Abstract
Nanobiotechnology involves the creation, characterization, and modification of organized nanomaterials to serve as building blocks for constructing nanoscale devices in technology and medicine. Living systems contain a wide variety of nanomachines and highly ordered structures of macromolecules. The novelty and ingenious design of the bacterial virus phi29 DNA packaging motor and its parts inspired the synthesis of this motor and its components as biomimetics. This 30-nm nanomotor uses six copies of an ATP-binding pRNA to gear the motor. The structural versatility of pRNA has been utilized to construct dimers, trimers, hexamers, and patterned superstructures via the interaction of two interlocking loops. The approach, based on bottom-up assembly, has also been applied to nanomachine fabrication, pathogen detection and the delivery of drugs, siRNA, ribozymes, and genes to specific cells in vitro and in vivo. Another essential component of the motor is the connector, which contains 12 copies of a protein gp10 to form a 3.6-nm central channel as a path for DNA. This article will review current studies of the structure and function of the phi29 DNA packaging motor, as well as the mechanism of motion, the principle of in vitro construction, and its potential nanotechnological and medical applications.
Keywords: Bacteriophage phi29, DNA packaging motor, pRNA, Nanotechnology, Nanobiotechnology, Bionanotechnology, Gene delivery, Cancer therapy
INTRODUCTION
Essential components in living organisms are routinely and actively transported by molecular motors, such as F1-ATPase, kinesin, myosin, helicase, and the viral DNA packaging motor. Elucidation of their structure and function has inspired attempts to integrate these machines into synthetic devices.3,84,92,112,118 Current micro- and nanoscaled fabrication approaches employ diverse and robust materials. Fabrication of super-scaled materials can be accomplished by various physical or chemical methods, including replication,37 crosslinking,6,12 deposition,23,26,141 complementation,4,87,89 donor/receptor interaction,101 self-assembly,28,78,125,151 colloidal crystallization,94,95,97,137,138 and nanoimprint lithography.70
In double-stranded (ds) DNA viruses, the viral genome is packaged into a preformed protein shell called a procapsid.47,53,55 The translocation of DNA into the procapsid is entropically unfavorable due to the extreme compact nature of DNA arrangement within the capsid.29 The DNA packaging task is accomplished by a DNA-packaging motor that uses ATP as energy.29,49,51,53 In phi29, the quantification of ATP consumption in DNA packaging has revealed that one ATP is needed for the translocation of two base pairs of DNA into the procapsid.51,120 The novel, ingenious design of the phi29 DNA packaging motor, one of the strongest biomotors studied to date,120 has inspired the synthesis of this motor and its components as biomimetics. An imitative motor has been successfully constructed in vitro using purified recombinant proteins and artificially synthesized RNA. The motor can be turned on and off at will. The whole motor complex, as well as each motor component, has the potential to be used as a building block in nanotechnology and nanomedicine. This review will summarize current studies on the structure, function, construction, and mechanism of the phi29 DNA packaging motor, as well as address the potential application of these artificial motors.
STRUCTURE OF PHI29 DNA PACKAGING MOTOR
