Abstract
Selective capture of glycopolypeptides followed by release and analysis of the former glycosylation-site peptides has been shown to have promise for reducing complexity of body fluids such as blood for biomarker discovery. In this work, a protocol based on capture of polypeptides containing a N-linked carbohydrate from human plasma using commercially available magnetic beads coupled with hydrazide chemistry was optimized and partially automated through the use of a KingFisher magnetic particle processor. Comparison of bead-based glycocapture at the protein-level vs. peptide-level revealed differences in the specificity, reproducibility and absolute number of former glycosylation-site peptides detected. Evaluation of a range of capture and elution conditions led to an optimized protocol with a 24% intraday and 30% interday CV, and a glycopeptide capture specificity of 99%. Depleting the plasma of 14 high abundance proteins improved detection sensitivity by approximately one order of magnitude compared to non-depleted plasma and resulted in an increase of 24% in the number of identified glycoproteins. The sensitivity of SPEG for detection of glycoproteins in depleted, non-fractionated plasma was found to be in the 10 to 100 pmol/ml range corresponding to glycoprotein levels ranging from 100’s of nanograms/ml to 10’s of micrograms/ml. Despite high capture specificity,the total number of glycoproteins detected and the sensitivity of SPEG in plasma is surprisingly limited.
Keywords: Solid phase extraction of N-linked glycoproteins, proteomics, glycoproteins, hydrazide chemistry, biomarker discovery, glycopeptide
Introduction
The protein profile of human blood plasma contains a wealth of information that is believed to reflect important processes occurring in the body at a given time, including the presence and stage of diseases such as cardiovascular disease and cancer. Over the past few years, there has been considerable interest in analyzing the human plasma proteome using state-of-the-art mass spectrometry technology to identify disease specific protein biomarkers of potential use as diagnostics or prognostic markers of disease 1–4. This has proven to be difficult owing to the enormous range in protein concentration and extensive complexity of the plasma proteome which limits detection to only a small portion of the total plasma proteome with current analytical technology 5, 6. Since many disease-specific biomarkers are expected to be present in relatively low abundance in blood, they are typically not among the proteins detected when using a straight-forward proteomics approach. In order to reduce the dynamic range in protein concentration and/or the complexity of a plasma sample prior to analysis by MS, various fractionation and enrichment strategies are widely employed that separate proteins and peptides based on properties like hydrophobicity, pI or size, and deplete high abundance proteins by immunoaffinity 7, 8.
Glycoproteins have been recognized as an important group of proteins in biomarker discovery 9–11, and more than half of all proteins are believed to be glycosylated, making it the most widespread of all post-translational modifications 12. Glycosylated proteins are typically present at the cell surface or secreted from cells to the extracellular environment and play a major role in molecular and cellular recognition and communication 13–15. Because of these properties, they also play an important role in immune responses and cancer progression. Indeed, many of the known disease markers are glycoproteins, including CA125 in ovarian cancer, prostate specific antigen in prostate cancer and alpha-fetoprotein used to monitor different cancer types. The two main types of glycosylation are N-linked and O-linked. For the N-linked type, the glyco-unit is attached to an asparagine residue followed by any amino acid (except proline) and a serine or threonine giving the following sequence motif N-^P-[ST]. N-linked glycosylation is particularly prevalent in proteins destined for the extracellular environment 15. In O-linked glycosylation, the carbohydrates are typically linked to serine or threonine residues. No reliable consensus sequence has been identified for O-linked sugars.
Lectin-affinity column chromatography and solid phase extraction of N-linked glycoproteins (SPEG) through hydrazide chemistry are the two main approaches that have been used to capture and enrich for N-linked glycoproteins from plasma or other body fluids. Lectins are a class of proteins that bind specific oligosaccharide structures, and both single lectin and serial lectin affinity chromatography using lectins of differing specificities have been used to capture glycoproteins from complex protein mixtures 16, 17. For SPEG based glycocapture, the cis-diol groups of the carbohydrates are oxidized into aldehydes, which then can form covalent hydrazone bonds with the hydrazide groups coupled to the solid support. Non-glycosylated proteins/peptides are removed by extensive washing, and the remaining glycopeptides are released by adding peptide-N-glycosidase F (PNGase F) which cleaves N-linked oligosaccharides from glycoproteins/peptides. In this process the attachment-site asparagine residue is deamidated to aspartic acid, leading to a mass difference of +0.9840 Da. Information on the structure of the carbohydrate is of course lost in this process. In a study by Pan et al., a direct comparison between lectin and hydrazide capture analysis of cerebrospinal fluid glycoproteins followed by LC-MS/MS showed that from a total of 216 identified glycoprotein, 86 were identified by both approaches, 53 only by hydrazide capture and 77 only by lectin capture 18. They found a glycoprotein specificity of 69% for the lectin approach.
Many different SPEG protocols have already been published and these have been mainly based on hydrazide residues linked to macroporous beads and manual sample processing 18–23. The variations of the protocols have been either coupling of the glyco-units to the beads before enzymatic cleavage (protein-level coupling) or after enzymatic cleavage (peptide-level coupling) of the protein content. Several different coupling solutions have been used during coupling of the glyco-units to the hydrazide groups, but the effect of these different buffers have never been systematically compared. Furthermore, reproducibility and detection limit are generally not reported in these studies. The effect of different bead washing procedures, PNGaseF incubation times and bead coupling times on the yield and glyco-capture specificity are among the parameters that have been scrutinized 22. Recently, Zou et al. reported the use of “in-house” synthesized magnetic beads with linked hydrazide groups to partially automate SPEG at the protein level in mouse plasma 23. Glycopeptide and glycoprotein specificity of 73.9% and 90.7%, respectively, were reported, with typically 167 unique glycopeptides and 68 unique glycoproteins identified in a single LC-MS/MS analysis. The reproducibility of the method was not calculated, but process replicates were reported to look identical from a 3D view of the LC-MS/MS runs.
Here we report the development and evaluation of a semi-automated protocol for the enrichment of N-linked glycopeptides from human plasma using commercially available magnetic beads modified with hydrazide. To optimize the protocol using magnetic beads, we systematically evaluated the effect of various coupling solutions, different washing conditions and the amount of magnetic beads on glycopeptide capture specificity, the number of glycopeptides identified and the glycopeptide yield. We also directly compared glyco-unit coupling to hydrazide groups before (protein-level) and after (peptide-level) enzymatic digestion, and evaluated the effect of abundant protein depletion prior to SPEG processing. The intraday and interday reproducibility of the optimized SPEG protocol, both for protein-and peptide-level capture were determined. Finally, a glycoprotein spike-in study was performed to evaluate if the relative amounts of glycosylated biomarkers in plasma could be estimated after sample processing with the optimized SPEG protocol and label-free LC-MS/MS analysis.
Experimental Section
Chemicals and solutions
Magnetic beads (1µm BcMag®Hydrazide-Modified Magnetic Beads) with hydrazide groups were purchased from Bioclone (San Diego). The glycerol free PNGase F solution was purchased from New England BioLabs. Water and acetonitrile (ACN) (J.T. Baker) were of HPLC quality. Unless otherwise stated, all other chemicals were purchased from Sigma Aldrich.
Sample preparation
A single pool of human plasma (purchased from Bioreclamation Inc. New York) with a concentration of ca. 66 mg/ml was used for all experiments described in this study except for the study where the proteins identified by SPEG were compared to the proteins identified without SPEG. The latter samples were collected from five healthy individuals for which IRB approval had been obtained. An aliquot of 1mg of protein in 15 ul of plasma (denoted as 1 mg/15 ul) was either subjected to SPEG without further processing, or 10 mg/150 ul of the plasma was depleted of 14 abundant proteins using a MARS Hu-14 multiple affinity removal LC column (10 × 100 mm) according to instructions from the supplier (Agilent Technologies). The depleted material was concentrated using an Amicon Ultra-4 3 kDa MWCO filter (Millipore), pre-rinsed with 0.1% n-octyl-beta-d-glucopyranoside (Anatrace) in MARS A buffer, through centrifugation at 3950g for one hour. The 0.6 mg of depleted plasma material was then subjected to SPEG.
Optimized peptide-level capture SPEG
Plasma proteins were reduced, alkylated, digested with trypsin, and cis-diols of carbohydrate groups on the glycopeptides oxidized as previously described 21 except that the sample was heated in 8 M urea denaturation buffer at 37 °C rather than 60 °C. In addition, a different reverse phase cartridge and elution conditions were used for desalting of the digest before and after the oxidation step and prior to bead loading (details, below). Depleted plasma samples were buffer exchanged with 8 M urea/0.4 M ammonium bicarbonate using 3 kDa MWCO filters prior to proteolytic digestion.
