information from package inserts, supplemental manufacturers' data, and five peer-reviewed articles (Data Supplement 1) (5, 6, 22-24). Top prescribed medications in the United States in 2007 (29) are also highlighted. Classification of all available cross-reactivity data yielded a total of 1961 datapoints – 162 strong true positives, 20 weak true positives, 20 strong false positives, 69 weak false positives, 1681 true negatives, and 9 false negatives. We then used two algorithms, MDL public keys and FCFP_6, to compute 2D similarity for the most common target compounds for the 18 immunoassay target compounds (d-amphetamine, secobarbital, diazepam, buprenorphine, 9-carboxy-11-nor-Δ9-tetrahydrocannabinol, carisoprodol, benzoylecgonine, cotinine, EDDP, 6-acetylmorphine, LSD, methadone, methaqualone, morphine, oxycodone, phencyclidine, propoxyphene, and desipramine) to compounds in the Expanded SCUT Database and, where applicable, to any additional compounds reported in the package inserts.
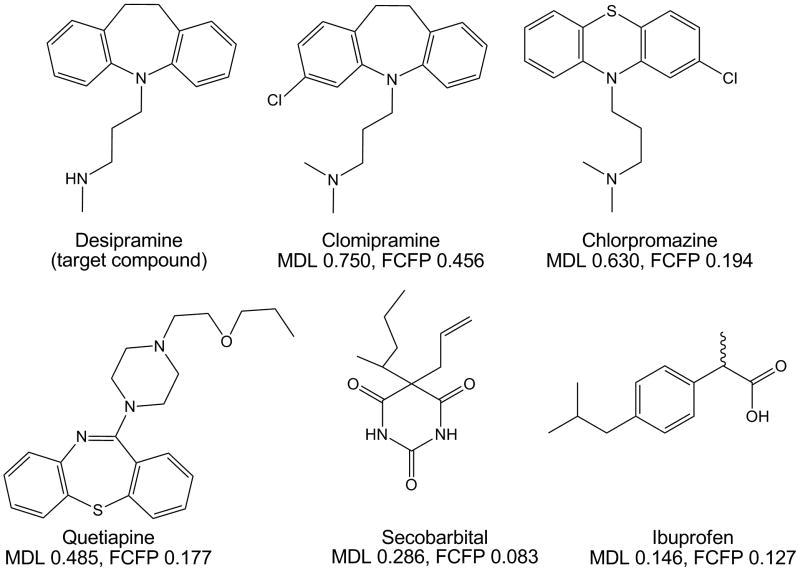
Fig. 1 shows the similarity of desipramine (target compound of several TCA assays) to five compounds. Desipramine has the highest similarity (in descending order) to clomipramine (another TCA), chlorpromazine (a phenothiazine antipsychotic), and quetiapine (another tricyclic). Desipramine has low similarity to secobarbital and essentially no 2D similarity to ibuprofen.
Fig. 1. Illustration of similarity measures.
Using desipramine (target compound of some TCA screening assays) as the target compound, 2D similarity was calculated using MDL public keys and FCFP_6 to five different compounds, three of which (clomipramine, chlorpromazine, quetiapine) are three-ringed molecules and two of which (secobarbital, ibuprofen) have a single ring in their structures. Of the five test compounds, clomipramine (a TCA like desipramine) has the highest similarity to desipramine, while ibuprofen has the lowest similarity.
The data analysis is complicated for some DOA/Tox screening assays in that the same target compound is not always used for all marketed versions (Data Supplement 1). This issue applies to amphetamine, benzodiazepine, and TCA assays. We chose d-amphetamine, diazepam, and desipramine as the target compound for amphetamine, benzodiazepine, and TCA screening assays, respectively, for the total data comparisons but also provide data using alternative target compounds (e.g., nordiazepam for benzodiazepines) in Data Supplement 2.