Due to its relatively simple structure, the phi29 DNA packaging motor is a good model to study the fundamental biological mechanisms of bionanomotors that transduce chemical energy from ATP hydrolysis into physico-mechanical motion of DNA.55,93 Exactly defined as in vitro phi29 DNA packaging,49 this system was developed by using synthetic nucleic acid and purified recombinant components, such as DNA-gp3, procapsid, gp16, pRNA, along with ATP as an energy source that can package up to 90% of the added DNA-gp3 into the procapsid. After DNA packaging, the in vitro assembly system can convert a DNA-filled capsid into infectious phi29 virions when purified proteins, such as tail protein (gp9), neck protein (gp11 and gp12), and morphogenic factor (gp13), are added. Recent single-molecule studies using an optical tweezers technique have demonstrated that the force required to prevent the DNA from being inserted in the phi29 DNA packaging motor is about 57 piconewtons (pN), indicating that the phi29 DNA-packaging motor is one of the strongest biomotors studied to date.123
Structural Components: Procapsid and Connector
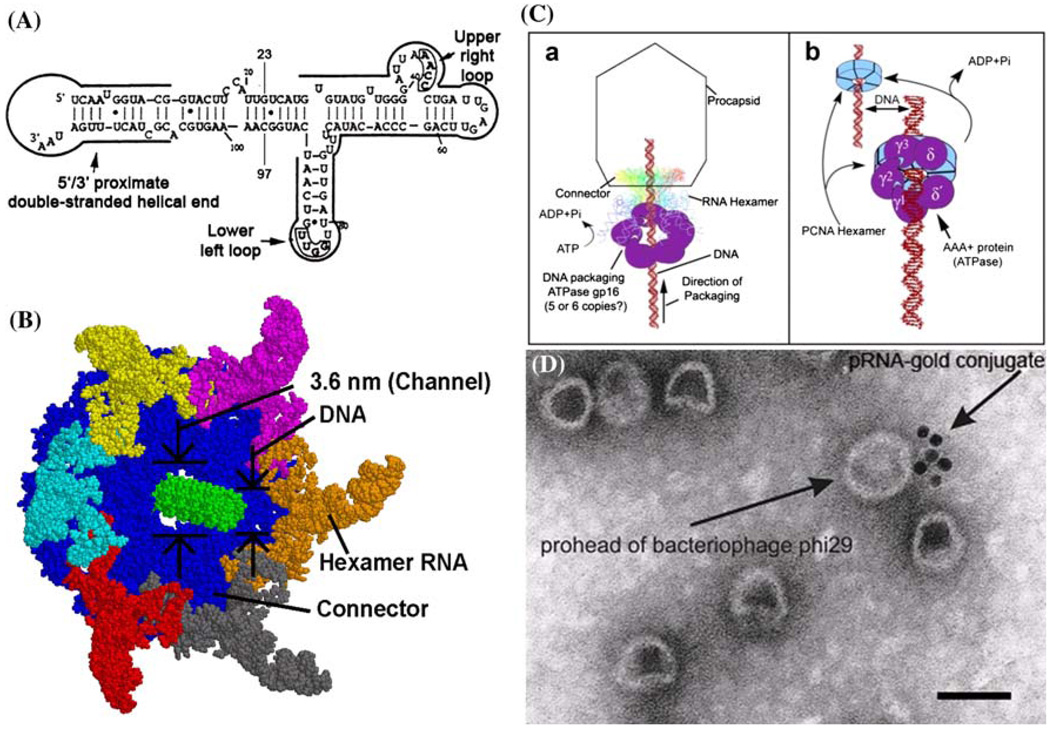
The procapsid is composed of three proteins: capsid, scaffolding, and connector.5,15,53 The three-dimensional structure of the phi29 procapsid, obtained by reconstruction of cryo-electron microscopy (cryo-EM) images120 revealed that the icosahedral procapsid of phi29 consists of 235 copies of the major capsid protein (gp8), 180 copies of scaffolding protein (gp7), and 12 copies of the head–tail connector protein (gp10).103,116,120 Scaffolding proteins are structural proteins that are required for correct procapsid assembly but are released from the procapsids at the initiation of or during DNA packaging. Phi29 scaffolding protein (gp7)5 is not required for the competence of procapsids in DNA packaging.53 Although the exact function of scaffolding proteins is not yet clear, they may form a core structure around which capsid proteins assemble. In some cases, they may serve as chaperones to promote the correct folding of capsid proteins. Additionally, they may be involved in mediating a putative capsid protein/connector protein interaction, excluding cellular proteins from the inside of the procapsid, or facilitating the early stages of DNA entrance into the procapsid.10,53,76,77
The procapsid contains a single portal vertex, called the connector (gp10) because of its role in DNA translocation.5,15 The connector is involved in the formation of the procapsid and is the base on which the procapsid is assembled. It is also involved in DNA packaging and in the binding of a tail protein to the mature head.10,53 Three-dimensional structural information for the phi29 connector has been obtained by using atomic force microscopy, cryo-EM, immuno-electron microscopy, and X-ray crystallography.43,135 The DNA-binding domain seems to be important for the recognition of DNA in the first steps of DNA packaging. The RNA-binding domain, rich in Arg-Lys residues, plays a critical role in pRNA binding and is essential for DNA packaging.7,48,63
The head fibers (gp8.5) radiate from the apical regions of the head. Twelve copies of the lower collar (gp11), 12 copies of the appendage (gp12), and 10 copies of the tail knob protein (gp9) are assembled onto the DNA-filled head in a single morphogenetic pathway to yield the mature virion.116,120 A morphogenetic factor (gp13) is also required for neck and tail (gp11–12) assembly. Recently, gp13 was found to be an enzyme aiding the degradation of the cell wall to help the virus attachment and DNA injection of phi29.22,142 However, these fiber and tail proteins are not required for DNA packaging/translocation.