For the magnetic beads approach, all glycopeptide capture and washing steps were conducted using the KingFisher magnetic particle processor (Thermo Scientific). KingFisher is constructed to work with 96-well plates covered by changeable plastic covers with 96 plastic pockets that go into each well. Up to eight 96-well plates can be placed on a rotating platform simultaneously and a magnet head with 96 magnets go into the corresponding plastic pockets to collect and transfer beads from one 96-well plate to another. For one sample, approximately two hours are saved when using magnetic beads in combination with KingFisher compared to manual processing of magnetic beads. When processing more than one sample, the increase in saved time would extrapolate linearly with the number of samples, with about 5–10 minutes added for every additional sample.
Magnetic hydrazide beads (4 mg of beads/mg of protein unless otherwise stated) were transferred to a 96 well plate compatible with the KingFisher processor and washed twice with 80% ACN/0.1% TFA. The beads were released into corresponding wells in a new 96-well plate that contained the digested and oxidized peptides, eluted in 400 ul of 80% ACN/0.1% TFA. The mixture of magnetic beads and oxidized peptides was incubated over night with vigorous shaking. After coupling of the glycopeptides, the beads were washed three times using KingFisher with each of the following solutions in this order: 80% ACN/0.1% TFA, 8 M urea/0.4 M ammonium bicarbonate/0.1% SDS, 100% N,N-Dimethylformamide (DMF, toxic) and 0.1 M ammonium bicarbonate. The washed beads were released in 100 ul 0.1 M ammonium bicarbonate, and 1.5 ul of PNGase F was added followed by incubation over night with shaking at 37 °C. The following settings on the Kingfisher were used for the steps described above: the magnetic heads where lowered into the sample 5 times during bead collection to ensure good recovery and transfer of beads between plates. For magnetic bead release and washing the magnets were removed from the plastic pockets followed by a 30 seconds fast speed shaking and 4.5 minutes medium speed shaking of the 96-well plates. Elution of peptides from the PNGaseF-treated beads and cleanup of the formerly glycosylated peptides was performed as previously described, using a 10 mg Oasis HLB cartridges instead of C18 SepPak, and 0.1% FA and 80% ACN/0.1% FA instead of 0.1% TFA and 80% ACN/0.1% TFA. Dried peptides were resuspended in 10 ul of 3% ACN/5% FA and 2 ul was analyzed by LC-MS/MS. While larger amounts of bead (up to 32 mg of beads/mg of protein) were found to provide somewhat higher glycocapture yield as described in the Results section, scaling the experiments to use these higher bead amounts during the method development phase would have increased the cost of the experiments significantly.
For the macroporous hydrazide resin approach, 50 ul of the 50% Affi-Prep® Hz Hydrazide Support slurry (BioRad) was washed once using 1 ml of deionized water and 5 minutes of shaking. Reduced, alkylated and oxidized plasma digests were loaded onto Oasis cartridges and peptides eluted in 400 ul of 80% ACN/0.1% TFA. The eluate was then coupled to the pre-washed beads and incubated over night with 1200 rpm shaking with an Eppendorf shaker. The unbound peptides were removed using the same washing strategy as described for the magnetic beads above, except that the wash steps were done manually. Each wash step consisted of 10 seconds of vortexing, followed by 5 minutes of shaking at 1200 rpm and centrifugation at 3000g. The elution and clean-up steps for formerly glycosylated peptides were the same as those used in the magnetic protocol, above. Dried peptides were resuspended in 10 ul of 3% ACN/5% FA and 2 ul was analyzed by LC-MS/MS.
Optimized protein-level capture SPEG
The undepleted plasma sample (15 ul/1 mg) was diluted to 40 ul with oxidation buffer (20 mM NaAc and 150 mM NaCl, pH 5.0), and oxidized with 15 mM sodium periodate (NaIO4) at room temperature (RT) in the dark with shaking for one hour. After oxidation, the excess sodium periodate was removed through a buffer exchange step with coupling solution using a 0.5 ml Zeba desalting spin column (Pierce). The oxidized protein mixture was then coupled to either magnetic hydrazide beads or macroporous hydrazide beads. The coupling solution consisted of 100 mM NaAc/1.5 M NaCl/0.2% CHAPS (w/v) when used in combination with the magnetic beads, whereas CHAPS was omitted for use with macroporous beads. The sample was diluted to 300 ul with coupling solution prior to coupling to the pre-washed beads.
For the protein-level coupling magnetic bead SPEG protocol, all washing steps were conducted using KingFisher with the same settings as for the peptide-level coupling. Four mg of magnetic beads (not optimized) was washed two times with coupling solution (100 mM NaAc/1.5 M NaCl/0.2% CHAPS (w/v)). The sample was resuspended in coupling solution, loaded onto the pre-washed beads and incubated over night at RT with shaking. Unbound proteins in the supernatant were saved for future analysis, while the beads with the bound glycoproteins were dissolved in denaturation buffer (8 M urea/0.4 M ammonium bicarbonate/0.1% SDS). Disulfides were reduced with 10 mM tris (2-Carboxyethyl) phosphine hydrochloride (Thermo Scientific) for one hour at RT and free thiols alkylated with 12 mM iodoacetamide for 30 minutes at RT. Unbound proteins were removed by washing 4-times with 1 ml of 8 M urea/0.4 M ammonium bicarbonate/0.1% SDS, with 5 minutes shaking for each wash step. Bound proteins were resuspended in 500 ul 0.1 M ammonium bicarbonate/0.2% CHAPS (w/v) before trypsin was added to a trypsin:protein ratio of 1:50, calculated from the amount of starting protein as determined by BCA. After over night incubation at 37 °C with shaking, the beads were washed three times using KingFisher with each of the following solutions in this order: 80% ACN/0.1% TFA, 8 M urea/0.4 M ammonium bicarbonate/0.1% SDS, 100% N,N-Dimethylformamide (DMF, toxic) and 0.1 M ammonium bicarbonate to remove the remaining non-glycopeptides. The washes were combined with the unbound peptide digest supernatant and saved for possible future analysis. Bound former glycosylation-site peptides were released by PNGase F and then desalted by Oasis cartridge as described in the peptide-level protocol above. Dried peptides were resuspended in 10 ul of 3% ACN/5% FA and 2 ul was analyzed by LC-MS/MS.
For the macroporous resin approach, 200 ul of the 50% resin slurry was washed once with 1 ml water during 5 minutes of shaking prior to adding the proteins in coupling solution. Coupling of protein to the beads, denaturation of proteins, reduction, alkylation and digestion were carried out as described for the magnetic bead approach above, except that the proteins were digested in 0.1 M ammonium bicarbonate. The rest of the procedure was performed as for the magnetic bead protein-level coupling approach described above, except that all washing steps were performed manually.
Quenching the oxidation reaction
We tested quenching of the sodium periodate oxidation step in the peptide-level capture protocol with sodium sulfite as an alternative to cleanup with the Oasis HLB cartridges. After one hour incubation with sodium periodate, sodium sulfite was added to a final concentration of 150 mM followed by 20 minute incubation with shaking at room temperature. After incubation, the solution was added directly to the pre-washed magnetic beads as previously described 20. This procedure has not been incorporated in the optimized protocol as further explained in section “The influence of different coupling solutions on the SPEG performance”.
Basic pH reverse phase HPLC fractionation
Basic pH reverse HPLC was used to fractionate digested plasma samples using a Narrow-Bore 2.1 × 150 mm capillary reverse phase column (Agilent: ZORBAX) packed with 3.5 µm beads, coupled to an Agilent 1100 HPLC system. The mobile phases were as follows: mobile phase A (20 mM Ammonium Formate in water, 2% ACN, pH 10) and mobile phase B (90%ACN/10% 20 mM Ammonium Formate, pH 10). The gradient was 0–5 mins 0% B, 5–55 mins 50% B, 55–57 mins 100% B, 57–61 mins 100% B, 61–80 mins 0% B at a constant flow rate of 0.200 ml/min. A total of 12 factions were collected in a linear fashion from 0–70 minutes. Fractions 1 and 2 and fractions 11 and 12 were combined, giving a total of 10 fractions to be analyzed by LC-MS/MS.
LTQ-FT analysis
All the LC-MS/MS analyses were done using an Agilent nanoflow HPLC system (Agilent, Palo Alto, CA). A PicoFrit column (New Objective, Woburn, MA) with an inner diameter of 75 µm packed with 12–14 cm of ReproSil-Pur C18 3 µm particles, was directly interfaced to an LTQ-FT mass spectrometer (Thermo Fisher, Waltham, MA) equipped with a custom nanoelectrospray ionization source. Analyses were of 90 min total duration, using the following mobile phases: mobile phase A (0.1% FA) and mobile phase B (0.1% FA/90% ACN). The gradient used was as follows: hold at 3% B at 0.6 µl/min from 0–13 min then reduce flow to 0.2 µl/min from 13–15 min. From 15–18 min 3–18% B, from 18–68 min 18–60% B, from 68–73 minutes 60–100% B. From 73–81 min hold at 100% B at a flow 0.6 µl/minute 81–82.5 min then ramp from 100%-3% B, and re-equilibrate column at 3% B from 82.5–100 min. For the MS method, nine scan events were conducted. The mass spectrometer was set to do one full FTMS scan at 100,000 resolution in profile mode followed by 8 data-dependent MS/MS scans at low-resolution in centroid mode in the LTQ on the top 8 most abundant peptide precursor ions. For the experiments described in sections “Comparison of proteins identified with and without glycocapture”, “Glycoproteins spiked into plasma” and “SPEG comparison between undepleted plasma and plasma depleted by MARS Hu-14” in the Results section, 3 MS/MS scans of the top 3 most abundant peptide precursors were acquired with an isolation width of 2 m/z. Charge state screening was enabled along with monoisotopic precursor selection and non-peptide monoisotopic recognition to prevent triggering of MS/MS on precursor ions with unassigned charge or a charge state of 1. Normalized collision energy was set to 35 with an activation Q of 0.25 and activation time of 30 ms. Dynamic exclusion parameters included a repeat count of 2, a repeat duration of 20 sec, and an exclusion duration of 30 sec.
Peptide Identification
MS/MS data was searched against the International Protein Index (IPI) database version 3.32 using the Spectrum Mill software package v4.0 beta (Agilent Technologies, Santa Clara, CA). The search parameters were: a maximum of two missed cleavages, precursor mass tolerance 0.035 Da, a product mass tolerance 0.7 Da and carbamidomethylation of cysteines as the fixed modifications. Allowed variable modifications were oxidized methionines and deamidation of asparagine. Identities interpreted for individual spectra were automatically designated as valid by applying the scoring threshold criteria provided below to all spectra derived from a particular experiment in a two step process. First, protein mode was used which requires 2 or more matched peptides per protein and while allowing a range of medium to excellent scores for each peptide. Second, peptide mode was applied to the remaining spectra allowing for excellent scoring peptides that are detected as the sole evidence for particular proteins. Protein mode thresholds: protein score >25, peptide (score, Scored Percent Intensity, delta rank1 – rank2) peptide charge +2: (>8, >65%, > 2) peptide charge +3: (>9, >65%, > 2) peptide charge +4: (>9, >70%, > 2) peptide charge +2: (>6, >90%,> 1). Peptide mode thresholds for all charge states: >13, >70, > 2, respectively.
The above criteria yielded a false discovery rate of <1% as estimated by target-decoy based searches using reversed sequences. In addition to the criteria described above, for a peptide to be assigned as a glycopeptide and matched to a glycoprotein, it also had to contain the consensus sequence motif for N-glycosylation. Furthermore, the precursor mass and relevant fragments for the former glycosylation-site peptide had to be shifted lower in mass by 0.9840 Da relative to the Asn-containing peptide. This is due to the conversion of the attachment-site Asn to Asp upon release of the carbohydrate by PNGase F. These additional criteria are filtering options that can be selected by the user in Spectrum Mill.
Extracted Ion Chromatograms (XICs)
The peak area for the XIC of each precursor ion in the intervening high-resolution MS1 scans of the data-dependent LC-MS/MS runs was calculated automatically by the Spectrum Mill software using narrow windows around each individual member of the isotope cluster. Peak widths in both the time and m/z domains are dynamically determined based on MS scan resolution, precursor charge and m/z subject to quality metrics on the relative distribution of the peaks in the isotope cluster vs. theoretical.
Calculation of CV for SPEG pipeline reproducibility study
The intraday and interday reproducibility for the SPEG process in both the peptide- and protein-level coupling modes were calculated using “protein XIC” values which are the sum of the observed XIC values of the constituent peptides for each specific protein. Intraday variation was determined from the variation in protein XIC values for five process replicate samples (a dataset) run on two different days (generating two data sets). The CV for each protein XIC value was calculated if the protein was observed in a minimum of two of the five replicates run on the same day. The median intraday CV of all proteins in a dataset was determined from the CVs of the protein XIC values. The median CVs for each of the two datasets is reported as an average value (Table 4). To determine the interday variation, the two intraday dataset were combined, and one CV for the XIC values for each protein over the runs was calculated if the protein was identified in two or more of the ten process replicates. The median CV was extracted from the resulting list of CVs (Table 4).
Table 4.
Calculated median CV values for the SPEG pipeline using either depleted or undepleted plasma with peptide-level coupling or undepleted plasma with protein-level coupling.
| Reproducibility | Intraday protein XIC (median CV) |
Interday protein XIC (median CV) |
|---|---|---|
| Depleted Plasma peptide-level |
24% | 30% |
| Neat plasma peptide-level |
25% | 32% |
|
aNeat plasma protein-level |
64% | 63% |
The last row shows the median CV when protein-level coupling was performed on undepleted plasma.
Calculation of SPEG pipeline performance
The principal criteria we used to evaluate the different SPEG pipelines and the effect of changes in the sample processing methods were the observed specificity for glycopeptide or glycoprotein enrichment and the total number of glycopeptide and glycoprotein identified. The definition of a glycopeptide in this context was that it had to be identified with the glyco-motif, and had to have a mass difference corresponding to deamidation. The definition of a glycoprotein was a protein that contained one or more identified glycopeptides. The specificity of the glycopeptide capture was defined as the percentage of unique glycopeptides among the total number of unique peptides identified. The specificity of glycoprotein capture was calculated from the percentage of unique proteins identified that contained identified glycopeptides.
Results and discussion
An overview of the reactions that occur during glycoprotein oxidation, coupling to the hydrazide groups on the magnetic beads and cleavage of the N-linked glyco-unit from the polypeptide using PNGase F is shown in Figure 1. A schematic overview of the peptide- and protein-level coupling approaches is shown in Figure 2.
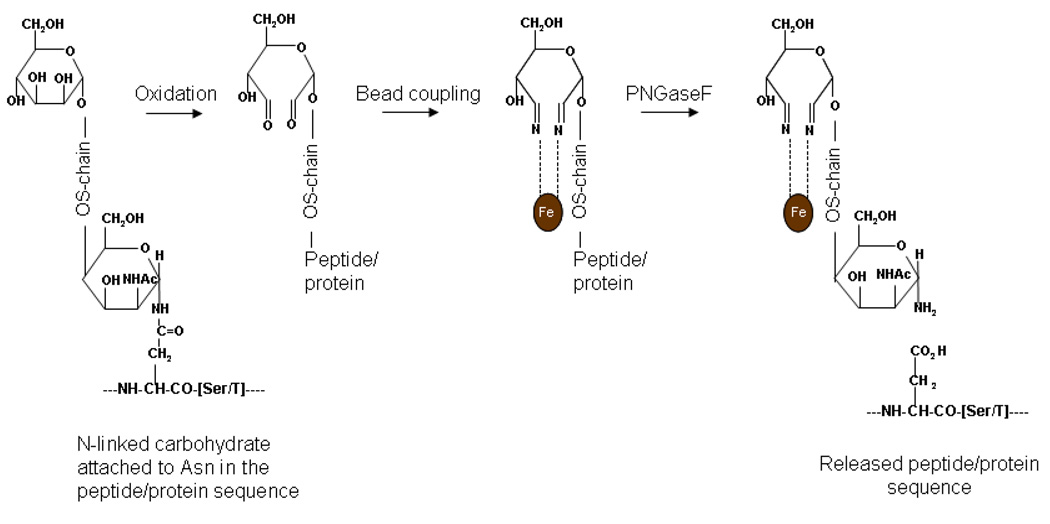
Figure 1.
Reactions occurring during oxidation, hydrazide magnetic bead coupling and cleavage of the oligosaccharide chain (OS-chain) from the peptide or protein by PNGaseF. The N-linked OS-chain is attached to the polypeptide through the side-chain of asparagine (Asn) which is found in the consensus sequence …Asn-X-Ser/Thr… in which X can be any amino acid other than Pro. During oxidation of the OS-chain with sodium periodate, hydroxyl groups on adjacent carbon atoms of the carbohydrates are converted to aldehydes. The hydrazide groups connected to the magnetic beads react spontaneously with aldehyde or ketone groups to form stable covalent hydrazone groups. PNGaseF hydrolyzes the bond between the OS-chain and the Asn on the amino acid backbone, converting the attachment-site asparagine to aspartic acid and releasing it from the bead-bound carbohydrate.
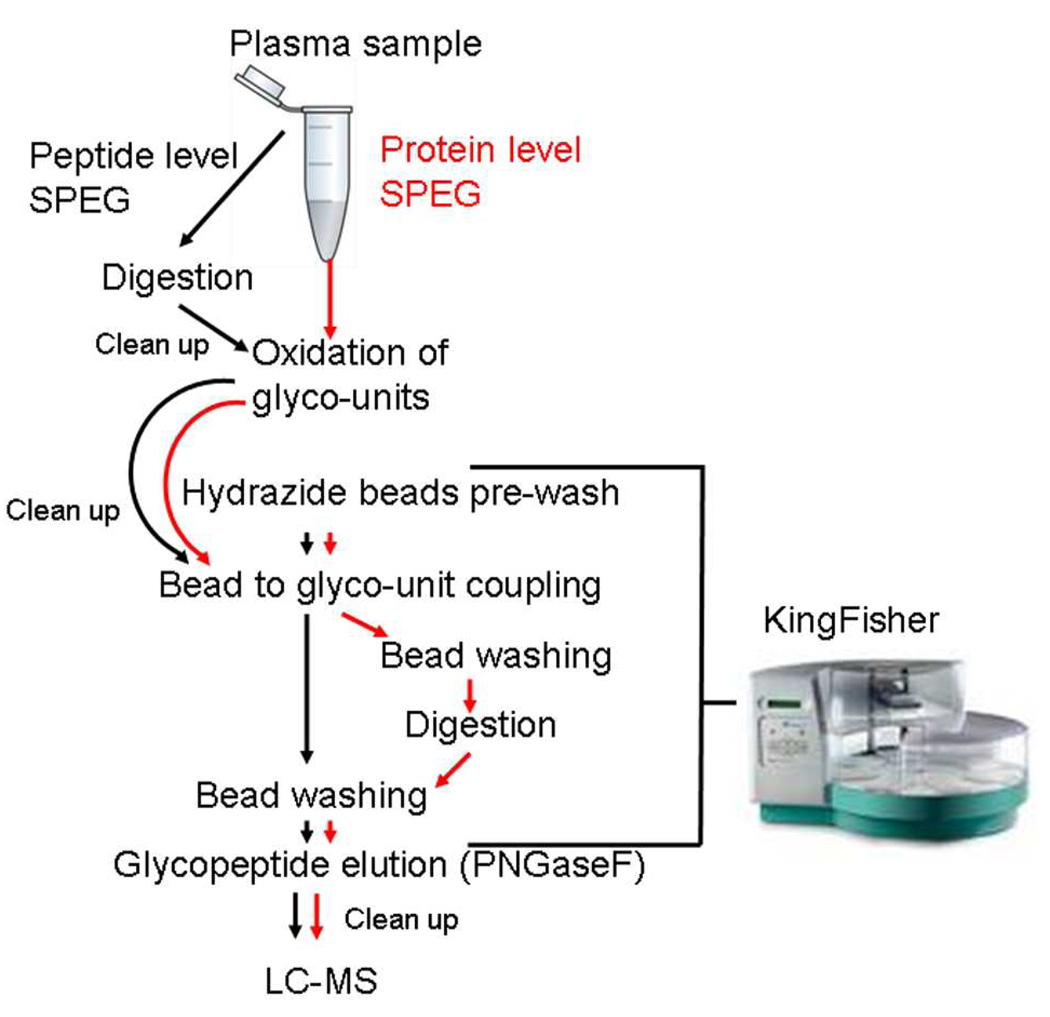
Figure 2.
Schematic overview of the peptide- and protein-level coupling approaches. The peptide-level path is indicated by black arrows, and the protein-level coupling path is indicated by red arrows. KingFisher (Thermo) was used for the magnetic bead handling steps, from the hydrazide beads pre-wash step throughout the glycopeptide elution step.
The influence of different coupling solutions on the SPEG performance
In previously published work, the following three different coupling solutions were used during coupling of the glyco-unit to the hydrazide groups when using macroporous beads: 0.1 M NaAc/150 mM NaCl pH 5.5 19, 0.1 M NaAc/1.5 M NaCl pH 4.5 22 and 80%ACN/0.1%TFA 21. However, there has not been a comparative evaluation of these different coupling solutions on the overall performance of the method. Yet another solution is recommended by the supplier of the magnetic beads (Bioclone) for coupling (0.1 M NaAc, pH 5.6) and for bead storage (PBS pH 7.4). To determine which of the coupling solutions gives the best performance in combination with peptide-level coupling and magnetic beads, each coupling solution was tested using the optimized core protocol and pipeline (Table 1 and Experimental Section). Supplementary Table 1 worksheet t1 lists all of the confidently assigned peptides and the corresponding proteins identified in these experiments. Process replicates for each coupling condition were prepared using a common human plasma pool and analyzed by LTQ-FT-MS/MS. From the results (Table 1), it was clear that the 80%ACN/0.1%TFA coupling solution yielded the best glycopeptide specificity. In addition, the number of unique glycopeptides that were identified was similar or better for this solution compared to all other coupling solutions tested. We also evaluated the use of sodium sulfite in 50%ACN/0.1%TFA prior to coupling to quench residual sodium periodate used for cis-diol oxidation and thereby avoid the need to desalt after oxidation as previously described 20. While this quenching approach gave similar numbers of glycopeptides as the 80%ANC/0.1%TFA coupling buffer combined with desalting, the specificity for glycopeptide enrichment was more variable, so we did not further pursue this option. Based on these experiments, 80%ACN/0.1%TFA with desalting to remove the oxidizing agent was selected as the optimal method for peptide-level coupling.
Table 1.
The effect of different coupling solutions on the SPEG performance when using peptide-level coupling and magnetic beads.
|
a0.1M NaAc/ 150mM NaCl |
a0.1M NaAc/ 1.5M NaCl |
a0.1M NaAc |
aPBS |
b80%ACN/ 0.1%TFA |
c50%ACN/0.1 % TFA, Sodium sulfite quenching |
|
|---|---|---|---|---|---|---|
| # of unique glycopeptides and (CV) |
105 (12%) | 100 (9%) | 80 (0%) | 82 (3%) | 101 (6%) | 101 (8%) |
| Glycopeptide specificity and (CV) |
68% (4%) | 67% (4%) | 58% (18%) | 61% (23%) |
99% (0.1%) | 94% (3%) |
Samples were analyzed as two process replicates using a LTQ-FTMS.
Samples were analyzed as three process replicates.
Three additional process replicates were included for the 50%ACN/0.1%TFA coupling condition with subsequent quenching.
For protein-level coupling to the magnetic beads, we found that 0.1 M NaAc/1.5 M NaCl/0.2% CHAPS gave much better specificity and number of glycopeptides identified than 80%ANC/0.1%TFA (results not shown). It was important to include 0.2% CHAPS in the coupling solution and in the 0.1 M ammonium bicarbonate solution during digestion to prevent the magnetic beads from clumping into larger complexes that potentially could non-specifically sequester peptides during bead washing. When three process replicates with and without CHAPS in the coupling solution were compared, the glycopeptide average specificity was 73% and 46%, respectively. Hence, 0.1 M NaAc/1.5 M NaCl/0.2% CHAPS was chosen as the protein-level coupling buffer of choice, and 0.1 M ammonium bicarbonate with 0.2% CHAPS was identified as the digestion buffer of choice.
Influence of the amount of beads on the SPEG performance
In clinical proteomics, maximizing the glycoprotein/glycopeptide yield is important as the sample material may be limited. To determine the optimal amount of beads to use in these experiments, we evaluated the influence of the amount of magnetic beads on the glycopeptide yield, specificity and number of identifications. Using the optimized peptide-level coupling SPEG procedure, we compared nine conditions by processing different amounts of a common pool of human plasma with varying amounts of beads (Figure 3). The estimate of peptide yield was based on the total peptide precursor XIC value reported by Spectrum Mill for all identified peptides for the middle 80% of the gradient containing matched MS/MS spectra. Supplementary Table 1 worksheet f3 lists all the confidently matched identifications from these experiments.
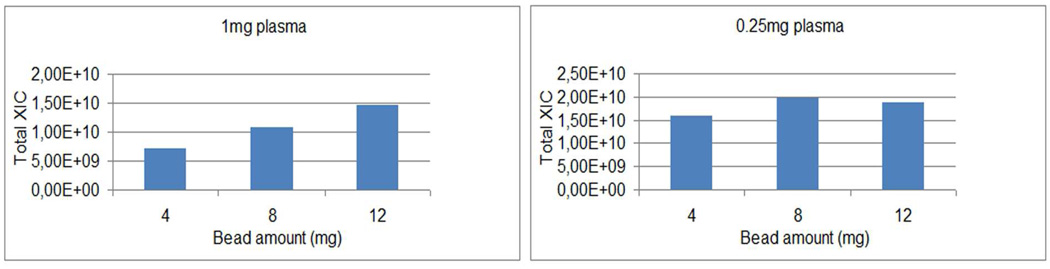
Figure 3.
The effect of magnetic bead amounts on the total XIC for different combinations of plasma and bead amounts in combination with peptide-level coupling. Twenty percent of the total sample amount was injected onto column for all samples. The estimate of peptide yield was based on the total precursor XIC value reported by Spectrum Mill, extracted from the XIC’s for all identified peptides for the middle 80% of the gradient containing matched MS/MS spectra.