Similarity Comparisons Combining Data from All Assays
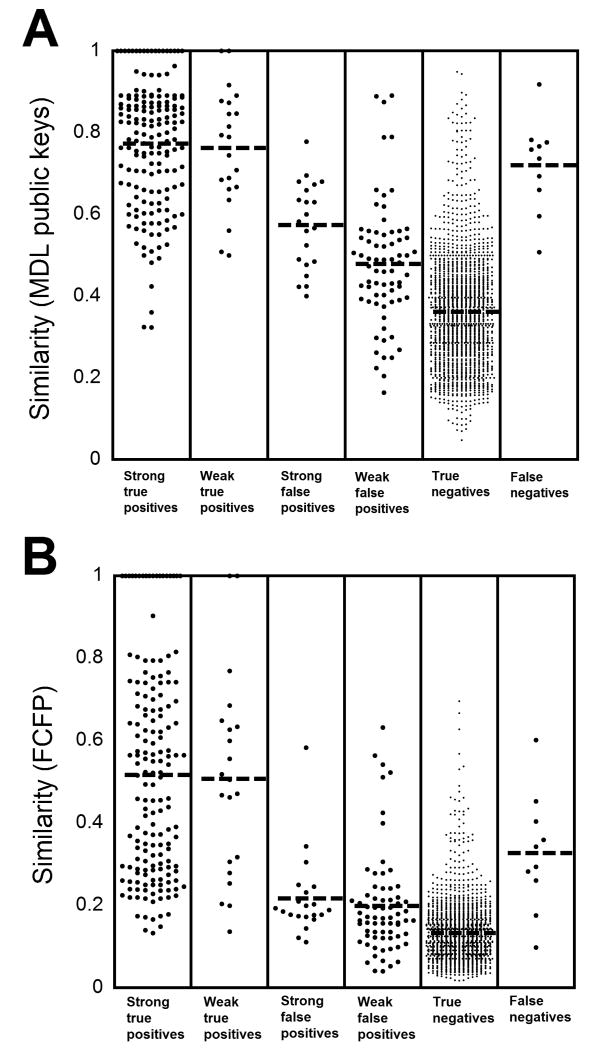
Plotting the similarity coefficients for all datapoints (Fig. 2) shows MDL public keys are generally higher than those calculated using FCFP_6. However, for either method, the mean similarity (mean±SD) is significantly higher (p < 0.001, unpaired t-test) using either MDL public keys or FCFP_6 for Strong True Positives (MDL: average similarity 0.773±0.154; FCFP: 0.517±0.254), Weak True Positives (MDL: 0.763±0.145; FCFP: 0.507±0.242), Strong False Positives (MDL: 0.574±0.106; FCFP: 0.217±0.100), or Weak False Positives (MDL: 0.478±0.151; FCFP: 0.199±0.124) compared to True Negatives (MDL: 0.362±0.138; FCFP: 0.133±0.071). Interestingly, the mean similarity for False Negatives (MDL: 0.720±0.113); FCFP: 0.328±0.142) was very close to that for True Positive using MDL public keys but in between Strong and Weak True Positives using FCFP_6 (Fig. 2).
Fig. 2. Plot of similarity of all data for DOA/Tox screening assays.
As described in “Materials and Methods”, cross-reactivity data for each DOA/Tox assay were used to classify compounds in one of six categories: “Strong True Positives”, “Weak True Positives”, “Strong True Positives”, “Weak True Positives”, “True Negatives”, and “False Negatives”. (A) Plot of data for all compounds using MDL public keys similarity and the Tanimoto coefficient. See text of “Results” for standard deviation values. (B) Plot of data for all compounds using FCFP_6 and the Tanimoto coefficient. See text of “Results” for standard deviation values.
Using MDL public keys, 45.6% of the strongly cross-reactive compounds (Strong True Positives and Strong False Positives) have similarity coefficients of 0.8 or higher relative to the target compound of the assay. For the True Negatives, only 24 of 1681 (1.4%) of compounds have similarity coefficients of 0.8 or higher to the target compounds. Thus, a cutoff of 0.8 would have a positive predictive value of 77.6% in distinguishing compounds capable of strong cross-reactivity from the True Negatives. Conversely, 65.9% of True Negatives have similarity coefficients of less than 0.4 to the target compounds whereas only 3 of 1681 (1.6%) of strongly cross-reactive compounds fit in this category. The three examples of strongly cross-reactive compounds with similarity coefficients of 0.4 or less were all from amphetamine assays (MDMA; 3,4-methylenedioxy-α-ethyl-N-methylphenethylamine, also known as MDBD; and 3,4-methylenedioxy-N-ethylamphetamine, also known as MDEA). Adopting a lower cutoff of 0.4 would have a negative predictive value of 99.8% in distinguishing compounds capable of strong cross-reactivity from the True Negatives.
Strongly cross-reactive compounds with MDL public keys similarity of 0.5 or less to the target compound were found for only 5 DOA/Tox screening assays (amphetamines, benzodiazepines, methadone, PCP, and TCAs). These compounds represented only 14 of 182 (7.6%) of the total strongly cross-reactive compounds. Ten of the 14 examples came from the benzodiazepines assays (5 compounds) and TCAs assays (5 compounds). In contrast, 1473 of 1681 (87.6%) of True Negatives had similarity coefficients of 0.5 or less.