Nonstructural Components: pRNA
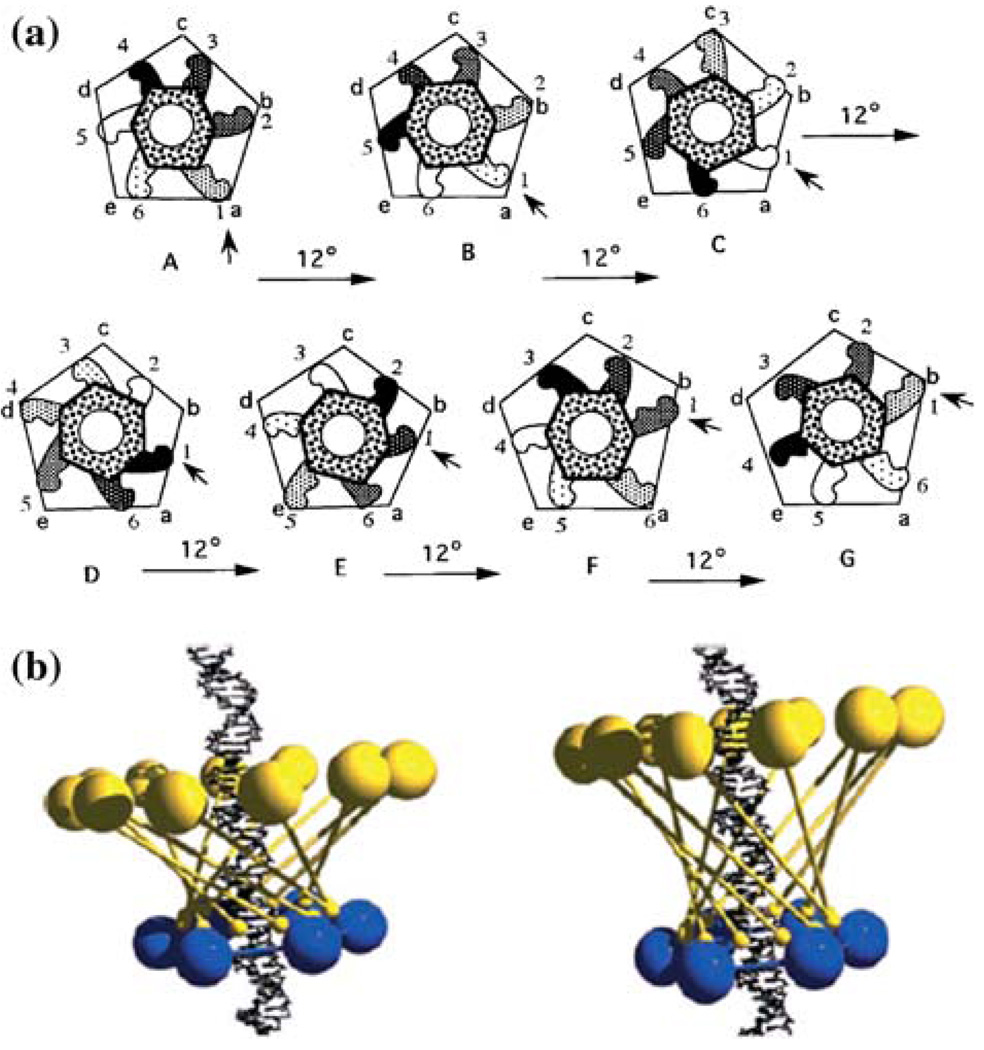
A unique feature of the phi29 DNA packaging motor is the presence of RNA. Small packaging RNA (pRNA), discovered in 1987,48 is vital for DNA packaging activity. Notably, the pRNA binds the connector and participates actively in DNA translocation but leaves the capsid after DNA packaging is completed. Two functional domains of pRNA have been identified (Fig. 1A): a procapsid-binding domain for binding to the N-terminus of the connector,143 located at its central region, and a DNA translocation domain for gp16 binding,73,79 located at the 5′/3′ paired ends. A three-base bulge (C18C19A20) located in the DNA translocating domain is critical for DNA translocation. It has been shown that pRNA can be redesigned by a two-module architecture approach, resulting in full activity in DNA packaging and assembly of infectious phi29 virions in vitro.32 The pRNA binds ATP114 and enhances the ATPase activity of gp16.42,80 The pRNA forms dimers which are the building blocks of pRNA hexamer conformation. Six phi29 pRNAs form a ring through intermolecular base-pairing between the right loop (bases 42–25) and the left loop (bases 82–85), as observed by transmission electron microscopy with nanogold-labeled pRNA (Fig. 1D)144,145 (on-line animation: http://www.eng.uc.edu/nanomedicine/newmovs.html, or http://nar.oxfordjournals.org/content/vol0/issue2008/images/data/gkn669/DC1/nar-01176-a-2008-File006.ppt). Cryo-AFM has also been used to directly visualize the tertiary structure of pRNA monomers, native and covalently linked dimers, and native trimers.118 The DNA translocating machine is geared by a pRNA hexameric complex (Fig. 1B), suggesting that the mechanism employed is similar to that of the consecutive firing of six cylinders of a car engine.20
FIGURE 1.
Phi29 DNA packaging motor. (A) Sequence and secondary structure of phi29 pRNA. (B) Schematic assembly model of phi29 DNA packaging motor. (C) Proposed phi29 DNA packaging motor structure with a pRNA hexameric ring and gp16 bound to a channel connector (a)79 in comparison with hexameric PCNA and clamp–loader complex (b). (D) Transmission electron micrograph of phi29 procapsid/pRNA-gold complexes. Bar equals 50 nm.144 All figures were adapted with permission from the Elsevier and Oxford Journals for the respective citation.
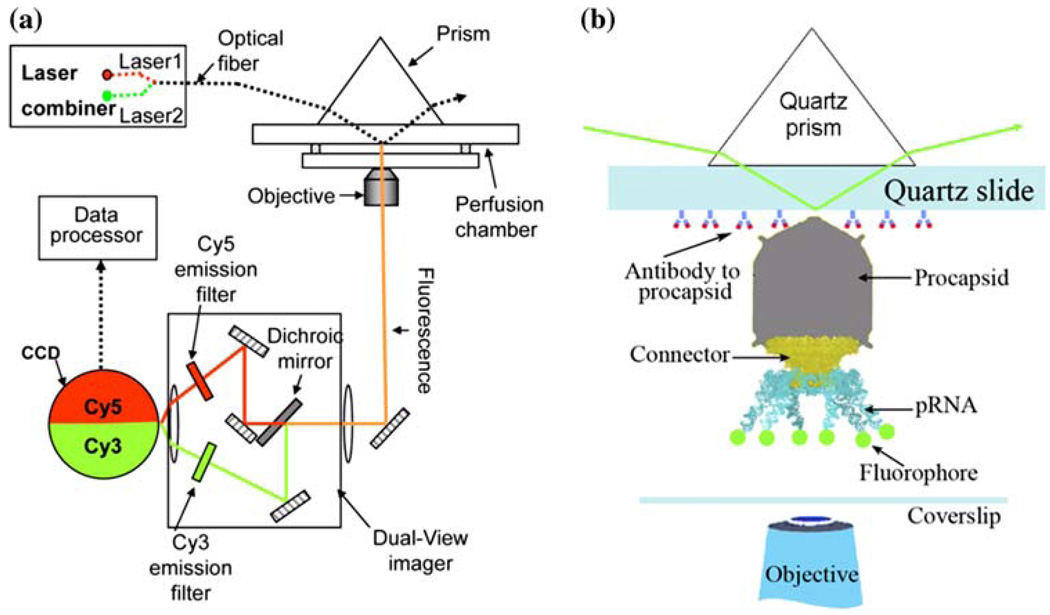
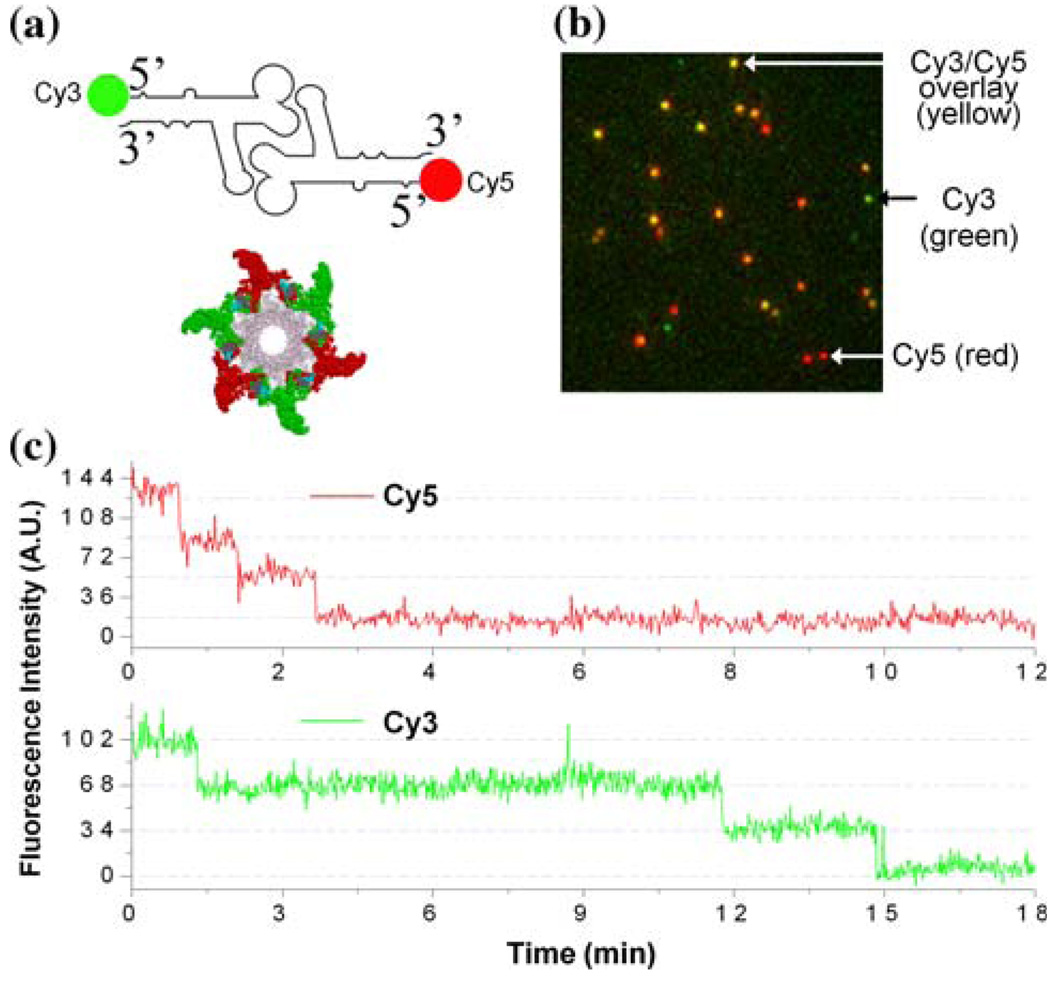
Substantial evidence supports the configuration of the pRNA hexamer. In 1997, extensive complementation analyses were published, showing the presence of a pRNA hexamer and inter-pRNA interactions.20 Again in 1998, two labs54,149 independently reported the hexameric nature of pRNA as it is bound to the motor. Following the approaches of biochemistry, mathematics, and genetics, the hexameric pRNA ring was observed in 2000 using cryo-EM.68 Recent high-sensitivity single-molecule counting with photobleaching technology revealed that each DNA packaging motor contains six copies of pRNA,119 a difficult task due to the limited resolution power and sensitivity of light microscopy. The newly customized single-molecule dual-viewing total internal reflection fluorescence (SMDV-TIRF) imaging system with a top-prism (Fig. 2a)119 produces stable signals with extremely low background for single fluorophore detection. Single-fluorophore imaging has enabled a clear identification of the quantized photobleaching steps in the phi29 pRNA labeled with a single fluorophore to conclude its stoichiometry within the motor (Fig. 2b). The packaging motors were stalled by nonhydrolyzable ATP, then restarted by adding fresh ATP to simulate the DNA packaging in real time. Dual-color detection in the stalled intermediates, containing both Cy3- and Cy5-pRNA, showed that most of the motor complexes contain six copies of pRNA before and during DNA translocation (Fig. 3a–3c).119 The hexameric structure was also supported by a DNA packaging model proposed by Fang et al.32 and by binding affinity quantification reported by Robinson et al.106 In addition, recent single-molecule imaging reports have revealed that the hexamer ring was formed by either a pure dimer or trimer alone.145 When two inactive pRNAs with intermolecular complementarity were mixed in a 1:1 molar ratio, the packaging motor was fully active to produce infectious virions, indicating that the stoichiometry of the pRNA should be a multiple of two. When the procedure was repeated with three inactive pRNAs, the motor was again fully active in production of virions, which suggests a multiple of three.116
FIGURE 2.
Methods for single-molecule studies on phi29 DNA packaging motor.119 (a) Setup of single-molecule dual-viewing total internal reflection fluorescence (SMDV-TIRF) microscopy. (b) Experimental design for detection of fluorescent labeled pRNA at its 5 end on phi29 DNA packaging motor. All figures were adapted with permission from the European Molecular Biology Organization.
FIGURE 3.
Dual-view imaging on phi29 DNA packaging motor.119 (a) pRNA dimer constructed with Cy3-pRNA and Cy5-pRNA, which are in alternating positions to form a hexamer. (b) Typical fluorescence image showing procapsids bound with the dual-labeled pRNA dimers. (c) