Abstract
This study describes the molecular and biochemical characterization of kynurenine aminotransferase (KAT) from Aedes aegypti mosquitoes. Through screening an A. aegypti larval cDNA library, a 1695-bp full-length cDNA clone with a 1434-bp open reading frame (ORF) was isolated. Its deduced amino acid sequence consists of 477 amino acid residues with a predicted molecular mass of 53,490 and the amino acid sequence shares 47 and 42% identity with KATs from Homo sapiens and Rattus norvegicus, respectively. This putative A. aegypti KAT (AeKAT) has four potential N-glycosylation sites (Asn-Xxx-Trp/Ser) and a typical mitochondrial leader sequence consisting of 49 amino acids at its NH2-terminus with a putative cleavage site between Met-49 and Ser-50. A consensus pyridoxal 5′-phosphate (PLP) binding domain (Gly-Ser-Ala-Gly-Lys-Thr-Phe-Ser) is present in the central region of the ORF. Using a baculovirus/insect cell expression system, a full-length AeKAT and a truncated AeKAT, lacking the mitochondrial leader sequence, were expressed. The full-length AeKAT expressed in Sf9 insect cells is insoluble and has no detectable KAT activity, but the truncated AeKAT is soluble and capable of catalyzing the transamination of kynurenine to kynurenic acid in the presence of pyruvate as an amino group acceptor. However, the expressed truncated protein is not active to 3-hydroxykynurenine. Northern analysis indicates that transcription of the AeKAT occurs at all stages during mosquito development, but higher levels of mRNA are observed during the pupal and adult stages. Results indicate that a specific KAT is present in mosquitoes and the enzyme may play an important physiological role in A. aegypti.
Keywords: Aedes aegypti, Baculovirus expression, Kynurenine aminotransferase, Kynurenic acid, Kynurenine
1. Introduction
Kynurenic acid (KA) is a metabolite in the tryptophan (Trp) oxidation pathway. In mammals, KA functions as a broad-spectrum antagonist at ionotropic excitatory amino acid receptors (NMDA receptors), which protects the central nervous system (CNS) from over-stimulation by excitatory cytotoxins (Beal et al., 1990, 1992; Moroni et al., 1991; Moroni, 1999; Stone, 2000a, b). For example, cotreatment of KA blocked quinolinic acid (a major endogenous excitatory cytotoxin) induced lesions in the CNS of rats (Moroni et al., 1991). Patients with neurodegeneration related diseases (i.e. Huntington’s and Parkinson’s diseases, and Down’s syndrome) often show a low concentration of KA and a high concentration of quinolinic acid (Beal et al., 1990, 1992). Because of the neuroprotective function of KA, the mammalian kynurenine aminotransferase (KAT, EC 2.6.1.7) that is responsible for the production of KA has attracted considerable attention (Stone, 2000a, b). Kynurenine is the direct precursor for the production of KA and the overall reactions involves an initial transamination of kynurenine to an α-keto acid intermediate by KAT and a subsequent non-enzymatic intramolecular cyclization of the intermediate to KA.
KAT should be present in other animals and play a role in maintaining physiological levels of KA. However, except for studies of human and rat KATs (Mosca et al., 1994; Alberati-Giani, et al., 1995; Buchli et al., 1995; Malherbe et al., 1995; Perry et al., 1995), there have been no detailed studies dealing with the characterization of KAT from other species. In our study of Trp metabolism in mosquitoes, we determined that the Trp to xanthurenic acid (XA) pathway via kynurenine and 3-hydroxykynurenine (3-HK) intermediates is the major catabolic pathway for Trp in Aedes aegypti mosquitoes. Our previous data suggest that A. aegypti has a transaminase that has a higher specific activity to 3-HK than to kynurenine (Li and Li, 1997). Because mammalian KAT catalyzes the transamination of both kynurenine and 3-HK, we presumed that a mosquito transaminase similar to mammalian KAT was present in A. aegypti, and that the enzyme was responsible for both maintaining physiological levels of KA and preventing accumulation of 3-HK in the mosquitoes.
To characterize the enzyme responsible for the transamination of 3-HK and kynurenine in mosquitoes, we designed degenerate primers based on the conserved regions of the mammalian KATs. We amplified a cDNA fragment from a mosquito larval cDNA pool and then isolated a putative KAT clone through the screening of an A. aegypti larval cDNA library with the amplified cDNA fragment as a probe. Subsequent expression of the isolated clone in a baculovirus/insect expression system resulted in the production of a recombinant protein capable of catalyzing the transamination of kynurenine to KA. However, the expressed protein unexpectedly showed no 3-HK transamination activity. In this report, we provide data that describe the isolation of the putative AeKAT clone, its functional identification as a KAT, and its expression in mosquitoes during development. We also discuss the likely physiological role AeKAT plays in mosquitoes.
2. Materials and methods
2.1. A. aegypti rearing and maintenance
A. aegypti used in this study were reared according to the methods described (Li et al., 1996). All mosquitoes were maintained at 25 ± 0.5°C, 60% RH and on a 16 h light:8 h dark cycle with a 90 min crepuscular period at the beginning and end of each light cycle.