Similarity Comparisons Within Individual Doa/Tox Screening Assays
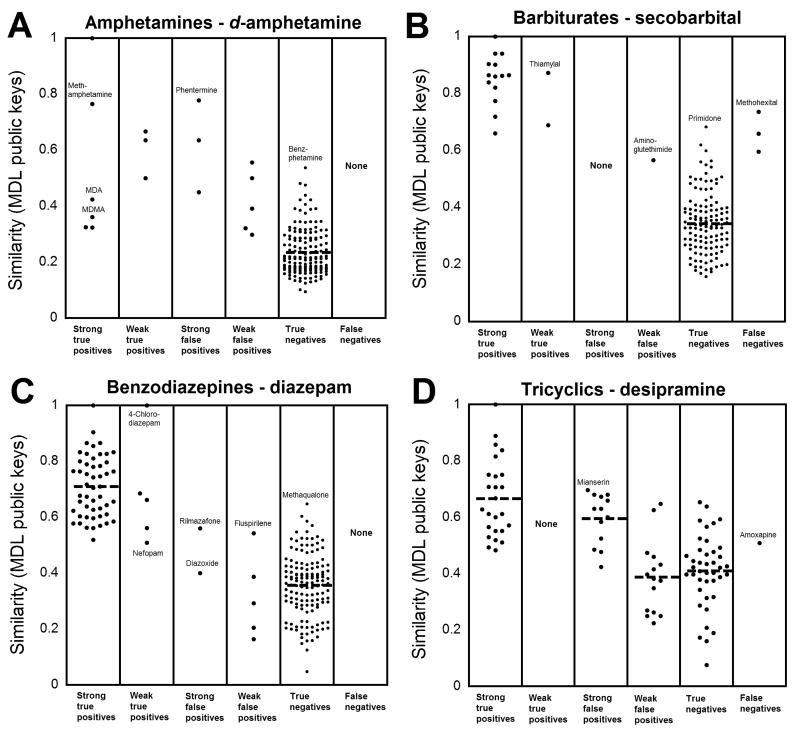
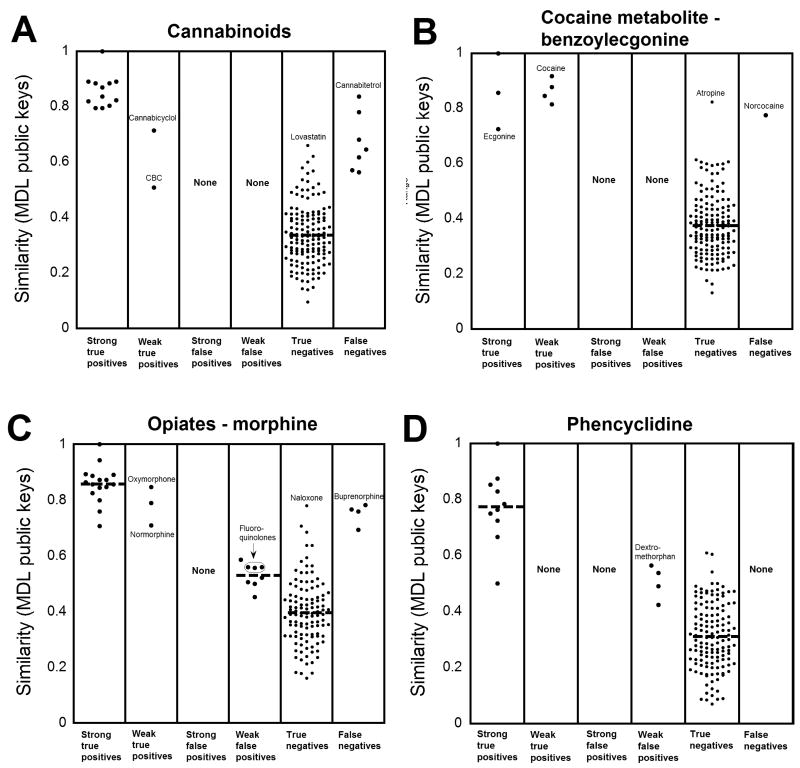
Fig. 3 shows data using MDL public keys for four broad-specificity assays (amphetamines, barbiturates, benzodiazepines, and TCAs). These data show the challenge in identifying weakly cross-reactive compounds for broadly specific assays, in that the similarity for weakly cross-reactive compounds can overlap with that for True Negatives. Fig. 4 shows data using MDL public keys for cannabinoid, benzoylecgonine, opiate, and PCP assays. For these four assays, all cross-reactive compounds (strong or weak) have similarity coefficients to the respective targets compounds higher than the average similarity of all True Negatives. For the opiate assays (Fig. 4C), several fluoroquinolone antibiotics, identified in the literature as cross-reacting with some opiate screening assays (5, 6), have similarity coefficients that overlap with those of the True Negatives but that are higher than the majority of compounds classified as True Negatives. In addition, fentanyl has relatively high similarity (MDL: 0.621, FCFP: 0.152) to LSD, consistent with studies indicating cross-reactivity of fentanyl with LSD immunoassays (10, 11). Plots for DOA/Tox screening assays not shown in Figs. 3 or 4 are found in Supplemental Data Figs. 1 and 2. These plots show the same general trends for DOA/Tox screening assays shown in Figs. 3 and 4.
Fig. 3. Similarity of drugs and drug metabolites relative to the target compounds for four broadly specific DOA/Tox assays.
As described in the legend to Fig. 2, cross-reactivity data for four DOA/Tox assays were sorted into six categories. The similarity (using MDL public keys and the Tanimoto coefficient) of each tested compound to the target compound of the DOA/Tox assay was plotted. (A) Amphetamine assays (using d-amphetamine as the target). (B) Barbiturate assays (using secobarbital as the target compound). (C) Benzodiazepine assays (using diazepam as the target compound). (D) TCA assays (using desipramine as the target compound).
