Abstract
Verification of candidate biomarkers relies upon specific, quantitative assays optimized for selective detection of target proteins, and is increasingly viewed as a critical step in the discovery pipeline that bridges unbiased biomarker discovery to preclinical validation. Although individual laboratories have demonstrated that multiple reaction monitoring (MRM) coupled with isotope dilution mass spectrometry can quantify candidate protein biomarkers in plasma, reproducibility and transferability of these assays between laboratories have not been demonstrated. We describe a multilaboratory study to assess reproducibility, recovery, linear dynamic range and limits of detection and quantification of multiplexed, MRM-based assays, conducted by NCI-CPTAC. Using common materials and standardized protocols, we demonstrate that these assays can be highly reproducible within and across laboratories and instrument platforms, and are sensitive to low µg/ml protein concentrations in unfractionated plasma. We provide data and benchmarks against which individual laboratories can compare their performance and evaluate new technologies for biomarker verification in plasma.
Proteomic technologies based on mass spectrometry (MS) have emerged as preferred components of a strategy for discovery of diagnostic, prognostic and therapeutic protein biomarkers. Because of the stochastic sampling of proteomes in unbiased analyses and the associated high false-discovery rate, tens to hundreds of potential biomarkers are often reported in discovery studies. Those few that will ultimately show sufficient sensitivity and specificity for a given medical condition must thus be culled from lengthy lists of candidates— a particularly challenging aspect of the biomarker-development pipeline and currently its main limiting step. In this context, it is highly desirable to verify, by more targeted quantitative methods, the levels of candidate biomarkers in body fluids, cells, tissues or organs from healthy individuals and affected patients in large enough sample numbers to confirm statistically relevant differences1,2. Verification of novel biomarkers has relied primarily on the use of sensitive, specific, high-throughput immunoassays, whose development depends critically on the availability of suitable well-characterized antibodies. However, antibody reagents of sufficient specificity and sensitivity to assay novel protein biomarkers in plasma are generally not available. The high cost and long development time required to generate high-quality immunoassay reagents, as well as technical limitations in multiplexing immunoassays for panels of biomarkers, is strong motivation to develop more straightforward quantitative approaches exploiting the sensitivity and molecular specificity of mass spectrometry.
Recently, multiple reaction monitoring (MRM) coupled with stable isotope dilution (SID)-MS for direct quantification of proteins in cell lysates as well as human plasma and serum has been shown to have considerable promise3–10. With SID-MRM-MS, up to tens of candidate proteins can be nearly simultaneously targeted and quantified in plasma by detecting ‘signature’ peptides, those that are diagnostic for each protein8,9. These reports suggest that this technology may be suitable for use in preclinical studies to rapidly screen large numbers of candidate protein biomarkers in the hundreds of patient samples necessary for verification2. Widespread acceptance and adoption of SID-MRM-MS methods are presently limited because the reproducibility and transferability of protein-based MRM assays across different instrument platforms and laboratories have yet to be demonstrated. To address this issue, the Clinical Proteomic Technology Assessment for Cancer network of the National Cancer Institute (NCI-CPTAC) evaluated intra- and interlaboratory analytical performance of SID-MRM-MS assays for quantifying seven target proteins added to human plasma. Our study demonstrates that targeted, quantitative and multiplexed MS-based assays can be rapidly configured and deployed in multiple laboratories to reproducibly measure proteins present at moderate to high abundance (>2 µg/ml), with a linear dynamic range spanning three orders of magnitude, in nondepleted, nonfractionated plasma, the most complex of all biological matrices.
RESULTS
Study design
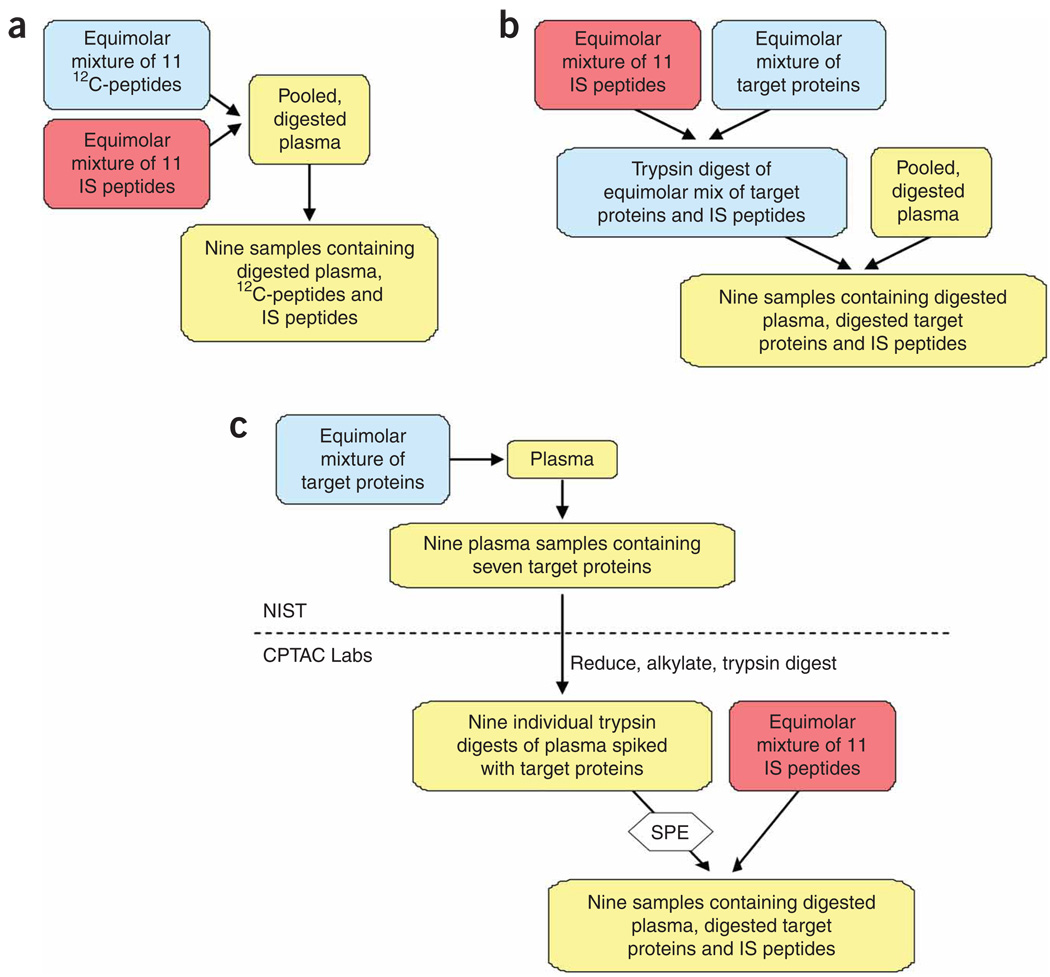
A series of interrelated studies was designed to assess the reproducibility and quantitative characteristics of MRM assays across the eight participating laboratories for measurement of peptides and proteins in the context of human plasma. The studies (I–III) sequentially introduced additional sources of variability in sample preparation and instrumental analyses, thereby enabling assessment of their impact on the quantitative measurements (Fig. 1 and Table 1). In studies I and II, samples were prepared centrally at the National Institute of Standards and Technology (NIST) and then distributed to the laboratories for SID-MRM-MS analyses. Variability arising from digestion of the target proteins was bypassed in study I by spiking a common pool of reduced, alkylated and trypsin-digested plasma with 11 unlabeled signature peptides derived from the target proteins at nine different concentrations. In study II, seven target proteins were digested separately, mixed with a stock solution of labeled peptides and digested plasma, then diluted serially with a labeled peptide/digested plasma stock to generate the same nine concentrations. Study III, which encompassed nearly all potential sources of analytical variability normally encountered, most closely simulated an actual biomarker verification experiment. Specifically, we produced an equimolar mixture of the same seven proteins in undiluted plasma at the same nine concentrations. Then, aliquots were distributed to the eight sites where the samples were denatured, reduced, alkylated, digested and desalted according to a standard operating procedure (SOP, Supplementary Methods). Labeled internal standard peptides were added immediately before SID-MRM-MS analysis. In all three studies, four technical replicates were performed at each concentration; in study III, three independent process replicates (IIIa, IIIb and IIIc) assessed intralaboratory and interlaboratory variability.
Figure 1.
Sample preparation workflow for studies I, II and III. (a) Study I. Pooled, digested plasma was spiked with 12C and 13C/15N peptides to generate a nine-point standard curve. (b) Study II. An equimolar mixture of the seven target proteins was digested separately and spiked with an equimolar mixture of IS peptides. The digest of target proteins plus IS peptides was added to pooled, digested plasma. A nine-point standard curve was prepared with pooled, digested plasma spiked with an equimolar mixture of IS peptides as the diluent. Study I and study II samples were prepared centrally at NIST. (c) Study III. Undiluted plasma was spiked with an equimolar mixture of the target proteins, then diluted with plasma to generate a nine-point standard curve. Three aliquots of these samples (prepared at NIST) were then shipped to the eight participating sites where reduction, alkylation, digestion and desalting were carried out before SID-MRM-MS analysis. IS, internal standard; SPE, solid phase extraction.
Table 1.
Target proteins and their signature peptides
| MRM transitions (m/z) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Protein | Abbrev | Species | Signature peptide | MH+ (mono) | Q1 | Q3 | ||
| Aprotinin | APR-AGL | Bovine | AGLCQTFVYGGCR | 1493.7 | 747.3 | 863.4 | 964.5 | 1092.5 |
| Leptin | LEP-IND | Mouse | INDISHTQSVSAK | 1407.3 | 469.9 | 590.8 | 647.8 | 728.4 |
| Myoglobin | MYO-LFT | Horse | LFTGHPETLEK | 1279.7 | 427.2 | 510.3 | 583.8 | 724.4 |
| Myelin basic protein | MBP-HGF | Bovine | HGFLPR | 732.4 | 366.7 | 391.3 | 538.3 | 595.4 |
| Myelin basic protein | MBP-YLA | Bovine | YLASASTMDHAR | 1328.6 | 443.5 | 491.2 | 526.8 | 823.4 |
| Prostate-specific antigen | PSA-IVG | Human | IVGGWECEK | 1082.5 | 541.7 | 808.3 | 865.4 | 969.4 |
| Prostate-specific antigen | PSA-LSE | Human | LSEPAELTDAVK | 1280.7 | 640.8 | 783.4 | 854.5 | 951.2 |
| Peroxidase | HRP-SSD | Horseradish | SSDLVALSGGHTFGK | 1483.8 | 495.3 | 711.4 | 798.4 | 982.5 |
| C-reactive protein | CRP-ESD | Human | ESDTSYVSLK | 1136.6 | 568.8 | 617.4 | 704.4 | 805.4 |
| C-reactive protein | CRP-GYS | Human | GYSIFSYATK | 1144.6 | 572.8 | 724.4 | 837.5 | 924.5 |
| C-reactive protein | CRP-YEV | Human | YEVQGEVFTKPQLWP | 1826.9 | 914.0 | 1053.5 | 1181.6 | 1525.8 |
Preselected MRM transitions are listed with further details in Supplementary Table 1. Bold face amino acids are stable, isotopically labeled residues. Cysteines (underlined) are carbamidomethylated. Q1, Q3, first and third quadrupoles.
The MRM assay configuration (including gradient development, selection of MRM analyte transitions for each signature peptide and general instrument settings) was performed at a single site using a nanoflow liquid chromatography (LC) (Eksigent NanoLC-2D) system coupled to a hybrid triple quadrupole/linear ion trap (AB/MDS Analytical Technologies 4000 QTRAP) mass spectrometer. These methods and parameters were transferred to all laboratories regardless of instrument platform to minimize variability arising from data acquisition (Online Methods and Supplementary Methods). All sites monitored three transitions per peptide, and precursor m/z values were consistent across all laboratories. Seven of the laboratories used 4000 QTRAP mass spectrometer instruments; the eighth site used a ThermoFisher TSQ Quantum Ultra triple quadrupole. Each laboratory tested and, if necessary, further optimized instrument parameters to maximize MS responses for the selected fragment ions on individual instruments. For the TSQ Quantum Ultra instrument, not all preselected transitions were ideal for achieving maximum sensitivity. For this subset of peptides, the site selected and optimized a substitute MRM transition for the signature peptide and its corresponding isotopically labeled analog (Supplementary Table 1b). Peptide YEVQGEVFTKPQLWP from C-reactive protein (CRP)-YEV did not ionize well and was detected with very low signals in the tuning mixtures or in the QC samples circulated to each site. Although MRM transitions for this peptide were included for data acquisition, subsequent data were not analyzed.