2.2. Chemicals
L-aminoadipate, 2-amino-2-methyl-1-propanol, formic acid, 3-hydroxy-DL-kynurenine (3-HK), α-ketoadipate, α-ketoglutarate, α-keto-γ-methiolbutyrate (α-KMB), DL-kynurenine, kynurenic acid (KA), L-kynurenine, β-mercaptoethanol (β-ME), oxalacetate, phenyl-methylsulfonyl fluoride (PMSF), pyridoxal 5′-phosphate (PLP), pyruvic acid and xanthurenic acid (XA) were purchased from Sigma (St Louis, MO). Protein molecular weight markers, DEAE sepharose and Bio-Sil gel filtration columns (7.8 × 300mm2) were from Bio Rad (Hercules, CA).
2.3. PCR amplification
A forward degenerate primer (5′-GGV GAY GAG GTS ATM ATY ATT GAA CC-3′) and a reverse degenerate primer (5′-CCA NCC NAN YTT CCA NCC NGT) were designed based on the conserved regions (GDEVIIIEP and TGWKIGW, respectively) of human and rat KATs, and used for PCR amplification of the first strand cDNA synthesized from total RNA of 3-day-old A. aegypti larvae using a cDNA synthesizing kit (Life Technologies). A 430-bp fragment was amplified, cloned into a PCR2.1-TOPO TA cloning vector (Invitrogen) and then sequenced. Blast search of Gen-Bank databases showed that the 430-bp partial cDNA shared about 45% sequence homology with mammalian KATs.
2.4. cDNA cloning and sequencing
An A. aegypti larval cDNA library was constructed using a library construction kit (Stratagene) according to the manufacturer’s instructions. The 430-bp fragment was labeled with α-32P-dCTP (NEN Life Science) and used for cDNA library screening. A total of 5 × 105 plaques were screened at high stringency. Ten clones were isolated and three of them were sequenced. DNA sequencing reactions were performed using the BigDye™ Terminator Cycle Sequencing Kit (PE Applied Biosystems) from both ends with primer walking. Sequence assembly was carried out with SeqEdit software (V1.0.3). Sequence data were analyzed using the Biology WorkBench 3.2 program (http://workbench.sdsc.edu) and the software package from Genetics Computer Group, Inc (GCG, University of Wisconsin, Madison).
2.5. Northern hybridization
Total RNA was isolated (using Trizol reagent, Life Technologies) from larvae at 1–6 days after hatching, from pupae at 0.5 and 12 h after pupation, from adults at 3-day after emerging, and from ovaries collected at 24 h post-bloodfeeding. Total RNA was electrophoresed in a 1% agarose-formadehyde gel in 20 mM MOPS buffer containing 4 mM sodium acetate and 1 mM EDTA at 5 V/cm for 3 h. RNA was transferred to a positive charge nylon membrane (Ambion) and crosslinked using a Bio-Rad UV crosslinker. The blot was hybridized sequentially with the 32P-dCTP labeled 430-bp fragment from AeKAT cDNA and then with a 500-bp probe generated from the 18S ribosomal RNA of A. aegypti as a loading control. After hybridization at 42°C for 16 h, the blots were washed with increasing stringency (twice in 2 × SSC containing 0.1% SDS at room temperature for 25 min, and twice with 0.1 × SSC containing 0.1% SDS at 68°C for 30 min), and exposed to X-ray films at −80°C.
2.6. Recombinant transfer vector construction
A 1434-bp full-length open reading frame (ORF) was amplified from the full-length AeKAT cDNA using a forward primer (5′-GGCCTCGAGATGATGTTTCTC CGT-3′) containing an Xho I site, and a reverse primer (5′-CCGGAATTCTTACGATGAACCTTTCC-3′) containing an EcoR I site. A 1290-bp truncated AeKAT cDNA fragment lacking the NH2-terminal mitochondrial leader sequence (amino acids 1–49) was amplified using a forward primer (5′-GGCCTCGAGATGTCCTCTA-CATCC-3′) containing an Xho I site and the same reverse primer as above (the nucleotides of restriction sites are underlined). Both the 1434-bp full-length ORF and the 1290-bp truncated AeKAT (coding amino acids 50–477) were inserted into a PCR2.1-TOPO TA cloning vector and then subcloned into a baculovirus transfer vector pBlueBac4.5 (Invitrogen) between the Xho I and the EcoR I restriction sites.