Fig. 4. Similarity of drugs and drug metabolites relative to the target compounds for four DOA/Tox assays.
As described in the legend for Fig. 2, cross-reactivity data for four DOA/Tox assays were sorted into six categories. The similarity (using MDL public keys and the Tanimoto coefficient) of each tested compound to the target compound of the DOA/Tox assay of the DOA/Tox assay was plotted. (A) Cannabinoid assays (using 9-carboxy-11-nor-Δ9-tetrahydrocannabinol as the target compound). (B) Cocaine metabolite (benzoylecgonine) assays. (C) Opiate assays (using morphine as the target compound). (D) Phencyclidine assays.
Another way to examine the data is to analyze how the 2D structural similarity for the cross-reactive compounds relates to that for True Negatives. Using MDL public keys data, the mean similarity for the True Negatives (indicated as a dashed line in Figs. 3 and 4 and Supplemental Data Fig. 1) is lower than the similarity for nearly all cross-reactive compounds. The only exceptions are the assays for benzodiazepines (ketoprofen, lovastatin, modafanil are weak false positives in one marketed assay each), propoxyphene (methaqualone is a weak false positive in one marketed assay), and TCAs (six compounds are weak false positives in a single marketed assay). The similarity for True Negatives overlapped more with cross-reactive compounds when using FCFP_6, with four assays (benzodiazepine, opiate, propoxyphene, and TCA) having cross-reactive compounds that had similarity coefficients lower than the average similarity for all True Negatives. Consequently, using MDL public keys or FCFP_6, Strong True Positives with similarity lower than the mean similarity coefficient for True Negatives are rare.
The 2D similarity results suggest that the targets of DOA/Tox screening immunoassays can be categorized on a continuum based on whether they have low to high similarity to other compounds that can be encountered clinically. At one extreme would be benzoylecgonine, which has high similarity to cocaine and other cocaine metabolites, but low predicted similarity to other clinically encountered drugs (Fig. 4B). This may explain why marketed benzoylecgonine screening immunoassays have very few documented cross-reacting substances. At the other extreme would be the TCA desipramine. There is substantial overlap between the similarity of despiramine to other TCAs and to other three-ringed molecules such as cyclobenzaprine, phenothiazines, or quetiapine (Fig. 3D), suggesting that cross-reactivity is likely to be a problem with TCA screening immunoassays no matter which TCA is chosen as the target.
We also explored 3D similarity classification approaches using three- or four-point pharmacophore fingerprints. However, we found that even by varying cutoff settings, these algorithms were too restrictive and missed many cross-reactive compounds, including some Strong True Positives (e.g., the four-point pharmacophore method assigned zero similarity to some Strong True Positives; Supplemental Data Fig. 3 found in Data Supplement 2).
Cross-Reactivity Testing Guided by Similarity Predictions
As evidenced in published reports, some cross-reactive compounds are recognized post-marketing. We used similarity analyses to predict additional cross-reactive compounds for ten assays: amphetamine, barbiturate, benzodiazepine, benzoylecgonine, cannabinoid, methadone, opiate, PCP, propoxyphene, and TCA (Supplemental Data Fig. 4 in Data Supplement 2). We then tested 46 such compounds on two different platforms (Biosite Triage, Syva EMIT assays on Siemens Viva-E analyzers), identifying additional cross-reactive compounds for amphetamines, barbiturates, opiates, PCP, and TCA assays (Table 2). Eight of the cross-reactivities we identified have not, to our knowledge, yet been reported in the published literature or package inserts to cross-react with any marketed version of a particular DOA/Tox immunoassay (footnote 4, Table 2). These eight new cross-reactivities were: atropine (Biosite Triage opiates assay), citalopram (Syva EMIT TCA assay), dextromethorphan (Syva EMIT opiates and propoxyphene assays), escitalopram (Syva EMIT TCA assay), mirtazapine (Syva EMIT TCA assay), oxcarbazepine (Biosite Triage barbiturates assay), and selegiline (Syva EMIT amphetamines assay). Cetirizine was previously identified as cross-reacting with fluorescence polarization immunoassays (30). Quetiapine was previously identified as cross-reacting with some marketed TCA screening immunoassays (7-9).
Table 2.
Results of cross-reactivity testing.
| Compound | Closest similarity to target compounds (MDL, FCFP_6)1 | Cross-reactivity positives1 |
|---|---|---|
| Acetaminophen | AMPH 0.310, 0.132 | None |
| Atropine | OPI 0.581, 0.159 | OPI 500,000 ng/mL2,4 |
| Azithromycin | PROP 0.532, 0.121 | None |
| Buspirone | BENZO 0.522, 0.088 | None |
| Caffeine | BENZO 0.524, 0.082 | None |
| Carbamazepine | TCA 0.460, 0.306 | None |
| Carisoprodol | BARB 0.508, 0.184 | None |
| Cetirizine | TCA 0.429, 0.120 | TCA 250,000 ng/mL3 |
| Chlorambucil | TCA 0.442, 0.177 | None |
| Chloroquine | TCA 0.686, 0.060 | None |
| Chlorzoxazone | BARB 0.508, 0.184 | None |
| Citalopram | PROP 0.534, 0.107 | TCA 500,000 ng/mL3,4 |
| Citalopram metabolite (N-desmethyl) | TCA 0.517, 0.176 | None |
| Clotrimazole | BENZO 0.473, 0.171 | None |
| Clozapine | BENZO 0.650, 0.174 | None |
| Dexamethasone | CANN 0.542, 0.098 | None |
| Dextromethorphan | PCP 0.565, 0.157 | OPI 250,000 ng/mL3,4 PCP 500,000 ng/mL2 25,000 ng/mL3 PROP 500,000 ng/mL3,4 |
| Digoxin | CANN 0.621, 0.071 | None |
| Escitalopram | PROP 0.534, 0.107 | TCA 200,000 ng/mL3,4 |
| 5-Fluorouracil | BARB 0.625, 0.220 | None |
| Fluoxetine | TCA 0.434, 0.288 | None |
| Gemfibrozil | CANN 0.604, 0.188 | None |
| Ipratropium | OPI 0.529, 0.105 | None |
| Maprotiline | TCA 0.659, 0.250 | TCA 10,000 ng/mL3 |
| Meprobamate | BARB 0.500, 0.190 | None |
| Mexiletine | AMPH 0.500, 0.286 | AMPH 20,000 ng/mL3 |
| Mirtazapine | TCA 0.653, 0.115 | TCA 500,000 ng/mL3,4 |
| Nifedipine | BARB 0.333, 0.090 | None |
| Norclozapine | BENZO 0.581, 0.179 PCP 0.519, 0.091 |
None |
| Oxcarbazepine | BARB 0.375, 0.067 | BARB 500,000 ng/mL2,4 |
| Phenytoin | BARB 0.542, 0.190 | None |
| Propylthiouracil | BARB 0.593, 0.312 | None |
| Quetiapine | TCA 0.485, 0.177 | TCA 100,000 ng/mL3 |
| Rapamycin | BARB 0.534, 0.073 | None |
| Reserpine | OPI 0.597, 0.108 | None |
| Rifampicin | OPI 0.587, 0.132 | None |
| Selegiline | AMPH 0.407, 0.333 MTD 0.556, 0.177 |
AMPH 40,000 ng/mL3,4 |
| Sibutramine | MTD 0.610, 0.231 | None |
| Simvastatin | CANN 0.604, 0.188 | None |
| Succinylcholine | PROP 0.549, 0.123 | None |
| Tamoxifen | MTD 0.578, 0.233 PROP 0.617, 0.235 |
None |
| Terbinafine | MTD 0.579, 0.156 | None |
| Thioridazine | PCP 0.490, 0.171 | TCA 100,000 ng/mL3 |
| Venlafaxine | PROP 0.696, 0.179 | None |
| Vinblastine | OPI 0.640, 0.133 | None |
| Warfarin | CANN 0.639, 0.198 | None |
Abbreviations and target compounds: AMPH, amphetamines (d-amphetamine); BARB, barbiturates (secobarbital); BENZO, benzodiazepines (diazepam); CANN, cannabinoids (9-carboxy-11-nor-Δ9-tetrahydrocannabinol); MTD, methadone; OPI, opiates (morphine); PCP, phencyclidine; PROP, propoxyphene; TCA, tricyclic antidepressants (desipramine)
Biosite Triage
Syva Emit
Previously unreported cross-reactive drug for a DOA/Tox screening immunoassay
Similarity-Based Predictions for Assays in Development
There are at least four other drugs with potential abuse liability for which immunoassays are in development: carisoprodol, fentanyl, ketamine, and meperidine. We calculated similarity of these four drugs to the compounds in the Expanded SCUT database (Data Supplement 1). Carisoprodol showed high similarity to its active metabolite meprobamate (MDL=0.949; FCFP=0.724) but low similarity to other compounds. The next closest similarities for carisoprodol are to barbiturates (MDL∼0.500-0.575; FCFP 0.2 or less). For fentanyl, the compounds with the closest similarity by MDL public keys are bupivacaine (0.712), imatinib (0.684), meperidine (0.673), buspirone (0.667), and meclizine (0.646). Ketamine has generally low similarity to other compounds but is moderately similar to several antidepressants or their metabolites (sertraline, bupropion, desmethylcitalopram).
For meperidine, a number of common drugs are close in similarity: loperamide (MDL: 0.726), fentanyl (0.696), linezolid (0.696), buspirone (0.667), ketoconazole (0.667), meclizine (0.648), risperidone (0.639), and aripiprazole (0.632). Cross-reactivity testing for a meperidine immunoassay could focus on these drugs.