Intralaboratory reproducibility and precision of MRM assays
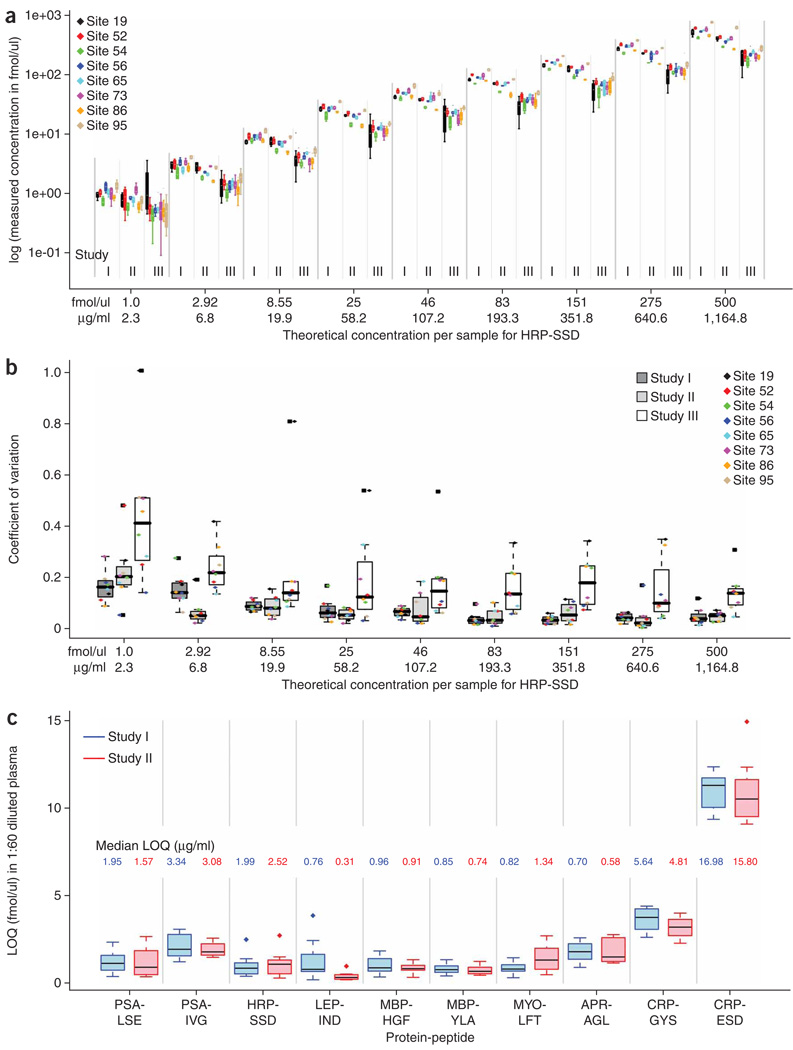
Intralaboratory variability and reproducibility in studies I–III were evaluated by comparing the measured concentrations to the actual concentrations across the range of spiked-in analytes and determining the coefficient of variation (CV) for these quantitative measurements. Figure 2a shows measured log concentration (y axis) versus theoretical (spiked-in) concentration (x axis) for the SSDLVALSGGHTFGK peptide derived from horseradish peroxidase (HRP-SSD; for all other peptides, Supplementary Fig. 1). Data for each site are color-coded, and organized by study and concentration. A linear trend is observed in the measured concentrations for studies I–III as spiked-in analytes increase across the concentration range. However, measured concentrations decrease as laboratories progress from study I to II to III. This trend is a result of apparent peptide loss from incomplete digestion of HRP protein and variability in sample handling at each site, as study complexity was increased (Fig. 1). Study I represents the optimum assay performance, as synthetic peptides (not proteins) were used as analytes. Protein digestion in study II (at a central location in the absence of plasma) and study III (at individual sites and in the presence of plasma) introduces potential sources of sample loss that decrease analyte recovery and reduce measured concentrations for studies II and III.
Figure 2.
Box plots of variation in MRM quantitative measurements, interlaboratory CV, intralaboratory CV and LOQ. (a) Intralaboratory assay CV. Box plots showing measured log concentration (y axis) versus theoretical (spiked-in) concentration (x axis) for HRP-SSD across the entire concentration range in diluted plasma. Protein concentration in µg/ml is mg protein equivalent in 1 ml undiluted plasma. The box plots for studies I and II are based on four replicate measurements, whereas those for study III summarize 12 measurements (four each from III a, b and c). Each of the eight sites was assigned a random numerical code (19, 52, 54, 56, 65, 73, 86, 95) for anonymization. (b) Interlaboratory assay CV. Values are shown for studies I–III for the entire range of HRP-SSD final analyte concentrations in plasma. Within each box plot, actual intralaboratory CV values for individual laboratories are shown with color-coded markers. The CV values are calculated based on the single best performing transition (lowest combined CV) across studies I and II. This same transition is also used for study III. (c) Interlaboratory assay LOQ. Values determined in studies I and II for the peptides indicated (see Table 1 for protein-peptide pair abbreviations). The inset values display the conversion of median LOQ to µg/ml (µg protein equivalent per 1 ml undiluted plasma) for each peptide. All measurements were made in 60-fold diluted plasma. Median is shown as a heavy horizontal line in all box plots. The box spans the interquartile range (IQR), with the whiskers extending to 1.5 × IQR. Values > 1.5 × IQR are deemed outliers, and shown as separate points.
Intralaboratory CVs for studies I and II constitute a measure of the technical variation due to instrument and data acquisition, as all sample preparation was performed centrally. The intralaboratory CVs at each analyte concentration point are shown in Figure 2b for the HRP-SSD peptide with color coded markers representing individual laboratories. Equivalent figures for all other peptides are shown in Supplementary Figures 2 and 3. Table 2 summarizes the range of median intralaboratory CVs observed across studies I, II and III, and Supplementary Table 2a–c shows the intralaboratory CVs calculated for each analyte at each of the nine final concentrations in plasma. Intralaboratory CVs are color coded in Supplementary Table 2a–c to facilitate visualization of the increasing variability from studies I–III. For all ten peptides in study I, median intralaboratory CVs were ≤15% across the concentration range (Supplementary Fig. 1 and Supplementary Table 2a). The median intralaboratory CVs for study II were very similar to those found in study I, with most intralaboratory CVs ≤15% across the concentration range (Supplementary Fig. 1 and Supplementary Table 2b). Finally, the intralaboratory CVs for study III were a measure of variation of the sample processing across replicates in addition to the technical variation of data acquisition. Increased variability is observed across the laboratories as individual sites were responsible for all sample handling and preparation (Fig. 2b). Although the intralaboratory CVs were elevated relative to studies I and II, >60% of the median intralaboratory CVs were still ≤25% across all concentrations, demonstrating very good reproducibility for sample processing (Supplementary Fig. 1 and Supplementary Table 2c).
Table 2.
Summary of results for studies I, II, and III
| Study I |
Study II |
Study IIIa |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Signature Peptide |
Interlaboratory CVb |
Intralaboratory CVc |
Linear slope |
Recov. (%)d |
Interlaboratory CVb |
Intralaboratory CVc |
Linear slope |
% Recov.d |
Interlaboratory CVb |
Intralaboratory CVc |
Linear slope |
Recov (%)d |
| APR-AGL | 9.2% | 3.9–11.2% | 1.157 | 114.5 | 13.1% | 2.0–7.8% | 0.575 | 57.5 | 13.7% | 7.3–45.2% | 0.738 | 79.4 |
| CRP-ESD | 5.9% | 2.2–5.9% | 1.124 | 118.4 | 10.5% | 3.1–8.4% | 0.573 | 61.4 | 16.7% | 8.5–18.1% | 0.439 | 48.9 |
| CRP-GYS | 5.4% | 1.4–10.2% | 1.324 | 140.5 | 5.6% | 1.2–6.4% | 0.546 | 56.0 | 18.5% | 6.6–35.0% | 0.159 | 18.5 |
| HRP-SSD | 14.1% | 4.0–8.9% | 1.198 | 120.4 | 5.5% | 4.6–7.3% | 0.794 | 82.3 | 21.9% | 8.4–21.4% | 0.430 | 45.7 |
| LEP-IND | 12.5% | 2.9–10.3% | 1.163 | 119.1 | 29.5% | 2.6–15.3% | 0.152 | 14.9 | 50.4% | 11.7–54.9% | 0.242 | 25.6 |
| MBP-HGF | 4.3% | 1.7–6.3% | 1.161 | 118.6 | 9.3% | 1.5–7.8% | 0.758 | 77.3 | 21.8% | 7.4–32.8% | 0.238 | 23.8 |
| MBP-YLA | 5.1% | 2.1–9.3% | 1.275 | 130.3 | 4.1% | 1.5–14.1% | 0.806 | 83.8 | N.M. | N.M. | N.M. | <1.0 |
| MYO-LFT | 4.9% | 1.6–5.7% | 1.518 | 154.4 | 3.8% | 2.0–6.3% | 1.012 | 101.3 | 23.1% | 8.9–21.6% | 0.504 | 60.4 |
| PSA-IVG | 6.9% | 1.3–14.7% | 1.658 | 165.4 | 5.5% | 2.0–11.2% | 0.848 | 81.9 | 17.2% | 7.9–20.3% | 0.587 | 58.0 |
| PSA-LSE | 8.9% | 1.2–6.9% | 1.098 | 111.4 | 5.3% | 2.0–4.6% | 1.524 | 151.3 | 10.3% | 7.6–13.7% | 0.918 | 92.7 |
Combined results for process replicates a, b, c for each peptide across sites for interlaboratory CV, intralaboratory CV, linear slope and percent recovery.
Interlaboratory CV was calculated from all replicates for each peptide using a single transition. The interlaboratory CV represented here is the median value across all sites for each peptide by study at the 2.92 fmol/µl concentration point. This concentration is at or near the LOQ for all peptides except those derived from CRP.
Intralaboratory CV was calculated from all replicates for each peptide using a single transition. The range of the median intralaboratory CV (over all concentrations) is reported here. Outlier laboratories (with CVs >1.5 times the interquartile range) have been excluded; in all three studies, the majority of the sites (seven or greater) are included in the intralaboratory CV range.
Percent recovery was determined from the mid-concentration point, 46 fmol/µl. The value shown is the average percent recovery across the eight sites using the same single transition as in CV and LOQ calculations. Recov., recovered; N.M., not measured.
Interlaboratory reproducibility and precision of MRM assays
The interlaboratory reproducibility and precision of the quantitative measurements was evaluated by calculating the CV of the quadruplicate analyses at each of the nine final analyte concentrations in plasma. The median interlaboratory CVs for HRP-SSD across studies I, II and III for the entire concentration range of 1–500 fmol/µl were predominantly ≤15% for this peptide in all three studies (Fig. 2b). As expected, interlaboratory CVs decreased as the concentration of spiked-in analyte increased to the upper range (Fig. 2b). However, even at lower analyte concentrations, the precision of the quantitative measurements across sites was very good. Table 2 summarizes the interlaboratory CVs at the 2.92 fmol/µl concentration for all peptides. This concentration is at or near the limit of quantification (LOQ) for most analytes in diluted plasma, except the two peptides derived from CRP (see below). Box plots of median interlaboratory CVs for all other peptides are shown in Supplementary Figure 2 (comparison of CVs across studies I, II and III) and Supplementary Figure 3 (comparison of CVs across process replicates for studies IIIa, IIIb and IIIc).
For study I, the interlaboratory CVs ranged from 4.3 to 14.1% at 2.92 fmol/µL, with eight of ten peptides in excellent agreement with values ≤ 10%. Because the interlaboratory CVs decreased at higher analyte concentrations, the median interlaboratory CVs across the entire concentration range was ≤5% (Supplementary Fig. 2 and Supplementary Table 2a). These results demonstrate excellent precision and reproducibility of the MRM assays for the signature peptides between laboratories when the major analytical variable is limited to the LC-MS system. Study II introduced new sources of variability attributable to sample loss during reduction, alkylation and trypsin digestion of the target proteins and desalting of the resulting peptide mixtures (Fig. 1 and Online Methods). The median interlaboratory CVs at 2.92 fmol/µl for study II ranged from 3.8% to 30% for all peptides, with nine of ten peptides having interlaboratory CVs ≤ 15%. Median interlaboratory CVs were predominantly ≤ 10% over the entire concentration range for study II (Supplementary Fig. 2 and Supplementary Table 2b), indicating that reproducibility of the assay across sites was not hampered by decreased recovery of target peptides. Finally, study III introduced the potential for the largest variability as each of the laboratories reduced, alkylated and trypsin digested the target proteins in plasma and desalted the subsequent peptide mixtures in three process replicates. Despite these additional sources of variability, average interlaboratory CVs for study III across process replicates IIIa, IIIb and IIIc ranged from 10.3–50% at 2.92 fmol/µl for nine of ten peptides (Table 2). Eight peptides had interlaboratory CVs ≤25%. Across the concentration range, the median interlaboratory CV was predominantly ≤20% (Supplementary Fig. 2 and Supplementary Table 2c).
Limits of detection and quantification
For studies I and II, inter- and intralaboratory measurement reproducibility of the ten signature peptides was determined at their estimated limits of detection (LOD) (Supplementary Fig. 4) and LOQ (Fig. 2c). The LOQ values represented in the box plot are based on the amount of peptide (in fmol) detected in plasma that was diluted 60-fold to a final protein concentration of 1 µg/µl for SID-MRM-MS analysis. The corresponding LOQ values for measurement of the proteins in undiluted plasma (in mg/ml) were also calculated (Fig. 2c). LOD and LOQ values calculated for each peptide at each site are shown in Supplementary Table 3.
The reproducibility of the LOQ estimations across sites was very good. For example, in study I, eight of ten peptides had median LOQ values between 0.66 and 2.0 fmol/µl when peptides were added into 1:60 diluted plasma (equivalent to a range of 0.70–3.34 µg/ml protein in plasma; Fig. 2c). The remaining two CRP peptides were detected at endogenous levels in the blank and 0 fmol/µl spiked plasma samples. A commercial enzyme-linked immunosorbent assay (ELISA) was performed on the plasma stock and yielded a concentration of 6 µg/ml of this protein (data not shown), which is equivalent to 4 fmol/µl of CRP in the diluted plasma. The LOQ values obtained in study II, which were similar to those obtained in study I, ranged between 0.31 and 1.8 fmol/µl for the same eight of ten peptides. The LOD/LOQ values for studies I and II were similar in magnitude for a majority of the signature peptides and showed acceptable variation across all eight laboratories.
Reproducibility of linear response and peptide recovery
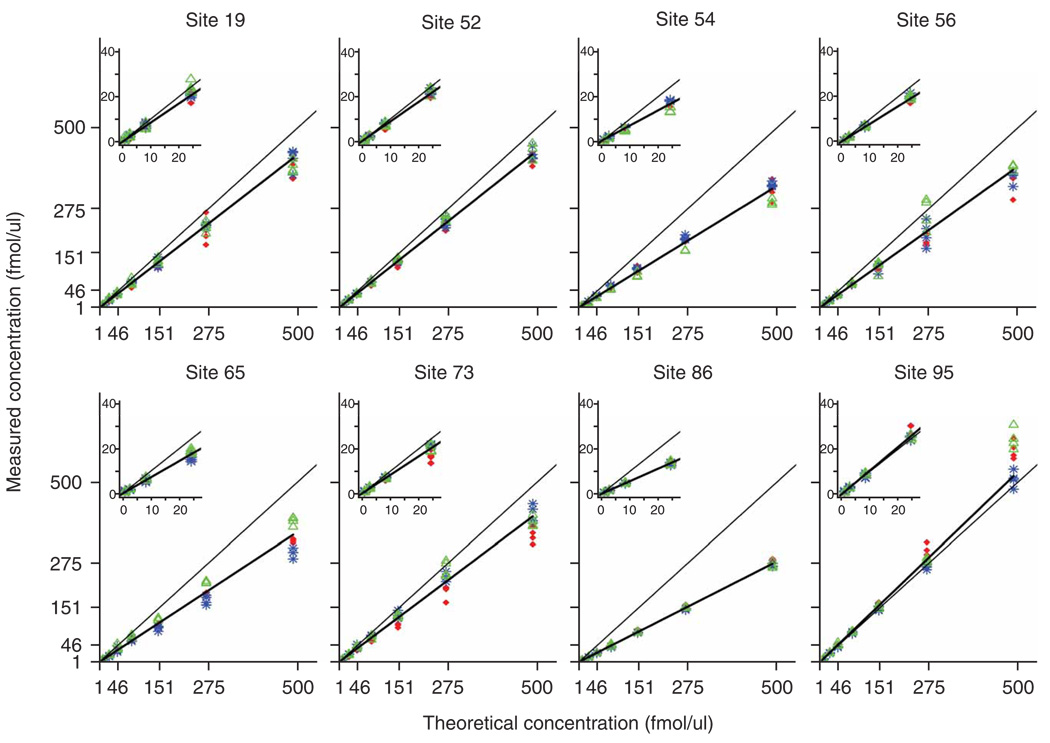
Figure 3 shows a compilation of response curves (study II) obtained at the eight sites and plotted on a linear-linear scale for the HRP-SSD peptide. Response curves are plots of experimentally determined concentrations versus theoretical concentrations of the target analyte, and provide useful visual representations of reproducibility and linearity. Quadruplicate replicates are shown at every concentration for all three MRM transitions. Interlaboratory reproducibility of linear responses and quantitative measurements across all laboratories and all three studies was, in general, very good (Table 2). The fitted slopes presented in Table 2 demonstrate the consistency in the linear response with a change in actual peptide (study I) or protein (studies II–III) concentration across the measurements made in each laboratory, and are also an estimation of peptide recovery. A slope of 1.0 is equal to the theoretical slope in which measured concentration is proportional to analyte concentration and recovery is equal to 100%. Slopes <1 indicate <100% recovery, whereas slopes >1 indicate >100% recovery (the latter likely a result of errors in the initial concentrations of the peptide or protein stock solutions). For the representative peptide, HRP-SSD, the average slope in study I was 1.2 with an interlaboratory CV of 15.6% (Table 2 and Supplementary Table 4a), showing excellent reproducibility between sites and highly consistent linear responses across laboratories and instrument platforms as indicated by the slopes being close to the theoretical line. As an estimation of the average percent recovery across the concentration range, the average slope for the HRP-SSD peptide agrees well with the calculation of percent recovery determined at the mid-concentration point of the response curve (46 fmol/µl; Table 2).
Figure 3.
Interlaboratory reproducibility of linear calibration curve slopes for study II. The eight plots display the concentration curves for the detection of HRP-SSD in study II across all laboratories. Each of the eight sites was assigned a random numerical code (19, 52, 54, 56, 65, 73, 86, 95) for anonymization. Comparison of the plots demonstrates good linearity, with the slopes falling close to the diagonal, black line (theoretical slope = 1), and good agreement between the three transitions at each concentration point. Four replicate measurements are represented at each concentration point. Analyte transitions: red diamond, transition 1, (m/z 492.6→703.4); blue asterisk, transition 2, (m/z 492.6→790.4); green triangle, transition 3, (m/z 492.6→974.5). In some cases, the data points overlay such that transition 1 is not visible. Inset plots show more detail of lower end of the concentration range. The mean slope calculation across all laboratories in this example is 0.794 with an interlaboratory CV of 18.7%. Final concentrations of heavy and light peptides and added proteins were adjusted according to the gravimetric measurements described in Supplementary Table 6a–f.
Response curves for all other peptides and proteins generated by each laboratory in all three studies are plotted on the linear-linear scale with scale-expansion insets to facilitate visualization of the lower concentration range (Supplementary Fig. 5). A weighted robust linear regression on the linear-linear scale was used to determine slope and percent recovery. In addition, the response curves are plotted on the log-log scale (Supplementary Appendix) without regression lines to facilitate data visualization. Individual parameters for slope, y intercept and their associated standard errors for each peptide across all sites are shown in Supplementary Tables 4a–e. Altogether, peptide responses in study I had an average slope ranging from 1.1 to 1.6 with an interlaboratory CV ≤ 10% for most of the peptides (Table 2 and Supplementary Tables 4a). The average slope value was more variable in study II, with a range of 0.15 to 1.5 across all peptides. Interlaboratory CV for slope in study II was ≤15% for nine of ten peptides (Supplementary Table 4b). Study III exhibited the lowest average slope values, which ranged from 0.16 to 0.92 for nine of ten peptides, and interlaboratory CVs for slope were ≤25% for the majority of peptides across the process replicates (Supplementary Table 4c–e). One peptide, MBP-YLA, was not detected by any site in any process replicate of study III. Overall, the responses were reproducible as indicated by the low interlaboratory CVs, and the measurements of the three transitions were highly uniform such that the replicates often overlaid at each concentration (Fig. 3 and Supplementary Fig. 5).
Because the slope is an estimation of percentage recovery, the decrease and variability in the slopes of the response curves observed across these studies (Supplementary Fig. 6) correlate with the increasing level of sequential experimental complexity, from the introduction of protein digestion in study II and protein digestion in the presence of plasma in study III (Fig. 1). Again, the average slopes for all peptides agree well with the calculation of percent recovery at the mid-point of the concentration range (Table 2). For study I and two of the ten peptides in study II, recovery ≥100% was observed for many peptides. This could most likely be attributed to the effect of errors in quantification of the protein or peptide stock concentrations by amino acid analysis, and inaccuracies associated with sample preparation, such as pipetting and freeze-thawing. In study III, six of the nine peptides detected had percent recoveries ≥40%, which is within an acceptable range for verification assays2,9. Four peptides (CRP-GYS, LEP-IND, MBP-HGF and MBP-YLA) had recoveries ≤25%, and would not be considered useable for verification or clinical validation assay purposes. No significant differences in peptide recovery were observed across the concentration range or between studies II and III (Supplementary Table 5 for two representative examples). Although <100% recovery of the target peptides limits the sensitivity of the assays, these results show very good reproducibility for recovery of most peptides and demonstrate the large role sample handling has in the variability of peptide recovery.
Common sources of variance and their detection
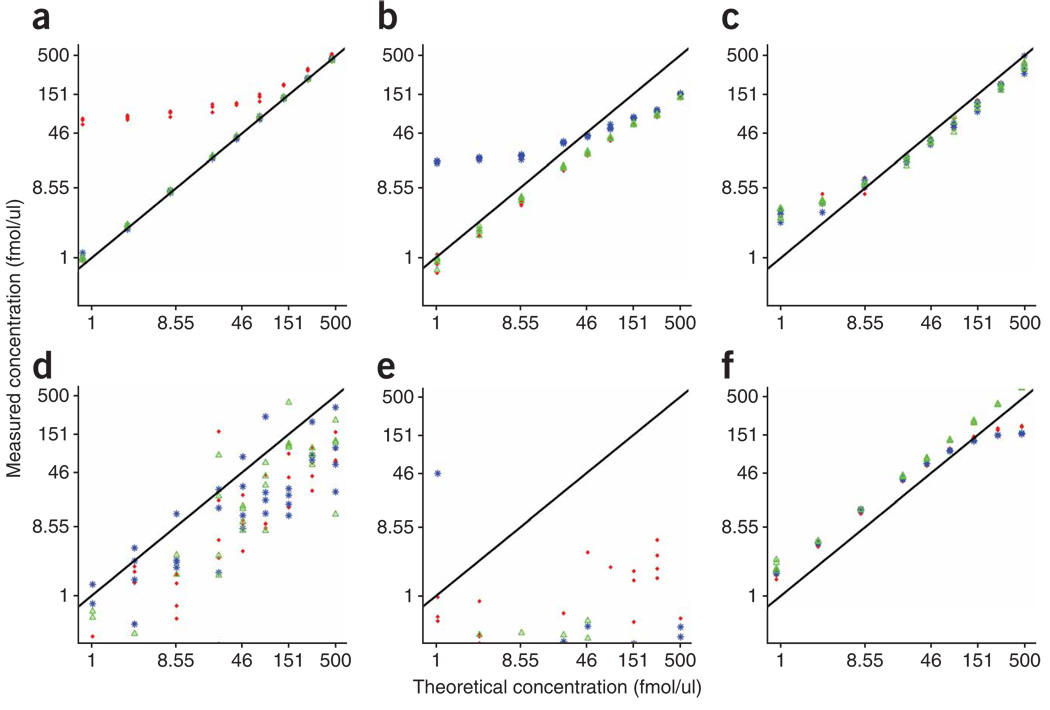
Although most of the signature peptides exhibited excellent reproducibility within and between laboratories (Supplementary Fig. 5), deviations from the trend lines were observed for some peptides at one or more sites. Typical problems that can arise in developing and applying MRM assays to quantify proteins in plasma are illustrated in Figure 4. The most common problem related to the appearance of ‘outliers’ was interference in one or more of the fragment-ion transitions monitored for either the light (12C/14N) peptides or heavy (13C/15N)-labeled internal standard peptides. Figure 4a,b illustrates interferences in transition 1 and 2 of the light peptides for MBP-HGF and MYO-LFT, respectively, at two analysis laboratories. In both cases, the relative ratios of the transitions were altered from those observed in the absence of plasma during assay configuration, resulting in considerable deviation from linearity for the respective product ions. Monitoring multiple transitions for each peptide, as done in our study, enables reliable quantification, which is accomplished by using the other unaffected transitions. In the case of CRP-ESD (Fig. 4c), obvious and highly consistent deviation from linearity was observed for all three transitions monitored at the lower end of the response curves. This flattening of the curves was due to the presence of endogenous levels of the protein within the measurable range of the MRM assays. We confirmed the level of CRP present in the plasma by ELISA. Other issues, such as unstable electrospray conditions, lack of recovery during sample processing and saturation of the MS detector were also observed and gave rise to recognizable patterns of misbehavior (Fig. 4d–f). Instability of the LC system and deterioration of the LC column are also common problems that are readily recognized. If not corrected, they can cause large shifts in peptide retention time and chromatographic peak broadening or tailing, particularly for early-eluting hydrophilic species, resulting in decreased reproducibility for peptide detection and quantification.
Figure 4.
Response curves representing deviations from the trend line. Red diamond, transition 1; blue asterisk, transition 2; green triangle, transition 3. (a) Study I, site 52, MBP-HGF: interference in transition 1 of the analyte. (b) Study IIIb, site 95, MYO-LFT: interference in transition 2 of the analyte, which was also observed in study I, II and IIIa for this laboratory. (c) Study II, site 86, CRP-ESD: endogenous protein level increased the estimated protein concentration at the low end of the concentration range of spiked-in proteins, resulting in flattening of slope. (d) Study IIIa, site 56, LEP-IND: unstable electrospray conditions resulted in a substantial increase in interlaboratory CV to 99%. (e) Study IIIa, site 19, MBP-YLA: no detection of MBP-YLA peptide at any site. (f) Study I, site 86, PSA-IVG: saturation at highest two concentrations. Site codes are identical to those given in Figures 2 and 3.
DISCUSSION
Targeted MRM assays have been used very successfully for quantifying small molecules (e.g., hormones, drugs and their metabolites) in pharmaceutical research and in clinical laboratories in applications such as screening newborns for disease11. More recently, the merits of SID-MRM-MS for quantifying peptides derived from proteins in plasma have been demonstrated in several laboratories4–9,12. These studies have, however, only addressed assay performance at a single laboratory, and thus were not able to demonstrate the multisite robustness needed in large-scale biomarker research and ultimately in preclinical and clinical applications. The main purpose of this study was to provide such a demonstration by performing an assessment of the analytical characteristics of a multiplexed, SID-MRM-MS assay across eight laboratories using seven target proteins with which to spike human plasma. A three-tiered experimental protocol was used that progressively introduced sample preparation variables likely to affect inter- and intralaboratory reproducibility, transferability, precision and sensitivity. Our results demonstrate that reproducible, quantitative measurements of proteins in plasma can be made by SID-MRM-MS in multiple laboratories using different instrument platforms through use of standardized protocols for sample preparation, data acquisition and data analysis. The robustness of such a targeted assay approach compensates for the greater variability in protein measurements inherent in shotgun (‘discovery’ proteomics) methods13,14, enabling the development of an effective biomarker pipeline1.
Reproducibility and precision of the quantitative measurements for nine of ten peptides tested across eight laboratories ranged from 4–14%, 4–13% and 10–23% interlaboratory CVs at or near the estimated LOQ for study I, II and III, respectively. Intralaboratory CVs were predominantly <15% and <25% at the identical concentration for studies I/II and III, respectively (Supplementary Table 2). Although the current assay performance under real biomarker conditions (study III) is below that generally stated for clinical assays (typically <10–15%), the performance achieved is sufficient for the verification of candidate biomarkers2 present at more than ~2–6 µg/ml in plasma, with a linear dynamic range spanning three orders of magnitude. In all cases, interlaboratory and intralaboratory CVs improved with increasing analyte concentration. Such modest differences between interlaboratory and intralaboratory CVs underscore the excellent agreement between the eight participating laboratories. Likewise, the progressive increases in CVs from studies I to III indicate convincingly that sample preparation contributes more to assay variability than instrumental variability, further highlighting the data quality obtainable from SID-MRM-MS. Although most important parameters were governed by detailed SOPs, the transfer of MRM assays across LC-MS platforms did require optimization of the transitions being monitored to compensate for differing instrument-specific ion source and collision-induced dissociation parameters, and to ensure that each platform achieved optimum sensitivity (Supplementary Tables 1a–e). Despite these variations concerning a small number of analyte peptides, interlaboratory variability and specificity of the assay were not affected (Table 2).
Differences emerged in assay performance for different peptides. Most peptides performed well at all eight sites, whereas a few exhibited variable or poor behavior. This result highlights the dependence of MRM assay performance in plasma on specific properties of the peptides selected as surrogates for the target proteins. Ideally the final selection of signature peptides for SID-MRM-MS biomarker assays should be based on multisite studies so as to ensure the most robust performance.
The most frequent cause of poor peptide performance was the presence of interference from the background plasma digest matrix, in either the analyte or internal-standard channels, which altered the ratios of these transitions. Monitoring a minimum of three transitions per analyte is critical in maintaining assay selectivity and recognizing such interferences when they occur. Most participating sites observed interferences in one or more peptides over the course of the three studies. In the case of CRP, we were able to establish that the flattening of the response curves was due to the presence of endogenous levels of CRP as all three transitions monitored were affected equally and the expected ratios of the transition-ion abundances to one another were maintained. Other interferences arose from problems with chromatography (e.g., large peak widths, shifting retentions times, or early elution and consequent sensitivity to intermittent or unstable electrospray conditions), which can be addressed by further refinement of protocols, particularly in LC operation and data acquisition.
Recovery of signature peptides generally decreased from study I to III, as proteolytic digestion and subsequent sample handling, such as desalting, were introduced into the experimental workflow. Digestion efficiency of proteins in the plasma matrix has only recently begun to be studied15. If a signature peptide is not detected in an MRM assay, it is often unclear if this is because of (i) losses from sample handling, such as fractionation or desalting, (ii) poor enzymatic digestion, (iii) concentration below LOD, (iv) post-translational modification such as glycosylation and phosphorylation, (v) artifactual modifications to reactive amino acids, such as oxidation or carbamylation, or (vi) some combination thereof. The effect of decreasing control of sample preparation was reflected in the increased variability and lower peptide recoveries for a majority of peptides as sites progressed from study II to III (Table 2 and Supplementary Fig. 6). In study III, one peptide was not recovered in any process replicate performed at all participating laboratories, and four peptides had <25% recovery (Table 2 and Supplementary Fig. 6). Addition of labeled internal standard (IS) peptides at an early stage in sample processing (e.g., during enzymatic digestion) could help to account for peptide loss. However, lower recovery of signature peptides does not impede the use of these assays for verification where the goal is to precisely define the relative difference in abundance for candidate proteins between cases and controls rather than to determine the absolute concentration of each protein. Absent a general method ensuring stoichiometric digestion, absolute concentration measurements would likely require addition of isotopically labeled, recombinant protein standards at the start of sample processing.
The purpose of the present study was not to define the ultimate sensitivity possible for proteins by SID-MRM-MS, but rather to evaluate the transferability and robustness of the technology within and between laboratories. For this first study, we made no attempt to reduce the complexity of the plasma matrix by either depletion of abundant proteins or fractionation. The sensitivity of protein quantification by SID-MRM-MS in plasma is severely limited by the complexity and 1011 dynamic range of protein abundances in blood, and the susceptibility to interference from other peptides and their fragment ions is greatest in this matrix16. Typical LODs and LOQs observed in prior studies of unfractionated plasma are in the high 100s of ng/ml to low µg/ml range of target protein6,8,17. Results described here are consistent with these reports across sites and instrument platforms (Fig. 2c and Supplementary Fig. 4). Although emphasis is often placed on discovery and verification of low-abundance candidate biomarkers (≤ ng/ml levels in serum), high-abundance serum proteins, such as CRP, transferrin, complement components, immunoglobulin classes and lipoproteins, are clinically relevant markers of disease and their levels in blood make them directly accessible by SID-MRM-MS using the approaches described here. The LODs and LOQs of MS-based assays have been extended into the low ng/ml range in plasma by using immunoaffinity depletion of high-abundance proteins, limited protein or peptide fractionation, or immunoaffinity enrichment at the protein or peptide level before SID-MRM-MS9,17–24. The additional processing steps used are likely to introduce new sources of experimental variation that will have to be assessed in interlaboratory studies similar to those described here. Nevertheless, the assay performance reported in the present studies, measured at maximum levels of interfering high-abundance peptides in unfractionated plasma digests, suggests that similar or better intra- and interlaboratory assay performance may be achievable for quantitative, multiplexed measurement of proteins in the low ng/ml range in plasma by MS.
Our study demonstrates that targeted, quantitative and multiplexed MS-based assays can be rapidly configured and deployed in multiple laboratories to yield robust and reproducible assays for proteins down to low µg/ml levels in the context of unfractionated plasma. This is a critical first step toward potential widespread implementation of SID-MRM-MS assays for verification of novel protein biomarker candidates. The SID-MRM-MS technology has the potential to become the critical filter used to assess candidate biomarker performance in a sufficient number of patient samples before committing the very substantial time and resources required to create clinical-grade immunoassays. The performance required of such assays2 is not as stringent as that currently required for US Food and Drug Administration–approved clinical assays25. Beyond candidate verification, SID-MRM-MS assays may eventually have potential to replace certain clinical immunoassays, especially in cases where interferences are known to exist23 or multiplex measurements are needed. By detecting a structural component of the protein, the signature peptide, with near-absolute structural specificity, SID-MRM-MS should avoid inter-assay differences that occur when different immunoassays for the same protein detect distinct, potentially labile epitopes. Furthermore, the simplicity of producing and characterizing peptide-based reference materials for SID-MRM-MS could help overcome well-known problems with ELISA assay standardization, which lead to varying results across multiple clinical laboratories26,27.
The methods, reagents and multilaboratory data sets presented here should facilitate testing and implementation of MRM-based multiplex assays for quantifying target proteins in plasma by the proteomics community. Our results should foster greater acceptance by the clinical community of SID-MRM-MS technology as a generally applicable approach to verify candidate biomarkers in large clinical sample sets, and thus provide a critical component for a systematic biomarker-development pipeline.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturebiotechnology/.
Data accession
A password-protected website was developed to manage the large number of data files generated for the described interlaboratory studies. This website, hosted at NIST, was designed as a portal used by the teams for initiating uploads and downloads of large data files. The data transfers were performed using Tranche (http://trancheproject.org/) an open source, secure peer-to-peer file-sharing tool. A customized user interface employed by the participating laboratories was developed and added to the Tranche code base. This tool allowed the website and database to communicate tracking information with Tranche by employing custom URLs. The Tranche hash (a unique data identifier) and pass-phrase, for each website, was automatically recorded into the website’s database when file uploading was complete. These stored links allow subsequent retrieval of data files using the Tranche download tool. The Tranche hashes and passphrases provide a simple and portable mechanism to access data sets and can be easily associated with supporting annotation. The data associated with this manuscript may be downloaded from the ProteomeCommons.org Tranche network using the following hash: CKpfN0bl2ULLwCaIovXn/spuw4rYfJF6H/L+/6sHAKGzCsj4fzTD0Rau JjAwf9baB8tI36HQ0izji2tupYAPM29P2cAAAAAAAT0iw==. The hash may be used to show exactly what files were published as part of this manuscript’s data set, and the hash may also be used to check that the data have not changed since publication. Accessible information includes all raw data files, all processed data export files, 4000 QTRAP MultiQuant results files, as well as detailed data submission sheets and file annotation legends for studies I–III from the eight participating laboratories.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Cancer Institute (NCI) (U24 CA126476, U24 126477, U24 126480, U24 CA126485, and U24 126479), part of NCI Clinical Proteomic Technologies for Cancer initiative. A component of this initiative is the Clinical Proteomic Technology Assessment for Cancer (CPTAC) Network and teams, which include the Broad Institute of MIT and Harvard (with the Fred Hutchinson Cancer Research Center, Massachusetts General Hospital, the University of North Carolina at Chapel Hill, the University of Victoria and the Plasma Proteome Institute), Memorial Sloan-Kettering Cancer Center (with the Skirball Institute at New York University), Purdue University (with Monarch Life Sciences, Indiana University, Indiana University-Purdue University Indianapolis and the Hoosier Oncology Group), University of California, San Francisco (with the Buck Institute for Age Research, Lawrence Berkeley National Laboratory, the University of British Columbia and the University of Texas M.D. Anderson Cancer Center) and Vanderbilt University School of Medicine (with the University of Texas M.D. Anderson Cancer Center, the University of Washington and the University of Arizona). A full listing of the CPTAC Team Network can be found at http://proteomics.cancer.gov/programs/CPTAC/networkmembership.
Footnotes
Note: Supplementary information is available on the Nature Biotechnology website.
AUTHOR CONTRIBUTIONS
The CPTAC Network contributed collectively to this study. The following CPTAC Network investigators contributed significant intellectual contributions to work described in this paper.
S.E.A., T.A., N.L.A., D.M.B, S.C.H., A.-J.L.H., H.K., D.R., B.S., S.J.S., L.J.Z. and S.A.C. contributed to study design and SOP development. D.M.B and N.G.D. prepared and shipped samples. S.E.A., T.A., S.A., H.L.C., J.M.H., A.J., E.B.J., H.K., D.S., T.J.T., J.R.W., A.W., S.W., L.Z., and L.J.Z. contributed to generation of data. M.P.C., J.L., D.R.M., R.K.N., S.J.S., T.C.P., P.A.R., C.H.S., D.L.T., A.M.V., and L.J.V.-M. contributed to bioinformatics and statistical analysis. S.E.A, T.A., H.K., D.R.M., S.J.S. and L.J.Z. centrally reviewed data. S.E.A., T.A., N.L.A., S.A.C., S.J.F., S.C.H., A.-J.L.H., H.K., D.R.M, B.S., S.J.S., and L.J.Z. wrote and prepared the manuscript. R.K.B., C.B., C.H.B., S.A.C., S.J.F., B.W.G., T.H., C.R.K., D.C.L., M.M., T.A.N., A.G.P., H.R., F.E.R., P.T., and M.W. contributed to experimental design. S.C.H. chaired the CPTAC Experimental Design and Statistics Verification Studies Working Group that designed interlaboratory studies and generated data.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 2.Paulovich AG, Whiteaker JR, Hoofnagle AN, Wang P. The interface between biomarker discovery and clinical validation: The tar pit of the protein biomarker pipeline. Proteomics Clin. Appl. 2008;2:1386–1402. doi: 10.1002/prca.200780174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr JR, et al. Isotope-dilution mass spectrometric quantification of specific proteins: model application with apolipoprotein A-1. Clin. Chem. 1996;42:1676–1682. [PubMed] [Google Scholar]
- 4.Barnidge DR, et al. Absolute quantification of the G protein-coupled receptor rhodopsin by LC/MS/MS using proteolysis product peptides and synthetic peptide standards. Anal. Chem. 2003;75:445–451. doi: 10.1021/ac026154+. [DOI] [PubMed] [Google Scholar]
- 5.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem mass spectrometry. Proc. Natl. Acad. Sci. USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnidge DR, Goodmanson MK, Klee GG, Muddiman DC. Absolute quantification of the model biomarker prostate-specific antigen in serum by LC-MS/MS using protein cleavage and isotope dilution MS. J. Proteome Res. 2004;3:644–652. doi: 10.1021/pr049963d. [DOI] [PubMed] [Google Scholar]
- 7.Kuhn E, et al. Quantification of C-reactive protein in the serum of patients with rheumatoid arthritis using multiple reaction monitoring mass spectrometry and 13C-labeled peptide standards. Proteomics. 2004;4:1175–1186. doi: 10.1002/pmic.200300670. [DOI] [PubMed] [Google Scholar]
- 8.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aguiar M, Masse R, Gibbs BF. Mass spectrometric quantitation of C-reactive protein using labeled tryptic peptides. Anal. Biochem. 2006;354:175–181. doi: 10.1016/j.ab.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 11.Wilcken B, Wiley V, Hammond J, Carpenter K. Screening newborns for inborn errors of metabolism by tandem mass spectrometry. N. Engl. J. Med. 2003;348:2304–2312. doi: 10.1056/NEJMoa025225. [DOI] [PubMed] [Google Scholar]
- 12.Whiteaker JR, et al. Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. J. Proteome Res. 2007;6:3875–3876. doi: 10.1021/pr070202v. [DOI] [PubMed] [Google Scholar]
- 13.Anderson NL, et al. The human plasma proteome: a non-redundant list developed by combination of four separate sources. Mol. Cell. Proteomics. 2004;3:311–326. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Omenn GS, et al. Overview of the HUPO plasma proteome project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 2005;5:3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- 15.Kuzyk MA, et al. MRM-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol. Cell. Proteomics. 2009 May 1; doi: 10.1074/mcp.M800540-MCP200. published online, doi:10.1074/mcp.M800540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson NL, Anderson NG. The human plasma proteome: History, character, and diagnostic prospects. Mol. Cell. Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 17.Whiteaker JR, et al. Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. J. Proteome Res. 2007;6:3962–3975. doi: 10.1021/pr070202v. [DOI] [PubMed] [Google Scholar]
- 18.Berna M, et al. Quantification of NTproBNP in rat serum using immunoprecipitation and LC/MS/MS: a biomarker of drug-induced cardiac hypertrophy. Anal. Chem. 2008;80:561–566. doi: 10.1021/ac702311m. [DOI] [PubMed] [Google Scholar]
- 19.Berna MJ, Zhen Y, Watson DE, Hale JE, Ackermann BL. Strategic use of immunoprecipitation and LC/MS/MS for trace-level protein quantification: Myosin light chain 1, a biomarker of cardiac necrosis. Anal. Chem. 2007;79:4199–4205. doi: 10.1021/ac070051f. [DOI] [PubMed] [Google Scholar]
- 20.Labugger R, et al. Strategy for analysis of cardiac troponins in biological samples with a combination of affinity chromatography and mass spectrometry. Clin. Chem. 2003;49:873–879. doi: 10.1373/49.6.873. [DOI] [PubMed] [Google Scholar]
- 21.Nicol GR, et al. Use of an immunoaffinity-mass spectrometry-based approach for the quantification of protein biomarkers from serum samples of lung cancer patients. Mol. Cell. Proteomics. 2008;7:1974–1982. doi: 10.1074/mcp.M700476-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Anderson NL, et al. Mass spectrometric quantitation of peptides and proteins using stable isotope standards and capture by anti-peptide antibodies (SISCAPA) J. Prot. Res. 2004;3:235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 23.Hoofnagle AN, Becker JO, Wener MH, Heinecke JW. Quantification of thyroglobulin, a low-abundance serum protein, by immunoaffinity peptide enrichment and tandem mass spectrometry. Clin. Chem. 2008;54:1796–1804. doi: 10.1373/clinchem.2008.109652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhn E, et al. Developing multiplexed assays for troponin I and interleukin-33 in plasma by peptide immunoaffinity enrichment and targeted mass spectrometry. Clin. Chem. 2009 April 16; doi: 10.1373/clinchem.2009.123935. published online, doi:10.1373/clinchem.2009.123935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biopharmaceutics Coordinating Committee, Center for Drug Evaluation and Research, Center for Veterinary Medicine, US Food and Drug Administration (FDA) Rockville, MD: FDA; Guidance for Industry: Bioanalytical Method Validation. 2001 May; 〈 http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm070107.pdf〉.
- 26.Slev PR, Rawlins ML, Roberts WL. Performance characteristics of seven automated CA 15−3 assays. Am. J. Clin. Pathol. 2006;125:752–757. doi: 10.1309/G6X6-PR75-26FA-KV0E. [DOI] [PubMed] [Google Scholar]
- 27.Sapin R. Insulin immunoassays: fast approaching 50 years of existence and still calling for standardization. Clin. Chem. 2007;53:810–812. doi: 10.1373/clinchem.2006.084012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.