2.7. Recombinant baculoviruses isolation
All recombinant transfer vectors were sequenced and verified that the inserted genes were in-frame and controlled downstream of a polyhedrin promoter. The full-length AeKAT or truncated AeKAT recombinant pBlue-Bac4.5 transfer vectors were cotransfected with linearized Bac-N-Blue™ (AcMNPV, Autographa californica multiple nuclear polyhedrosis virus) viral DNA in the presence of InsectinPlus™ insect cell-specific liposomes to Spodoptera frugiperda (Sf9) insect cells (Invitrogen). The recombinant baculoviruses were purified by a plaque assay. Blue putative recombinant plaques were transferred to 12-well microtiter plates and amplified in Sf9 cells. Viral DNA was isolated for PCR analysis to determine the purity of recombinant viruses. High-titer viral stocks (HTS) of a pure recombinant virus were generated through amplification in suspension cultured Sf9 cells. The titer of HTS and the time-course at a multiplicity of infection (MOI) of 6 was established according to the manufacturer’s instructions (Invitrogen).
2.8. Recombinant AeKAT expression and purification
Sf9 cells were cultured in spinner flasks in TNM-FH medium containing 10% fetal bovine serum (Gibco BRL). HTS of the pure recombinant virus with an MOI of 6 was inoculated into the culture at a cell density of 2.5 × 106cells/ml. Sf9 cells were harvested at the fourth day after AeKAT recombinant virus inoculation by centrifugation (800g for 15 min at 4°C), and the cell pellets were dissolved in a lysis buffer (0.1% Triton X-100 in PBS) containing 1 mM PMSF. After incubation on ice for 30 min, cell lysates were centrifuged at 18,000g for 20 min at 4°C and the supernatant was collected. The insoluble cell lysate sediments were treated with 2% SDS for 30 min at room temperature and then centrifuged at 18,000g for 20 min to obtain the SDS-solubilized protein. To determine the presence and the relative levels of the expression of the recombinant AeKAT (rAeKAT), both soluble and SDS-solubilized protein samples from cells infected with the full-length AeKAT recombinant virus or truncated AeKAT recombinant virus were analyzed by SDS-PAGE.
Initial AeKAT activity assays indicated that the truncated AeKAT protein expressed by the Sf9 cells was active in the transamination of kynurenine to KA; therefore, the rAeKAT lacking the NH2-terminal mitochondrion leader sequence was purified from the soluble cytosolic fractions for biochemical assays. DEAE sepharose chromatography, native preparative PAGE, and gel filtration chromatography were used to purify the recombinant protein, and purity of the protein was assessed by SDS-PAGE analysis. Protein concentration was determined by a Bio-Rad protein assay kit using bovine serum albumin as a standard.
2.9. Biochemical characterization
KAT activity assay was based on methods described earlier (Okuno et al., 1990; Li and Li, 1997). Briefly, a reaction mixture of 50 μl containing 10 mM L-kynurenine or 5 mM 3-HK, 12 mM pyruvate, 70 μM PLP and a varying amount of protein sample was prepared using 200 mM tris buffer, pH 8.5. The mixture was incubated for 10 min at 50°C, and the reaction was stopped by adding an equal volume of 0.8 M formic acid. Supernatant was obtained by centrifugation of the reaction at 15,000g at 4°C for 10 min and analyzed by HPLC-UV at 330 or 340 nm for KA or XA, respectively. To determine the cosubstrate specificity for the rAeKAT, pyruvate was replaced with either α-ketoglutarate, α-ketoadipate, α-KMB or oxalacetate in the above reaction mixture and the capacity of these keto acids to serve as amino acceptors for the enzyme was compared to that of pyruvate. To determine the effect of temperature on AeKAT activity, the substrate preparation of 45 μl, containing 11.1 mM kynurenine, 12 mM pyruvate, and 70 μM PLP in 100 mM tris buffer, pH 8.5, was incubated at 35, 40, 45, 50, 55, 60, 65 or 70°C for 5 min prior to the addition of 5 μl of the enzyme preparation and continuously incubated for 10 min after addition of purified rAeKAT. The amount of KA formed in the reaction mixture was determined by HPLC-UV analysis. The effect of pH on AeKAT activity was determined by preparation of the typical reaction mixtures in 200 mM acetate buffer (pH 5.5), phosphate buffer (pH 6.0–7.5), tris buffer (pH 8–9) or 2-amino-2-methyl-1-propanol buffer (9.5–11), respectively. The amount of KA produced in the reaction mixture was quantified by HPLC-UV analysis.
To evaluate the correlation between the mosquito KAT activity and AeKAT mRNA expression, crude larval, pupal and adult protein was assayed for KAT activity with either kynurenine or 3-HK as the amino group donor and pyruvate as the amino group acceptor. Four-day-old larvae, 12 h pupae or 3-day-old adults were homogenized in 100 mM phosphate buffer (pH 7.0) containing 1 mM PMSF, 1 mM PTU and 10 mM β-ME, and supernatant was obtained by centrifugation of the homogenates at 20,000g for 10 min at 2°C. Protein in the supernatant was precipitated by the addition of solid ammonium sulfate to 60% saturation. The precipitated larval, pupal or adult protein was used for KAT activity assays. The specific activity was expressed as nmol/min/mg crude larval, pupal or adult protein.