Discussion
Cross-reactivity by structurally related compounds remains a challenge in the design and clinical use of DOA/Tox screening immunoassays (3). In this study we applied similarity analysis as a new tool to classify compounds that are likely to cross-react with common DOA/Tox screening tests. Using our predictions, we performed cross-reactivity testing and identified eight assay cross-reativities not previously reported. Of the three molecular descriptors evaluated, MDL public keys were shown as the most useful for this purpose. The similarity coefficients generated by the MDL analysis are well distributed with clear separation (on average) between cross-reactive compounds and those that do not cross-react. FCFP_6 and pharmacophore fingerprints are best suited towards identifying very close structural analogs but not compounds with lower degrees of similarity. There are other molecular fingerprints that could be evaluated in the future (31).
There are several screening strategies that could be employed using the techniques we report in this study. One approach could be to test all clinically relevant compounds with similarity coefficients of 0.8 or higher to the target compound and avoid any testing of those with a coefficient of 0.4 or less. For compounds with similarity coefficients between 0.4 and 0.8, additional selection criteria could be used such as pharmacokinetics and frequency of overdoses.
A limitation of the similarity approaches is that these cannot account for the complex 3D molecular interactions inherent in antibody-antigen binding. To our knowledge, a 3D structure of an antibody used in a DOA/Tox screening assay and its antigen target has not been reported, although there has been structural determination of antibodies being evaluated as novel antidotes to DOA overdose (e.g., PCP (32) and cocaine (33, 34)), in which the antibody interacts with all portions of the target molecule. For DOA/Tox screening assays where similar antibody-drug interactions apply, whole molecule similarity measures (as used in this study) seem appropriate for prediction. However, a crystal structure of morphine bound to a monoclonal antibody showed the antibody interacting with the more hydrophobic portion of morphine, while the hydrophilic half was mostly exposed to solvent (35). The crystal structure of digoxin with a Fab fragment revealed the carbohydrate portions of the drug unbound by antibody and exposed to solvent (36). For target compounds like these, similarity searching using substructures may be worth evaluating.
An additional limitation of the similarity methods used in this study is that these do not account for the concentration-dependence of cross-reactivity. For instance, a substance with poor cross-reactivity may be problematic if present in serum/plasma or urine at much greater concentrations than the analyte. This is likely to especially be an issue for drugs used in low doses, resulting in low concentrations in body fluids. The synthetic opioid fentanyl would be an example of such a drug. For a drug such as fentanyl, a wider range of similarities may need to be considered in testing potentially cross-reactive substances.
Lastly, biological specimens may contain several cross-reacting substances. The situation can be quite complex for classes of drugs such as benzodiazepines that have multiple metabolites. Total cross-reactivity to a DOA/Tox immunoassay can be derived from multiple compounds, each cross-reacting to varying degrees. Future studies can be directed at predicting such total cross-reactivity by extensions of the similarity methods used in this study.
False positive DOA/Tox screening results present a challenge for clinical chemists and clinicians, as these assays are used by emergency departments, substance abuse treatment programs, transplant programs, and pain clinics, as well as other settings. Clinicians may use the results for a variety of decisions including antidote administration, prescribing of narcotic medications, and whether to proceed with or delay elective surgeries. Clinical chemists may be consulted as to the likely cause of an unexpectedly positive screening assay. The extensive similarity calculations we performed (summarized in the Data Supplement 1) highlight drugs or metabolites that have high similarity to the assay target antigen(s) but whose cross-reactivity has not been reported. These data can aid clinical chemists in determining what drugs or metabolites may contribute to assay positivity unexplained by known assay cross-reactivities, clinical history, or confirmatory testing. The similarity tools also provide a rational framework for manufacturers and regulators to focus cross-reactivity testing on drugs or metabolites most likely to cross-react.
Supplementary Material
Acknowledgments
The authors thank Darla Lower and Jacqueline Rymer for technical assistance. SE gratefully acknowledges Accelrys, Inc. for providing Discovery Studio.
Grant/funding support: This research was supported by National Institutes of Health grant K08-GM074238 to MDK.
Nonstandard abbreviations
- DOA
drug of abuse
- Tox
toxicology
- 6-AM
6-acetylmorphine
- LSD
lysergic acid diethylamide
- MDMA
3,4-methylenedioxymethampetamine (Ecstasy)
- EDDP
2-ethylidine-1,5-dimethyl-3,3-diphenypyrrolidine
- PCP
phencyclidine
- TCA
tricyclic antidepressant
- 2D
two-dimensional
- 3D
three-dimensional
- FCFP_6
long range functional class fingerprint description 6 keys
- FDA
Food and Drug Administration
- MDBD
3,4-methylenedioxy-α-ethyl-N-methylphenethylamine
- MDEA
3,4-methylenedioxy-N-ethylamphetamine
Footnotes
Publisher's Disclaimer: “This is an un-copyedited authored manuscript copyrighted by The American Association for Clinical Chemistry (AACC). This may not be duplicated or reproduced, other than for personal use or within the rule of ‘Fair Use of Copyrighted Materials' (section 107, Title 17, U.S. Code) without permission of the copyright owner, AACC. The AACC disclaims any responsibility or liability for errors or omissions in this version of the manuscript or in any version derived from it by the National Institutes of Health or other parties. The final publisher-authenticated version of the article will be made available at http://www.clinchem.org 12 months after its publication in Clinical Chemistry.”
References
- 1.Kricka LJ. Principles of immunochemical techniques. In: Burtis CA, Ashwood ER, Bruns DE, editors. Tietz textbook of clinical chemistry and molecular diagnostics. Vol. 4th. St. Louis, MO: Elsevier Saunders; 2006. pp. 219–43. [Google Scholar]
- 2.Moeller KE, Lee KC, Kissack JC. Urine drug screening: practical guide for clinicians. Mayo Clin Proc. 2008;83:66–76. doi: 10.4065/83.1.66. [DOI] [PubMed] [Google Scholar]
- 3.Kricka LJ. Interferences in immunoassays - still a threat. Clin Chem. 2000;46:1037–8. [PubMed] [Google Scholar]
- 4.Powers DM, Boyd JC, Glick MR. Interference testing in clinical chemistry (EP7-A) Villanova, PA: NCCLS; 1986. [Google Scholar]
- 5.Baden LR, Horowitz G, Jacoby H, Eliopoulos GM. Quinolones and false-positive urine screening for opiates by immunoassay technology. JAMA. 2001;286:3115–9. doi: 10.1001/jama.286.24.3115. [DOI] [PubMed] [Google Scholar]
- 6.Zacher JL, Givone DM. False-positive urine opiate screening associated with fluoroquinolone use. Ann Pharmacother. 2004;38:1525–8. doi: 10.1345/aph.1D632. [DOI] [PubMed] [Google Scholar]
- 7.Caravati EM, Juenke JM, Crouch BI, Anderson KT. Quetiapine cross-reactivity with plasma tricyclic antidepressant immunoassays. Ann Pharmacother. 2005;39:1446–9. doi: 10.1345/aph.1G107. [DOI] [PubMed] [Google Scholar]
- 8.Henrickson RG, Morocco AP. Quetiapine cross-reactivity among three tricyclic antidepressant immunoassays. J Toxicol Clin Toxicol. 2003;41:105–8. doi: 10.1081/clt-120019122. [DOI] [PubMed] [Google Scholar]
- 9.Sloan KL, Haver VM, Saxon AJ. Quetiapine and false-positive urine drug testing for tricyclic antidepressants. Am J Psychiatry. 2000;157:148–9. doi: 10.1176/ajp.157.1.148-a. [DOI] [PubMed] [Google Scholar]
- 10.Gagajewski A, Davis GK, Kloss J, Poch GK, Anderson CJ, Apple FS. False-positive lysergic acid diethylamine immunoassay screen associated with fentanyl medication. Clin Chem. 2002;48:205–6. [PubMed] [Google Scholar]
- 11.Ritter D, Cortese CM, Edwards LC, Barr JL, Chung HD, Long C. Interference with testing for lysergic acid diethylamide. Clin Chem. 1997;43:635–7. [PubMed] [Google Scholar]
- 12.Fitzgerald RL, Herold DA. Improved CEDIA benzodiazepine assay eliminates sertraline cross-reactivity. J Anal Toxicol. 1997;21:32–5. doi: 10.1093/jat/21.1.32. [DOI] [PubMed] [Google Scholar]
- 13.Nishikawa T, Ohtani H, Herold DA, Fitzgerald RL. Comparison of assay methods for benzodiazepines in urine. A receptor assay, two immunoassays, and gas chromatography-mass spectrometry. Am J Clin Pathol. 1997;107:345–52. doi: 10.1093/ajcp/107.3.345. [DOI] [PubMed] [Google Scholar]
- 14.Sena SF, Kazimi S, Wu AH. False-positive phencyclidine immunoassay results caused by venlafaxine and O-desmethylvenlafaxine. Clin Chem. 2002;48:676–7. [PubMed] [Google Scholar]
- 15.Bender A, Glen RC. Molecular similarity: a key technique in molecular informatics. Org Biomol Chem. 2004;2:3204–18. doi: 10.1039/B409813G. [DOI] [PubMed] [Google Scholar]
- 16.Ekins S, Mestres J, Testa B. In silico pharmacology for drug discovery: applications to targets and beyond. Br J Pharmacol. 2007;152:21–37. doi: 10.1038/sj.bjp.0707306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willett P. Similarity-based approaches to virtual screening. Biochem Soc Trans. 2003;31:603–6. doi: 10.1042/bst0310603. [DOI] [PubMed] [Google Scholar]
- 18.Gillet VJ, Willett P, Bradshaw J. Identification of biological activity using substructural analysis and genetic algorithms. J Chem Inf Comput Sci. 1998;38:165–79. doi: 10.1021/ci970431+. [DOI] [PubMed] [Google Scholar]
- 19.Hert J, Willett P, Wilton DJ, Acklin P, Azzaoui K, Jacoby E, Schuffenhauer A. Comparison of fingerprint-based methods for virtual screening using multiple bioactive reference structures. J Chem Inf Comput Sci. 2004;44:1177–85. doi: 10.1021/ci034231b. [DOI] [PubMed] [Google Scholar]
- 20.Hert J, Willett P, Wilton DJ, Acklin P, Azzaoui K, Jacoby E, Schuffenhauer A. Comparison of topological descriptors for similarity-based virtual screening using multiple bioactive reference structures. Org Biomol Chem. 2004;2:3256–66. doi: 10.1039/B409865J. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Q, Muegge I. Scaffold hopping through virtual screening using 2D and 3D similarity descriptors: ranking, voting, and consensus scoring. J Med Chem. 2006;49:1536–48. doi: 10.1021/jm050468i. [DOI] [PubMed] [Google Scholar]
- 22.ElSohly MA, Jones AB, elSohly HN. Cross-reactivity of selected compounds in the Abbott TDx cannabinoid assay. J Anal Toxicol. 1990;14:277–9. doi: 10.1093/jat/14.5.277. [DOI] [PubMed] [Google Scholar]
- 23.Mura P, Kintz P, Robert R, Papet Y. Buflomedil is a potent interfering substance in immunoassays of tricyclic antidepressants. J Anal Toxicol. 1998;22:254. doi: 10.1093/jat/22.3.254. [DOI] [PubMed] [Google Scholar]
- 24.Nebinger P, Koel M. Specificity data of the tricyclic antidepressants assay by fluorescent polarization immunoassay. J Anal Toxicol. 1990;14:219–21. doi: 10.1093/jat/14.4.219. [DOI] [PubMed] [Google Scholar]
- 25.Paolini GV, Shapland RH, van Hoorn WP, Mason JS, Hopkins AL. Global mapping of pharmacological space. Nat Biotechnol. 2006;24:805–15. doi: 10.1038/nbt1228. [DOI] [PubMed] [Google Scholar]
- 26.Chang C, Bahadduri PM, Polli JE, Swaan PW, Ekins S. Rapid identification of novel P-glycoprotein ligands. Drug Metab Dispos. 2006;34:1976–84. doi: 10.1124/dmd.106.012351. [DOI] [PubMed] [Google Scholar]
- 27.Ekins S, Johnston JS, Bahadduri P, D'Souza VM, Ray A, Chang C, Swaan PW. In vitro and pharmacophore-based discovery of novel hPEPT1 inhibitors. Pharm Res. 2005;22:512–7. doi: 10.1007/s11095-005-2505-y. [DOI] [PubMed] [Google Scholar]
- 28.Gomella LG, Haist S, Adams AG, Smith KM. Clinician's pocket drug reference 2008. New York: McGraw-Hill Companies; 2008. [Google Scholar]
- 29.Red Book. Montvale, NJ: Thomson Healthcare; 2008. [Google Scholar]
- 30.Dasgupta A, Wells A, Datta P. False-positive serum tricyclic antidepressant concentrations using fluorescence polarization immunoassay due to the presence of hydroxyzine and cetirizine. Ther Drug Monit. 2007;29:134–9. [PubMed] [Google Scholar]
- 31.Todeschini R, Consonni V. Handbook of molecular descriptors. Weinheim: Wiley-VCH; 2000. [Google Scholar]
- 32.Lim K, Owens SM, Arnold L, Sacchettini JC, Linthicum DS. Crystal structure of monoclonal 6B5 Fab complexed with phencyclidine. J Biol Chem. 1998;273:28576–82. doi: 10.1074/jbc.273.44.28576. [DOI] [PubMed] [Google Scholar]
- 33.Larsen NA, Zhou B, Heine A, Wirsching P, Janda KD, Wilson IA. Crystal structure of a cocaine-binding antibody. J Mol Biol. 2001;311:9–15. doi: 10.1006/jmbi.2001.4839. [DOI] [PubMed] [Google Scholar]
- 34.Pozharski E, Moulin A, Hewagama A, Shanafelt AB, Petsko GA, Ringe D. Diversity in hapten recognition: structural study of an anti-cocaine M82G2. J Mol Biol. 2005;349:570–82. doi: 10.1016/j.jmb.2005.03.080. [DOI] [PubMed] [Google Scholar]
- 35.Pozharski E, Wilson MA, Hewagama A, Shanafelt AB, Petsko G, Ringe D. Anchoring a cationic ligand: the structure of the Fab fragment of the anti-morphine antibody 9B1 and its complex with morphine. J Mol Biol. 2004;337:691–7. doi: 10.1016/j.jmb.2003.12.084. [DOI] [PubMed] [Google Scholar]
- 36.Jeffrey PD, Strong RK, Sieker LC, Chang CY, Campbell RL, Petsko GA, et al. 26-10 Fab-digoxin complex: affinity and specificity due to surface complementarity. Proc Natl Acad Sci U S A. 1993;90:10310–4. doi: 10.1073/pnas.90.21.10310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.