For peptide-level coupling from 0.5 (not shown) and 1 mg (Figure 3A and Supplementary Table 2A) of digested plasma protein, we found that increasing the amount of magnetic beads from 4 mg to both 8 and 12 mg increased the yield of glycopeptides based on the observed total ion current. When 0.25 mg of plasma was used (Figure 3B and Supplementary Table 2B), there was no increase in the total XIC value when increasing the bead amount from 8 mg to 12 mg, indicating that the binding capacity for the beads for this application was approximately 8 mg of beads / 0.25 mg of digested protein. This extrapolates to ~32 mg beads for processing 1 mg of digested plasma protein. In summary, increasing the bead amounts seemed to improve the peptide yield in general up to the level of 32 mg of beads / 1 mg of plasma proteins, but did not lead to capturing a higher number of unique glycopeptides. Interestingly, the number of uniquely identified glycopeptides and the glycopeptide specificity did not change significantly with increasing peptide yield, only the absolute amount of what had already been captured increased (results not shown). This could be due to the large dynamic abundance range of glycoproteins in blood, resulting in glycopeptides from abundant glycoproteins interfering with or suppressing detection of lower abundance glycopeptides despite the significant overall reduction in sample complexity provided by SPEG. Alternatively, it could be that lower abundance glycopeptides are not being efficiently captured and/or recovered. Both hypotheses are supported by results from the Smith lab in which ca. 300 glycoproteins were identified by depleting plasma of abundant proteins prior to SPEG processing and extensive fractionation of the resulting sample prior to LC-MS/MS 24.
For protein-level coupling the yield of peptides captured when using 0.25 mg of plasma and 2, 4, 8 or 12 mg of beads was compared. Supplementary Table 1 worksheet f3 lists all of the confidently assigned peptides and the corresponding proteins identified in these experiments. Starting with 0.25 mg of plasma protein, the apparent yield of glycopeptide continued to increase with increasing amounts of bead up to and including 12 mg of bead, the highest amount tested (Supplementary Figure 1 and Supplementary Table 2C). Hence it appeared that ~48 mg of beads / 1 mg of plasma proteins (extrapolated) was not enough beads to avoid saturation of the beads with bound glycoproteins. In conclusion there seems to be a need to go higher in bead amount when using protein-level coupling compared to when using peptide-level coupling to avoid bead saturation.
The influence of different washing conditions on the SPEG performance
Washing conditions used to remove non-specifically bound peptides from the beads were evaluated in an effort to further reduce background and potentially improve reproducibility. Several different combinations of solutions, number of repeated steps and washing times were tested and compared using a human plasma pool and various SPEG protocols for the different conditions tested. A recently published 5-solution washing protocol was used as a reference point 22. This 5-solution protocol consisted of three washes with each of the following five solutions: 1.5 M NaCl, ACN, MeOH, H2O and 0.1 M ammonium bicarbonate. From the different reagents tested we found that substituting methanol for DMF increased the glycopeptide specificity by about 5% (data not shown). Substituting the H2O and NaCl washing solutions with a solution containing 8 M urea/0.4 M Ammonium bicarbonate/0.1% SDS reduced the loss of magnetic beads during the washing. Substituting ACN with 80%ACN/0.1%TFA was based on the idea that an acidic wash would have positive impact on the removal of unbound peptides. Reducing the number of washing iterations from three to two gave a decrease in the resulting glycopeptide specificity (data not shown). The number of uniquely identified glycopeptides was not significantly influenced by the choice of washing procedure (results not shown). Of the different washing protocols tested, a 4-solution washing procedure consisting of 80%ACN/0.1%TFA, 8 M urea/0.4 M ammonium bicarbonate/0.1% SDS, DMF and 0.1 M ammonium bicarbonate provided high glycopeptide specificity with little loss of magnetic beads. We further tested the 4-solution washing procedure in comparison with the 5-solutions reference procedure (Table 2). Supplementary Table 1 worksheet t2 lists all the confidently matched identifications from this section. Glycopeptide specificity was consistently better for the 4-solution procedure vs. the 5-solution method for both magnetic beads and macroporous beads, and for both protein and peptide-level coupling options. The exception was for protein-level coupling and macroporous beads, where the result was similar for both washing procedures. The 4-solutions washing procedure was also more compatible with magnetic beads and the KingFisher magnetic particle processor, as significantly fewer beads were lost during washing. These advantages make the 4-solution washing procedure the clear choice, especially when using magnetic beads in combination with KingFisher.
Table 2.
The glycopeptide specificity for protein and peptide-level coupling to macroporous and magnetic beads using two different washing procedures.
| b4-solution wash | c5-solution wash | ||
|---|---|---|---|
| Hydrazide bead type |
Protein- or peptide- level bead coupling |
aGlycopeptide specificity and (CV) |
aGlycopeptide specificity and (CV) |
| Macroporous | Peptide | 94% ( 4%) | 87% ( 2%)d |
| Magnetic | Peptide | 99% ( 0.1%) | 94% ( 2%) |
| Macroporous | Protein | 94% ( 3%) | 94% ( 4%) |
| Magnetic | Protein | 46% ( 3%)e | 41% ( 17%)e |
All samples were analyzed by LC-MS/MS. The numbers are shown as average of 3 process replicates from a human plasma pool, with associated CV.
The 4-solutions washing procedure relied on the following solutions: 80%ACN/0.1%TFA, 8 M urea/0.4 M Ammonium bicarbonate/0.1% SDS, DMF and 0.1M Ammonium bicarbonate.
The 5-solutions protocol consisted of three washes with each of these five solutions: 1.5M NaCl, ACN, MeOH, H2O and 0.1M Ammonium bicarbonate.
Only two process replicates were used for this condition.
Magnetic beads protein-level coupling was performed without CHAPS in the coupling solution.
Comparing SPEG performance for macroporous and magnetic beads for both the protein and peptide-level coupling options
After optimizing the magnetic bead SPEG protocol as described above, a comparison between the performance of these beads and the macroporous beads was conducted. If the performance was similar or better for the magnetic bead approach, these beads would be preferred due to the ease of automation. We also compared the performance of the optimized peptide-level and protein-level coupling protocols for both bead types. To accomplish these comparisons, three aliquots of a pooled human plasma sample were processed for each of the four conditions: protein and peptide-level coupling for both macroporous and magnetic beads. The optimized SPEG procedures (described in detail in the Experimental Section) were used, and the processed samples were analyzed by LC-MS/MS. The performance of each condition was evaluated based on glycopeptide and glycoprotein specificity and number of identifications, as summarized in Table 3. Supplementary Table 1 worksheet t3 lists all the confidently matched identifications from this section. Supplementary Table 3 lists all the proteins identified from the four conditions (peptide- and protein-level and macroporous and magnetic beads) and for which condition they were observed. The typical plasma abundance for a selection of the identified proteins is reported based on the values given in the publication of Hortin et al.25. From these data, there is a tendency that the most abundant glycoproteins are identified under all conditions, whereas some of the lower abundant proteins are identified only when doing protein-level coupling.
Table 3.
Glycoprotein and glycopeptide specificity and number of unique glycopeptides/glycoproteins for protein and peptide-level coupling to macroporous and magnetic beads.
| Hydrazide bead type |
Protein- or peptide- level bead coupling |
a# of unique peptides with N[^P][ST] motif; (CV) |
aGlycopeptide specificity; (CV) |
a# of unique proteins with N[^P][ST] motif; (CV) |
aGlycoprotein specificity (CV) |
| Macroporous | Peptide | 108 (8%) | 94% (4%) | 46 (9%) | 93.9% (2 %) |
| Magnetic | Peptide | 101 (6%) | 99% (0.1%) | 50 (11%) | 97.3% (1%) |
| Macroporous | Protein | 131 (7%) | 94% (3%) | 59 (11%) | 91.7% (1%) |
| Magnetic | Protein | 141 (3%) | 73% (1%) | 61 (1%) | 89.7% (0.1%) |
All samples were analyzed by LC-MS/MS. The numbers are shown as average of 3 process replicates from a human plasma pool with associated CV values.
When comparing glycoprotein- and peptide-level coupling, protein-level consistently provided more glycopeptide and glycoprotein identifications regardless of bead type. One possible explanation for this could be that highly hydrophilic glycopeptides are lost during the Oasis HLB clean-up prior to hydrazide bead coupling, as they do not bind to the reverse phase material. This step was not used in protein-level bead coupling, where a size exclusion column was used for clean-up instead. Peptide-level coupling gave a higher glycopeptide and glycoprotein capture specificity than protein-level coupling for the magnetic beads.
Macroporous beads showed significantly better glycopeptide specificity compared to the magnetic beads when using protein-level coupling, with a specificity of 94% and 73%, respectively. From the list of non-glycopeptides identified from the magnetic bead approach, it appeared that the majority of these peptides arose from the captured glycoproteins and not from non-glycoproteins (results not shown). This was also supported by the fact that the glycoprotein specificity for the two bead types was similar. Peptide-level coupling gave a different result, as both the glycopeptide and glycoprotein specificity was approximately 4% better for the magnetic bead approach. The number of identified glycopeptides and proteins was similar with both bead types.
These results indicate that the hydrazide SPEG approach allows for very high glycoprotein/peptide specificity over lectins (reported specificity of 69% for glycoproteins18), and in addition, the hydrazide approach can be automated for higher throughput. It is also our impression that it is easier to avoid background from non-glycoproteins/peptides when using the hydrazide approach as it allows for more stringent washing due to covalent attachment to the magnetic beads. This should also increase the reproducibility of the hydrazide method.
SPEG comparison between undepleted plasma and plasma depleted by MARS Hu-14
The relatively low number of identifications from the different SPEG experiments suggested that the high abundance and dynamic range of non-glycosylated peptides could be limiting the dynamic range for glycopeptide detection. We therefore compared SPEG on neat undepleted plasma and plasma depleted of the 14 most abundant proteins using a MARS Hu-14 antibody column. Eight samples each of a human plasma pool sample were processed by SPEG before and after depletion and the samples were subsequently analyzed by LC-MS/MS, each in technical replicate. Supplementary Table 1 worksheet f4 lists all the confidently matched identifications from this section.
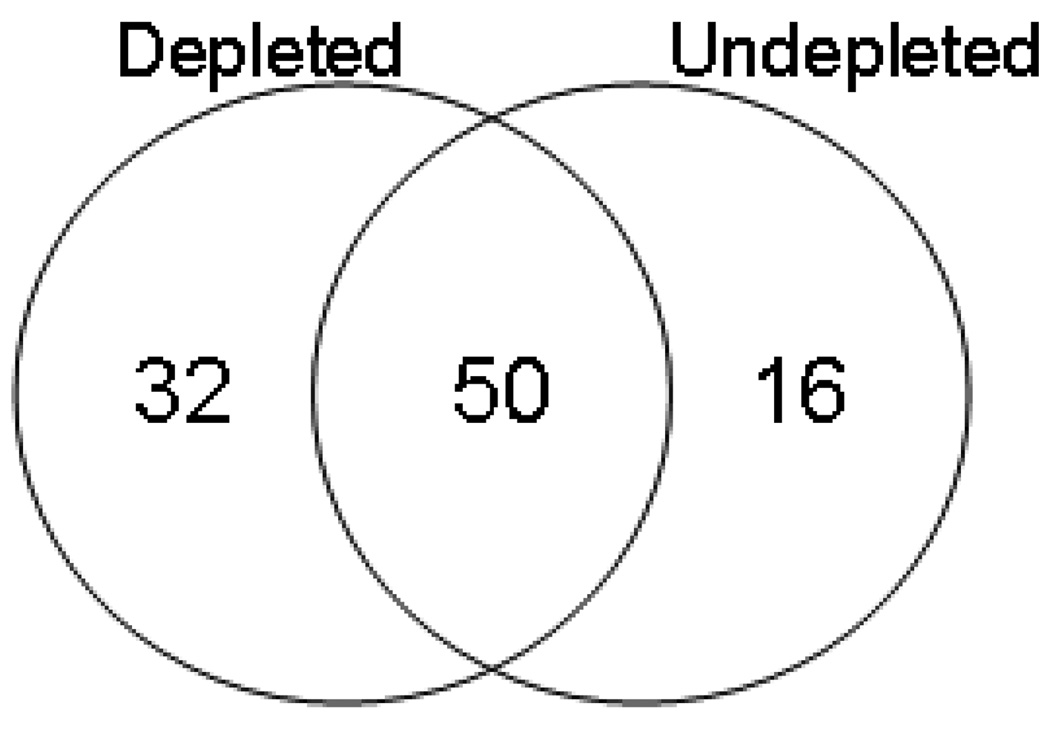
The Venn-diagram displayed in Figure 4 shows the overlap of glycoproteins identified in the undepleted and depleted samples. The MARS Hu-14 depletion of plasma prior to SPEG leads to identification of 32 new glycoproteins not detected prior to depletion, an increase of 24% in the total number of glycoproteins identified. Twelve of the 14 proteins in plasma targeted by the MARS Hu-14 depletion column are glycoproteins, albumin and apolipoprotein AII are the exceptions. Eleven of the twelve glycoproteins targeted for depletion, as well as immunoglobulin J chain, polymeric immunoglobulin receptor and aplipoprotein D, three glycoproteins known to bind proteins targeted for depletion (IgM/IgA, IgM/IgA and apolipoprotein AII, respectively), were detected in neat plasma following SPEG (Supplementary Table 4). In addition two different entries were given for IgG, IGHG3 and IGHG4. Therefore only 2 glycoproteins are uniquely detected in undepleted plasma that are not expected to have been removed by the depletion column, factor VII protein and coagulation factor XI.
Figure 4.
The Venn-diagram shows the number of glycoproteins identified from the depleted and undepleted plasma samples. Thirty-two additional glycoproteins were uniquely identified following depletion. Eleven glycoproteins targeted for removal by the MARS Hu-14 column, as well as three glycoproteins known to bind to one or more of the targeted proteins, are included in the total of 16 unique glycoproteins observed only in the undepleted plasma by SPEG.
Alpha2-macroglobulin was the only one of the 12 glycoproteins targeted by the MARS column that was detected following depletion. Albumin, the most abundant protein in plasma, was detected in neat plasma as a non-glycoprotein following SPEG (Supplementary Table 4). Following depletion, albumin and alpha2-macroglobulin were still detected, albeit with very low XIC values. Using a constant mass loading of ca. 1 ug of protein onto the LC-MS/MS, corresponds to the analysis of ten times more volume of depleted plasma than that of non-depleted plasma. The total XIC values obtained under these conditions gave nearly equivalent values (ca. 2.5 E+10), indicating that close to one order of magnitude in detection range is achieved by depleting the plasma samples prior to SPEG processing.
Evaluation of the reproducibility of the magnetic hydrazide protocol
Good reproducibility of proteomics discovery methods is important in order to allow for the discovery of small differences in protein abundance between samples. To address the reproducibility of the semi-automated, magneticbead based SPEG procedure, five aliquots of depleted and non-depleted human plasma from a common pool were processed by SPEG and analyzed by LC-MS/MS on two different days. Both peptide-level and protein-level coupling were evaluated. Supplementary Table 1 worksheet t4 lists all the confidently matched identifications from these experiments. The reproducibility of the different SPEG pipelines (Table 4) was calculated as described in the Experimental section under “Calculation of CV for SPEG pipeline reproducibility study”. From these calculations the reproducibility of the peptide-level glyco-unit coupling approach was significantly better than for protein-level coupling, with a CV of 31.9% vs. 62.9%, respectively. This is in line with the expectations that the higher glycopeptide capture specificity observed for peptide-level coupling approach contributes to better reproducibility, and that the level of non-specific background peptides varies more between samples when compared to the level of specifically bound glycopeptides. Other points that can be made from the data are that there is a small increase in CV when going from intraday to interday analysis (~6%) and that the depletion of plasma prior to SPEG processing did not have a detectable effect on the CV.
Comparison of proteins identified with and without glycocapture
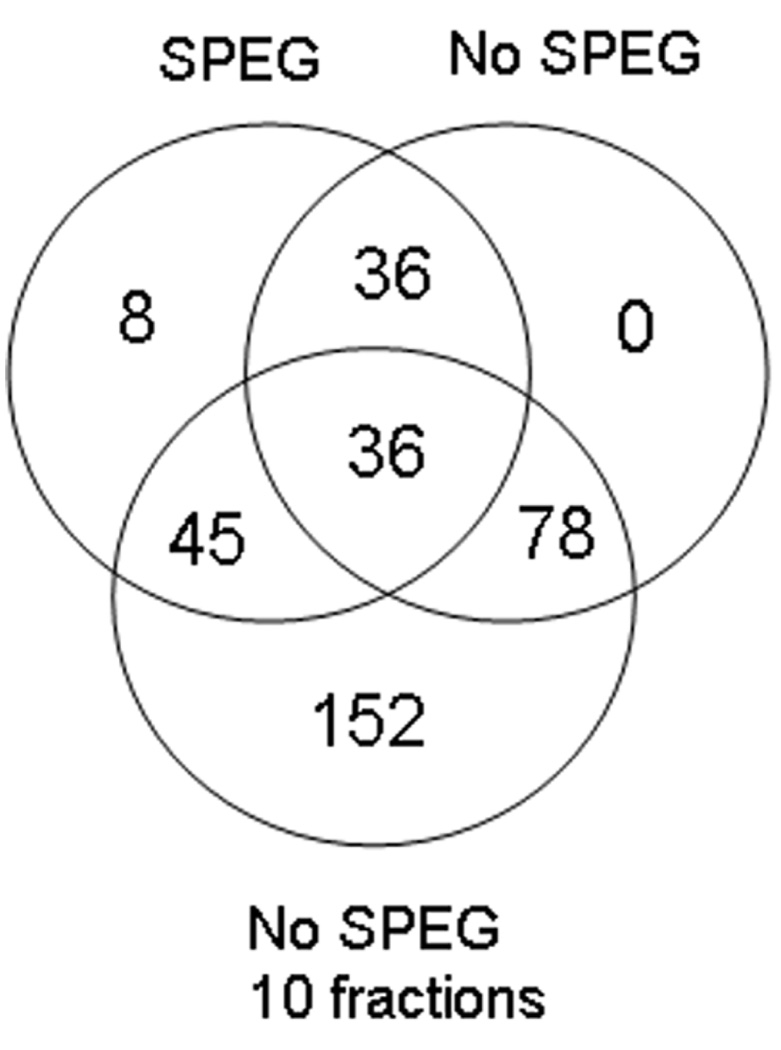
An important question is whether SPEG processing results in the identification of proteins that could not be identified by LC-MS/MS alone, even if the latter is carried out using fractionation to improve depth of detection. To address this, plasma samples were taken from five different healthy human individuals and depleted of the 14 most abundant proteins using MARS Hu-14. Each sample was then split into three aliquots, where one aliquot was processed by SPEG (250 µg), one was digested without SPEG (10 µg), and one was digested without SPEG (100 µg) and then separated into 10 fractions using basic pH reverse phase HPLC. After processing, all samples were analyzed by LC-MS/MS, loading ~1 µg from each sample/fraction onto column. Supplementary Table 1 worksheet f5 lists all the confidently matched identifications from this section. The resulting protein identification lists given as output by Spectrum Mill from the five patients was combined within each of the three sample processing approaches. The overlap in protein identifications between the different approaches is shown as a Venn diagram in Figure 5. A total of 53 unique proteins (51 of these were glycoproteins) were identified by SPEG. For samples not subjected to SPEG, a total of 78 proteins were identified in the sample that had not been pre-fractionated while a total of 239 proteins and glycoproteins were identified in the sample that had been fractionated prior to LC-MS/MS. Eight unique proteins were identified by SPEG when compared to both the non-SPEG approaches, and 17 proteins where uniquely identified by SPEG when compared only to the unfractionated non-SPEG process. These results show that SPEG prior to LC-MS/MS gives complementary information for glycoproteins present in a plasma sample even when compared to plasma fractionated into 10 fractions prior to LC-MS/MS.
Figure 5.
Venn-diagram of the protein identification lists from three sample sets, where the first was processed by SPEG (SPEG) prior to LC-MS/MS, the second was not processed by SPEG (No SPEG) prior to LC-MS/MS, and the third was not processed by SPEG, but fractionated into 10 fractions by offline basic pH=10 reverse phase (No SPEG 10 fractions) prior to LC-MS/MS. This is how the circle for the SPEG experiment should be read: 8 proteins were unique to this experiment, 45 proteins were common between SPEG and No-SPEG 10 fractions, 36 proteins were common between SPEG and No-SPEG, 36 proteins were common for all conditions. All the 36 proteins common between SPEG and No-SPEG were also common for all conditions. Hence, a total of 53 (8 plus 45) proteins were identified by the SPEG experiment.
Although not demonstrated here, fractionation will likely improve detection of glycoproteins in plasma by SPEG, but at the considerable cost of additional sample processing and analysis time. For example, in a prior study from the Smith laboratory 24, SPEG was used to capture glycopeptides from an Agilent MARS-7 depleted human plasma sample followed by SCX-based-LC separation into 30 fractions and LC-MS/MS, identifying 303 unique glycoproteins. 180 of these were unique for the SPEG approach when compared to a dataset of 938 proteins identified from a global shotgun approach by the same group. This further indicates that SPEG enrichment reveals new information from the plasma proteome, and that after further fractionation the number of glycoproteins identified will increase significantly.
Detection limits for glycoprotein standards spiked into plasma
In order to begin to determine if the SPEG pipeline could be used for biomarker discovery, two bovine glycoproteins, Fetuin-A and Fibronectin (Sigma), were spiked into identical aliquots of 150 ul neat plasma (purchased from Bioreclamation) in five different amounts, 0 pmol, 10 pmol, 100 pmol, 1 nmol and 10 nmol/ml of plasma. Each concentration was spiked into two aliquots of plasma that separately went through MARS Hu-14 depletion following peptide-level magnetic bead capture and analyzed two times each by LC-MS/MS giving a total of four data files per spike-in level. Supplementary Table 1 worksheet t5 lists all the confidently matched identifications from this section. The XIC values for one identified glycopeptide from each of the two spiked-in glycoproteins were manually calculated for each replicate based on the precursor peak area. These bovine glycopeptides had unique amino acid composition compared to the corresponding human sequences. The average XIC value from the four replicates for each spike-in level and the corresponding CV is reported in Table 5A. The fold difference between the spiked-in glycoproteins between neighboring spike-in levels based on the average XIC values in Table 5A is given in Table 5B. One expects a 10 fold difference between the spiked-in proteins from neighboring spike-in levels, whereas the observed fold difference varied from 9 to 12, with one exception where the fold change was 6. No difference (one fold difference) in Table 5B means that the spike-in level could not be distinguished from the background. The Fetuin-A glycopeptide was identified in all four replicates at the 100 pmol to 10 nmol spike-in levels, and in one replicate at the 10 pmol level. The Fibronectin glycopeptide was identified in all four replicates at the 10 nmol and 1 nmol levels, but not at the other levels. XIC values could, however, also be extracted from the 100 pmol/ml level. Based on these results, the relative amount of spiked-in proteins can be estimated with good accuracy after SPEG processing based on the detection of a single peptide down to a level of 10–100 pmol/ml, which corresponds to 384 ng-3.84 µg for Fetuin-A and 2.49 µg-24.9 µg for Fibronectin.
Table 5.
| Table 5A. Average XIC value and CV of one peptide from Fetuin-A and Fibronectin spiked into plasma at different levels. | |||||
|---|---|---|---|---|---|
| 10 nmol/ml aAverage XIC / CV |
1 nmol/ml aAverage XIC / CV |
100 pmol/ml aAverage XIC / CV |
10 pmol/ml aAverage XIC / CV |
0 pmol/ml aAverage XIC / CV |
|
|
cFetuin-A XIC |
7.09E+8 ( 5%) | 1.2E+8 ( 6%) | 1E+7 ( 24%) | 8.6E+5 ( 9%) | 9.9E+5 ( 16%) |
|
dFibronectin XIC |
1.34E+8 (13%) | 1.4E+7 ( 19%) | 1.4E+6 ( 18%) | bNot detected |
bNot detected |
| Table 5B. Fold difference between neighboring spike-in levels based on the average XIC values given in Table 5A. | ||||
|---|---|---|---|---|
| 10 nmol vs 1 nmol fold difference |
1 nmol vs 100 pmol fold difference |
100 pmol vs 10 pmol fold difference |
10 pmol vs 0 Pmolfold difference |
|
| Fetuin-A XIC | 6 | 12 | 12 | 1a |
| Fibronectin XIC | 9 | 10 | 1a | 1a |
The average value is based on a total of 4 replicate analysis for each spike-in level. The XIC values were manually extracted based on the peak area of one peptide from each of the two proteins in the table.
Not detected means that no peak area could be defined for that spike-in level.
The Fetuin-A peptide was VVHAVEVALATFNAESNGSYLQLVEISR and
the Fibronectin peptide was DQCIVDGITYNVNDTFHK.
A fold difference of 1, means that the spike-in level could not be distinguished from the background.
Conclusion
In this work we have optimized and compared different SPEG procedures and have concluded that commercially available magnetic hydrazide beads overall perform comparably or better than the commonly used macroporous beads, and are much simpler to automate for high throughput discovery projects. The choice of coupling solution, the magnetic bead amounts and washing procedure and the depletion of the most abundant protein in plasma prior to SPEG proved to be important factors that influenced the SPEG performance. The reproducibility of the peptide-level SPEG-LC-MS/MS pipeline is sufficiently high to allow this procedure to be used for discovery clinical proteomics, for example for biomarker discovery, with an interday CV of 30%. Peptide-level coupling to magnetic beads had better glycopeptide specificity and also better reproducibility compared to protein-level coupling protocol. On the other hand, protein-level coupling gave a higher number of identified glycopeptides/glycoproteins. Whether to use protein- or peptide-level coupling should be determined based on the need of the intended application (coverage versus reproducibility), as both of these procedures have advantages that could help increase the chances of finding novel glycoproteins in plasma and other body fluids. Importantly, the sensitivity of the SPEG method, even when coupled to abundant protein depletion, is relatively limited. Based on the glycoprotein spike-in experiments reported here, proteins present in the range of 10–100 pmol/ml (100’s of nanograms/ml to 10’s of micrograms/ml) in depleted plasma can be detected. In order for SPEG to detect glycoproteins at lower levels, a requirement for biomarker discovery in plasma, fractionation at the protein or peptide level will be required. The need to combine fractionation with the relatively complex sample processing involved in the SPEG method may limit adoption of SPEG for biomarker discovery in plasma.
Supplementary Material
Acknowledgment
This work was supported in part by grants to S.A.C. from the Bill and Melinda Gates Foundation as part of Grand Challenge 13, from the National Institutes of Health Grants 1U24 CA126476 as part of the NCI’s Clinical Proteomic Technologies Assessment in Cancer Program, from the National Heart Lung and Blood Institute, grant R01 HL096738-01 and from the Women’s Cancer Research Fund of the Entertainment Industry Foundation. The work was also supported in part by grants to F.S.B. from the Western Norway Regional Health Authority and The Leiv Eiriksson Mobility Program of the Norwegian Research Council. We would like to thank Margaret Pyle and Jacob Jaffe of the Broad Institute for their help with sample processing and data analysis, respectively. We also thank Hui Zhang, Department of Pathology, Johns Hopkins University for discussion of her previously published methodology and for sharing her reference mouse plasma samples and in-house developed magnetic beads for our initial protocol testing.
Footnotes
Supporting Information available: The raw mass spectral data in the original instrument vendor file format associated with this manuscript may be downloaded free of charge from ProteomeCommons.org Tranche, https://proteomecommons.org/tranche/, using the following hash: CDVFJW4RaxglQ0xIGsnXBq9zQ/1CaWNcJOZaZsPx32NIZnREQbartyLlSPDw1bKNuYtY7ltbwjSm vjP2yLv+e6t+ZcsAAAAAAAB7iw==. Additional material is available free of charge at http://pubs.acs.org. These materials include Supplementary Table 1, 2, 3, 4 and 5, Supplementary Figure 1 and 2. Supplementary Figure 1 shows the total yield of captured peptides when combining 0.25 mg of plasma and 2, 4, 8 and 12 mg of beads using protein-level coupling. Supplementary Figure 2 contains annotated mass spectra for each peptide that was the sole peptide observed for a particular protein within the SPEG experiments described in this manuscript. Supplementary Table 1 lists all the confidently matched identifications from all the runs included in all the Tables and Figures presented. Supplementary Table 2 lists all the proteins identified from different combinations of magnetic bead amounts and plasma amounts. Supplementary Table 3 lists all the proteins identified from the four different combinations of bead types and coupling conditions in addition to the abundance of some of these proteins in human plasma. Supplementary Table 4 lists all identified peptides from all 16 LC-MS/MS analyses for both the 8 depleted and 8 undepleted samples in the section “SPEG comparison between undepleted plasma and plasma depleted by MARS Hu-14”. Two bovine glycoproteins, Fetuin-A and Fibronectin, were spiked into these 16 samples to create equivalent dilution series between the depleted and undepleted sets as described in the section “Detection limits for glycoprotein standards spiked into plasma” and presented in Supplementary Table 3 and Figure 4. However, these Bovine proteins were not included in the data analysis where the samples were treated as 2 sets of 8 replicates and presented in Supplementary Table 4 and Figure 4, but they were included when the 8 depleted samples were analyzed as a dilution series for Table 5. The highest spike level is <1% of the total protein and thus should not significantly affect the results presented in Supplementary Table 4 and Figure 4, when the samples were treated as replicates. Supplementary Table 5 provides a non-redundant list of all unique former glycopeptides identified in at least one of the SPEG runs described in this manuscript. All the Supplementary Tables are supplied in Excel format. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Davis MA, Hanash S. High-throughput genomic technology in research and clinical management of breast cancer. Plasma-based proteomics in early detection and therapy. Breast Cancer Res. 2006;8(6):217. doi: 10.1186/bcr1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geho DH, Liotta LA, Petricoin EF, Zhao W, Araujo RP. The amplified peptidome: the new treasure chest of candidate biomarkers. Curr Opin Chem Biol. 2006;10(1):50–55. doi: 10.1016/j.cbpa.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24(8):971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 4.Pitteri SJ, Hanash SM. Proteomic approaches for cancer biomarker discovery in plasma. Expert Rev Proteomics. 2007;4(5):589–590. doi: 10.1586/14789450.4.5.589. [DOI] [PubMed] [Google Scholar]
- 5.Anderson L. Candidate-based proteomics in the search for biomarkers of cardiovascular disease. J Physiol. 2005;563(Pt 1):23–60. doi: 10.1113/jphysiol.2004.080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schenk S, Schoenhals GJ, de Souza G, Mann M. A high confidence, manually validated human blood plasma protein reference set. BMC Med Genomics. 2008;1:41. doi: 10.1186/1755-8794-1-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu S, Loo JA, Wong DT. Human body fluid proteome analysis. Proteomics. 2006;6(23):6326–6353. doi: 10.1002/pmic.200600284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajcevic U, Niclou SP, Jimenez CR. Proteomics strategies for target identification and biomarker discovery in cancer. Front Biosci. 2009;14:3292–3303. doi: 10.2741/3452. [DOI] [PubMed] [Google Scholar]
- 9.Durand G, Seta N. Protein glycosylation and diseases: blood and urinary oligosaccharides as markers for diagnosis and therapeutic monitoring. Clin Chem. 2000;46(6 Pt 1):795–805. [PubMed] [Google Scholar]
- 10.Freeze HH. Update and perspectives on congenital disorders of glycosylation. Glycobiology. 2001;11(12) 129R–143R. [PubMed] [Google Scholar]
- 11.Spiro RG. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12(4) doi: 10.1093/glycob/12.4.43r. 43R–56R. [DOI] [PubMed] [Google Scholar]
- 12.Van den Steen P, Rudd PM, Dwek RA, Opdenakker G. Concepts and principles of O-linked glycosylation. Crit Rev Biochem Mol Biol. 1998;33(3):151–208. doi: 10.1080/10409239891204198. [DOI] [PubMed] [Google Scholar]
- 13.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291(5512):2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 14.Lowe JB. Glycosylation, immunity, and autoimmunity. Cell. 2001;104(6):809–812. doi: 10.1016/s0092-8674(01)00277-x. [DOI] [PubMed] [Google Scholar]
- 15.Roth J. Protein N-glycosylation along the secretory pathway: relationship to organelle topography and function, protein quality control, and cell interactions. Chem Rev. 2002;102(2):285–303. doi: 10.1021/cr000423j. [DOI] [PubMed] [Google Scholar]
- 16.Kobata A, Endo T. Immobilized lectin columns: useful tools for the fractionation and structural analysis of oligosaccharides. J Chromatogr. 1992;597(1–2):111–122. doi: 10.1016/0021-9673(92)80101-y. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z, Harris LE, Palmer-Toy DE, Hancock WS. Multilectin affinity chromatography for characterization of multiple glycoprotein biomarker candidates in serum from breast cancer patients. Clin Chem. 2006;52(10):1897–1905. doi: 10.1373/clinchem.2005.065862. [DOI] [PubMed] [Google Scholar]
- 18.Pan S, Wang Y, Quinn JF, Peskind ER, Waichunas D, Wimberger JT, Jin J, Li JG, Zhu D, Pan C, Zhang J. Identification of glycoproteins in human cerebrospinal fluid with a complementary proteomic approach. J Proteome Res. 2006;5(10):2769–2779. doi: 10.1021/pr060251s. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21(6):660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 20.Sun B, Ranish JA, Utleg AG, White JT, Yan X, Lin B, Hood L. Shotgun glycopeptide capture approach coupled with mass spectrometry for comprehensive glycoproteomics. Mol Cell Proteomics. 2007;6(1):141–149. doi: 10.1074/mcp.T600046-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Tian Y, Zhou Y, Elliott S, Aebersold R, Zhang H. Solid-phase extraction of N-linked glycopeptides. Nat Protoc. 2007;2(2):334–339. doi: 10.1038/nprot.2007.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y, Aebersold R, Zhang H. Isolation of N-linked glycopeptides from plasma. Anal Chem. 2007;79(15):5826–5837. doi: 10.1021/ac0623181. [DOI] [PubMed] [Google Scholar]
- 23.Zou Z, Ibisate M, Zhou Y, Aebersold R, Xia Y, Zhang H. Synthesis and evaluation of superparamagnetic silica particles for extraction of glycopeptides in the microtiter plate format. Anal Chem. 2008;80(4):1228–1234. doi: 10.1021/ac701950h. [DOI] [PubMed] [Google Scholar]
- 24.Liu T, Qian WJ, Gritsenko MA, Camp DG, 2nd, Monroe ME, Moore RJ, Smith RD. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J Proteome Res. 2005;4(6):2070–2080. doi: 10.1021/pr0502065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hortin GL, Sviridov D, Anderson NL. High-abundance polypeptides of the human plasma proteome comprising the top 4 logs of polypeptide abundance. Clin Chem. 2008;54(10):1608–1616. doi: 10.1373/clinchem.2008.108175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.