3. Results
3.1. Molecular cloning and sequencing
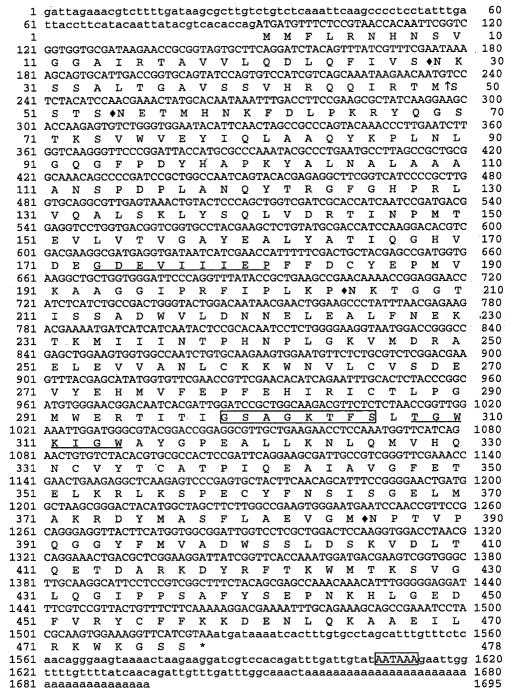
Ten positive clones were isolated after screening the cDNA library, and three clones were sequenced. Except for the difference in the number of nucleotides, the sequences of the three cDNA inserts were identical. The 1695-bp cDNA insert (longest among the three sequenced clones) has two in-frame stop codons at nucleotide number 4 and 58 of the 5′-untranslated region and a complete ORF of 1434-bp starting and ending at nucleotide numbers 91 and 1524, respectively. The 3′-untranslated region has a typical polyadenylation signal (AATAAA) located 44-bp upstream of the poly A tail (Fig. 1).
Fig. 1.
Nucleotide sequence and deduced amino acid sequence of AeKAT. The underlined sequences represent regions used for designing degenerate primers. Arrow indicates the potential cleavage site of the mitochondrion leader sequence. Diamond indicates the potential N-linked glycosylation sites. Boxed amino acid sequence represents the putative PLP binding site. Asterisk represents the stop condon (TAA). Boxed nucleotide sequence represents the polyadenylation signal (AATAAA) at the 3′-untranslated region. The nucleotide sequence of KAT presented in this report had been deposited in the GenBank database (accession number AF395204).
3.2. Deduced amino acid sequences
The ORF of the isolated clone codes for a protein of 477 amino acid residues with a predicted molecular mass of 53,490 (Fig. 1). Analysis of the deduced sequence with a TargetP program (http://www.cbs.dtu.dk) predicted a typical mitochondrial leader sequence consisting of 49 amino acids at the NH2-terminal region and a potential cleavage site between Met-49 and Ser-50. There are four potential N-glycosylation sites (Asn-Xxx-Trp/Ser) in the protein. A highly conserved PLP binding site (Gly-Ser-Ala-Gly-Lys-Thr-Phe-Ser) is located between Gly-299 and Ser-306 (Figs. 1 and 2).
Fig. 2.
Alignment of deduced amino acid sequence of AeKAT with the four most similar KAT sequences. The GenBank accession numbers for each of the protein sequences are as follows: AeKAT (A. aegypti KAT, AF395204), HsKAT (Homo sapiens KAT, XM_011753), RnKAT (Rattus norvegicus KAT, Z49696), FrKAT (Fugu rubripes putative KAT, Y17462), and DmKAT (Drosophila melanogaster putative KAT, AE003693). Boxed amino acid sequences represent the PLP binding site.
3.3. Amino acid sequence homologies
Using the CLUSTALW program of multiple sequence alignment, amino acid sequence comparison shows that the mosquito clone shares 47% identity with Homo sapiens KAT, 42% with Rattus norvegicus KAT, and 49% with Fugu rubripes putative KAT, respectively (Fig. 2). Blast search of Drosophila genomic database identified a putative KAT (NCB protein accession number: AAF54708) sharing 72% identity with the mosquito clone.
3.4. Expression profile during development
The expression profile of the AeKAT was evaluated by Northern analysis (Fig. 3). AeKAT mRNA (about 1.7 kb) was detected in all stages during mosquito development, including developing ovaries. Although the AeKAT cDNA clone was isolated from a larval cDNA library, levels of its transcript were low during larval development (lanes 1–6). Its mRNA quantities increased at the beginning of pupal development (lanes 7 and 8) and the highest transcript levels were observed during the adult stage (lane 9). When RNA was extracted separately from adult bodies and adult heads, the level of transcripts in the heads (lane 12) was greater than that in the bodies (lane 11).
Fig. 3.

Expression profiles of AeKAT gene during development. Total RNA was extracted from mosquitoes at different developmental stages. Total RNA from individual samples (5 μg per sample) was electrophoresed on 1% agarose-formadehyde gel, blotted on nylon membrane, and sequentially hybridized with a 430-bp 32P-dCTP labeled AeKAT probe (top panel) and a 500-bp 32P-dCTP labeled 18S rRNA probe (bottom panel). Total RNA samples were isolated from 1- to 6-day-old larvae (lanes 1–6), newly formed white pupae (lane 7), 12 h black pupae (lane 8), 3-day-old adult (lane 9), ovaries at 24 h post-bloodfeeding (lane 10), adult bodies (lane 11), and adult heads (lane 12), respectively.
3.5. rAeKAT expression and purification
Cotransfection of the linearized AcMNPV DNA and baculovirus transfer vectors containing either the full-length AeKAT or the truncated AeKAT sequence lacking the mitochondrial leader sequence resulted in the successful isolation of the recombinant viruses. When the soluble protein samples from Sf9 cells at the fourth day after inoculation were analyzed by SDS-PAGE, the full-length rAeKAT was not observed in the soluble sample (Fig. 4, lane B1), but the truncated rAeKAT with an Mr of 48,000 was clearly detected (Fig. 4, lane B2). In contrary, when SDS-solubilized protein samples (derived from SDS treatment of the insoluble cell lysate precipitate) were analyzed by SDS-PAGE, the full-length rAeKAT with an Mr of 53,000 was present (Fig. 4, lane B4), but the truncated rAeKAT was not observed (Fig. 4, lane B5).
Fig. 4.
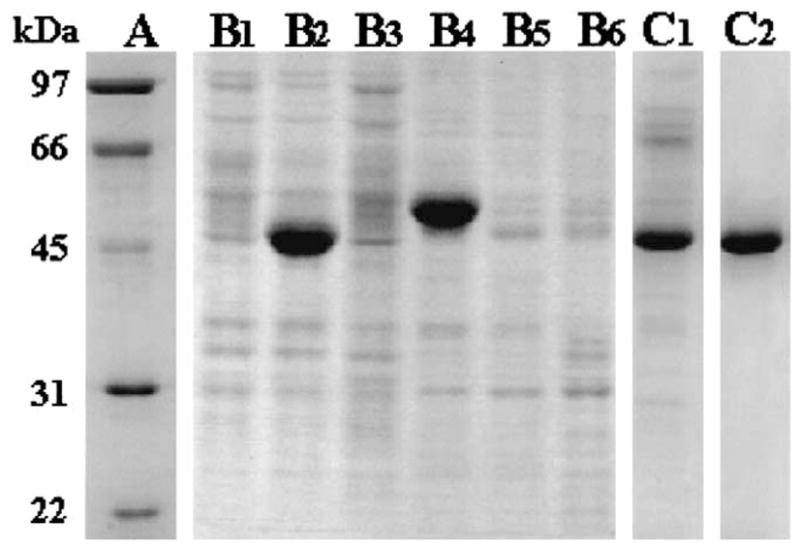
Recombinant AeKAT expression and purification. Sf9 cells were harvested at the fourth day after infection with the full-length, truncated AeKAT recombinant baculoviruses or AcMNPV wild type virus. Soluble cytosolic protein and SDS-solubilized protein were obtained as described in Section 2 and subjected to SDS-PAGE analysis. Lane A shows the protein molecular mass standard. Lanes B1, B2, and B3 represent soluble cytosolic protein from cells infected with full-length AeKAT recombinant virus, truncated AeKAT recombinant virus and wild type virus, respectively. Lanes B4, B5 and B6 are SDS-solubilized protein from the pellets of B1, B2 and B3 samples, respectively. Lanes C1 and C2 illustrate the remaining protein after DEAE sepharose chromatography and preparative native PAGE separation of truncated rAeKAT, respectively. The relative amount of protein is 20 μg (lane B1), 35 μg (lane B2), 25 μg (lane B3), 25 μg (lane B4), 10 μg (lanes B5 and B6), 12 μg (lane C1) and 8 μg (lane C2), respectively.
KA was not observed when either soluble protein or insoluble cell lysate precipitate, obtained from Sf9 cells infected with the full-length AeKAT recombinant virus, was incubated with kynurenine, PLP and pyruvate (not shown). In contrary, production of KA was observed when soluble protein, obtained from Sf9 cells infected with the truncated AeKAT recombinant virus, was incubated with the same substrate preparation (not shown). Therefore, the truncated rAeKAT, lacking the NH2-terminal mitochondrial leader sequence, was purified from the supernatant of cell lysate for biochemical characterization. Purification of the rAeKAT was achieved by DEAE sepharose and preparative native PAGE. A number of other proteins were eliminated from the supernatant of cell lysate after DEAE sepharose chromatography (Fig. 4, lane C1), and the target protein became the major one (>95%) after preparative electrophoresis (Fig. 4, lane C2). A single absorbance peak with a calculated molecular mass of 97,000 was observed when the concentrated protein sample from preparative native PAGE was chromatographed on a gel filtration column (not shown), which suggests that the truncated recombinant AeKAT is present as a homodimer under non-denaturing conditions.
3.6. KAT activity assay
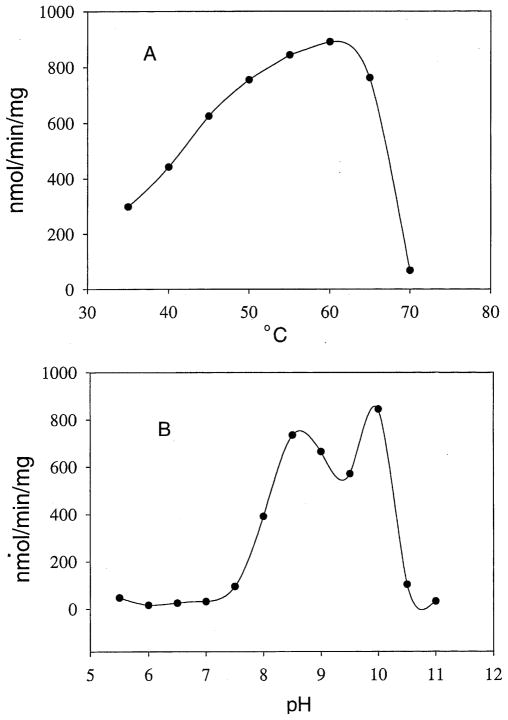
The truncated rAeKAT was capable of catalyzing the transamination of kynurenine in the presence of pyruvate and PLP; surprisingly, however, it showed essentially no activity when 3-HK was used as the amino group donor (not shown). Therefore, kynurenine was used as the amino group donor for subsequent biochemical characterization of the rAeKAT. It was found that the amino acceptors used in the reaction mixture had a major impact on the specific activity of rAeKAT. Among the tested α-keto acids, pyruvate was the most effective amino acceptor for the recombinant AeKAT (Table 1). Both temperature and pH also influenced KAT activity. The protein showed the highest activity at a temperature around 60°C (Fig. 5A). The rAeKAT was more active at basic conditions with two optimum pHs at 8.5 and 10, respectively (Fig. 5B).
Table 1.
The cosubstrate specificity of recombinant AeKAT
| Co-substrates | α-Ketoadipate | α-Ketoglutarate | α-KMB | Oxalacetate | Pyruvate |
|---|---|---|---|---|---|
| KAT activity, % | 25 | 31 | 11 | 90 | 100 |
Purified recombinant AeKAT was incubated in the presence of keto acids and L-kynurenine as described in Section 2. Data are expressed as a percentage of the activity of that with pyruvate as amino acceptor and are the means of two separate experiments.
Fig. 5.
Effect of temperature and pH on rAeKAT activity. The detailed assay methods are described in Section 2. (A) AeKAT activity at temperatures from 35 to 70°C, respectively. (B) AeKAT activity at pHs from 5.5 to 11, respectively.
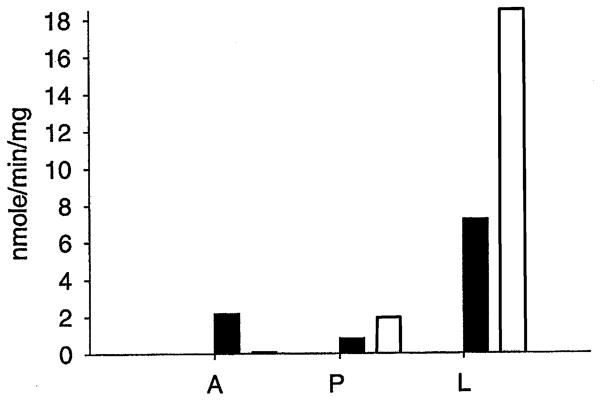
When crude protein from 4-day-old larvae was mixed with pyruvate, PLP, kynurenine or 3-HK, formation of KA or XA was observed in the corresponding reaction mixtures (Fig. 6L). Production of KA or XA was also detected when the crude pupal protein was mixed with the above reaction mixtures (Fig. 6P), but the specific activity of pupal protein to kynurenine and 3-HK was greatly decreased. Protein extracted from adult mosquitoes also catalyzed the transamination of kynurenine to KA, but had essentially no detectable activity to 3-HK (Fig. 6A).
Fig. 6.
Specific KAT activity of crude larval, pupal and adult protein. Protein from 4-day-old larvae, 12 h pupae and 3-day-old adult mosquitoes was extracted as described in Section 2. The reaction mixture of 100 μl consisting of 10 mM kynurenine or 5 mM 3-HK, 70 μM PLP, 12 mM pyruvate and 0.5 mg of crude larval, pupal or adult protein was prepared in 100 mM tris buffer (pH 8.5) and incubated at 50°C for 10 min. KA or XA produced in the corresponding reaction mixtures was quantified by HPLC-UV analysis. Solid bar and open bar illustrate the specific activity of crude larval (L), pupal (P) and adult (A) protein to kynurenine and 3-HK, respectively.
4. Discussion
In mammals, KA functions as a broad-spectrum antagonist at ionotropic excitatory amino acid receptors (NMDA receptors) in CNS, which protects these receptors from over-stimulation by excitatory cytotoxins. Because of that, KAT, the enzyme responsible for the production of KA in mammals, has attracted a considerable attention (Beal et al., 1990, 1992; Moroni et al., 1991; Moroni, 1999; Stone, 2000a, b). The high sequence similarity of the isolated A. aegypti clone with mammalian KATs and its functional expression in the recombinant baculovirus/insect expression system verified the presence of a specific KAT in A. aegypti. Our data suggest that the AeKAT may play a similar physiological role as that described for human and rat KATs (Okuno et al., 1990; Perry et al., 1993; Alberati-Giani et al., 1995; Buchli et al., 1995; Perry et al., 1995), especially during adult stage.
Although mammalian KAT catalyzes the transamination of both kynurenine and 3-HK, it has generally been accepted that the major physiological role for mammalian KAT is to maintain physiological levels of KA, which seems critical for mammals to counteract excitatory cytotoxins. Similar to mammalian KAT, the AeKAT may play a similar physiological role in the mosquitoes. However, the low levels of AeKAT transcript seen in mosquitoes during larval development raises a question: if the enzyme is critical for maintaining physiological levels of KA in mosquitoes, why the enzyme does not have to be as actively expressed in larvae as it does in adults. In a previous study, we showed that oxidation of Trp to 3-HK via kynurenine intermediate was a major pathway of Trp catabolism in mosquitoes during larval development and proposed that a specific transaminase, which was more active to 3-HK than to kynurenine, was present in A. aegypti and that this enzyme prevented the accumulation of 3-HK in mosquitoes during larval development (Li and Li, 1997). Recently, we just achieved the purification of the A. aegypti 3-HK transaminase and isolated its cDNA clone (GenBank database accession number AF 435806) from a larval cDNA library. Sequence comparison revealed 4% sequence similarity between the mosquito 3-HK transaminase and the KAT (unpublished data). Although the larval 3-HK transaminase is more active to 3-HK, it also catalyzes the transamination of kynurenine to KA (see Fig. 6 as a reference), which may explain the relatively low levels of AeKAT mRNA in mosquito larvae. In contrary, there is an extremely low level of 3-HK transaminase activity in pupae and adults; consequently, production of KA may depend heavily on AeKAT during these stages, which may explain the increased level of AeKAT mRNA in pupae and adults. Although mosquito KAT is more active at relatively high temperatures, the level of its activity at physiological temperature may be adequate for maintaining a physiological level of KA. Based on northern analysis, there is a relatively high level of AeKAT mRNA transcript in mosquito heads (Fig. 3, lane 12), which provides additional support for the notion that KA may be required for the CNS system in mosquitoes.
The mosquito larval 3-HK transaminase that is capable of catalyzing the transamination of both kynurenine and 3-HK (see Fig. 6) should be able to produce the required KA in mosquitoes. This leads to a question as to why the 3-HK transaminase has to be down-regulated, while the AeKAT is up-regulated for the production of KA during pupal and adult stages. Our previous data suggest that the Trp catabolism is quite different in mosquitoes as compared to that of mammals. Unlike mammals, mosquitoes cannot completely oxidize kynurenine or 3-HK to CO2 and H2O due to the absence of kynureninase. In mosquito larvae, production of 3-HK is a major pathway of Trp catabolism, but 3-HK oxidizes easily, which stimulates the production of reactive oxygen species (Li et al., 1999). Data from our previous study suggest that the transamination of 3-HK to chemically stable XA is the mechanism by which mosquitoes prevent the accumulation of the chemically reactive and potentially toxic 3-HK (Li and Li, 1997). Interestingly, however, 3-HK is also a substrate for the biosynthesis of ommochromes (major eye pigments in mosquitoes) during pupal and earlier adult stages (Li et al., 1999). Coincidently, the 3-HK transaminase activity becomes extremely low in pupae and essentially undetectable in adults, while at the same time accumulation of 3-HK in the compound eyes of pupae and adult mosquitoes becomes apparent (Li et al., 1999). It seems that the mosquito KAT as well as 3-HK transaminase is regulated in a manner that reflects the specific physiological requirements associated with different developmental stages.
In summary, results from this study demonstrate the presence of a specific AeKAT in A. aegypti mosquitoes, which shares high sequence identity with mammalian KATs and shows similar biochemical activities. AeKAT gene expression is up-regulated during pupal and adult stages. Both its substrate specificity and expression profile during mosquito development suggest the enzyme may be responsible for maintaining a physiological level of KA in mosquitoes during development. However, the AeKAT may play other physiological roles in mosquitoes. Recently, we have achieved the crystallization of AeKAT and anticipate gaining a better understanding of the enzyme upon completion of its 3-D structural analysis.
Acknowledgments
This work was supported by National Institutes of Health Grant (AI 44399).
Footnotes
The nucleotide sequence of KAT presented in this report had been deposited in the GenBank database (accession number AF395204).
References
- Alberati-Giani D, Malherbe P, Kohler C, Lang G, Kiefer V, Lahm H, Cesura AM. Cloning and characterization of a soluble kynurenine aminotransferase from rat brain: identity with kidney cysteine conjugate β-lyase. J Neurochem. 1995;64:1448–1455. doi: 10.1046/j.1471-4159.1995.64041448.x. [DOI] [PubMed] [Google Scholar]
- Beal MF, Matson WR, Storey E, Milbury P, Ryan EA, Ogawa T, Bird ED. Kynurenic acid concentrations are reduced in Huntington’s disease cerebral cortex. J Neurol Sci. 1992;108:80–87. doi: 10.1016/0022-510x(92)90191-m. [DOI] [PubMed] [Google Scholar]
- Beal MF, Matson WR, Swartz KJ, Gamache PH, Bird ED. Kynurenine pathway measurements in Huntington’s disease striatum: evidence for reduced formation of kynurenic acid. J Neurochem. 1990;55:1327–1339. doi: 10.1111/j.1471-4159.1990.tb03143.x. [DOI] [PubMed] [Google Scholar]
- Buchli R, Alberati-Giani D, Malherbe P, Kohler C, Broger C, Cesura AM. Cloning and functional expression of a soluble form of kynurenine/α-aminoadipate aminotransferase from rat kidney. J Biol Chem. 1995;270:29330–29335. doi: 10.1074/jbc.270.49.29330. [DOI] [PubMed] [Google Scholar]
- Li J, Beerntsen BT, James AA. Oxidation of 3-hydroxkynurenine to produce xanthommatin for eye pigmentation: a major branch pathway of tryptophan catabolism during pupal development in the Yellow Fever Mosquito, Aedes aegypti. Insect Biochem Mol Biol. 1999;29:329–338. doi: 10.1016/s0965-1748(99)00007-7. [DOI] [PubMed] [Google Scholar]
- Li J, Hodgeman BA, Christensen BM. Involvement of peroxidase in chorion hardening in Aedes aegypti. Insect Biochem Mol Biol. 1996;26:309–317. doi: 10.1016/0965-1748(95)00099-2. [DOI] [PubMed] [Google Scholar]
- Li J, Li G. Transamination of 3-hydroxykynurenine to produce xanthurenic acid: a major branch pathway of tryptophan metabolism in the mosquito, Aedes aegypti, during larval development. Insect Biochem Mol Biol. 1997;27:859–867. doi: 10.1016/s0965-1748(97)00068-4. [DOI] [PubMed] [Google Scholar]
- Malherbe P, Alberati-Giani D, Kohler C, Cesura AM. Identification of a mitochondrial form of kynurenine aminotransferase/glutamine transaminase K from rat brain. FEBS Lett. 1995;367:141–144. doi: 10.1016/0014-5793(95)00546-l. [DOI] [PubMed] [Google Scholar]
- Moroni F. Tryptophan metablism and brain function: focus on kynurenine and other indole metabolites. Eur J Pharmacol. 1999;375:87–100. doi: 10.1016/s0014-2999(99)00196-x. [DOI] [PubMed] [Google Scholar]
- Moroni F, Russi P, Gallo-Mezo MA, Moneti G, Pellicciari R. Modulation of quinolinic and kynureninc acid content in the rat brain: effects of endotoxins and nicotinylalanine. J Neurochem. 1991;57:1630–1635. doi: 10.1111/j.1471-4159.1991.tb06361.x. [DOI] [PubMed] [Google Scholar]
- Mosca M, Cozzi L, Breton J, Speciale C, Okuno E, Schwarcz R, Benatti L. Molecular cloning of a rat kynurenine aminotransferase: identity with glutamine transaminase K. FEBS Lett. 1994;353:21–24. doi: 10.1016/0014-5793(94)01003-x. [DOI] [PubMed] [Google Scholar]
- Okuno E, Du F, Ishikawa T, Tsujimoto M, Nakamura M, Schwarcz R, Kydo R. Purification and characterization of kynurenine-pyruvate aminotransferase from rat kidney and brain. Brain Res. 1990;534:37–44. doi: 10.1016/0006-8993(90)90109-o. [DOI] [PubMed] [Google Scholar]
- Perry S, Harries H, Scholfield C, Lock T, King L, Gibson G, Goldfarb P. Molecular cloning and expression of a cDNA for human kidney cysteine conjugate β-lyase. FEBS Lett. 1995;360:277–280. doi: 10.1016/0014-5793(95)00123-q. [DOI] [PubMed] [Google Scholar]
- Perry S, Schofield MA, Macfarlane M, Lock EA, King L, Gibson G, Goldfarb P. Isolation and expression of a cDNA coding for rat kidney cytosolic cysteine conjugate β-lyase. Mol Pharmacol. 1993;43:660–665. [PubMed] [Google Scholar]
- Stone TW. Development and therapeutic potential of kynurenic acid and kynurenine derivatives for neuroprotection. Trends Pharmacol Sci. 2000a;21:149–154. doi: 10.1016/s0165-6147(00)01451-6. [DOI] [PubMed] [Google Scholar]
- Stone TW. Inhibitors of the kynurenine pathway. Eur J Med Chem. 2000b;35:179–186. doi: 10.1016/s0223-5234(00)00121-5. [DOI] [PubMed] [Google Scholar]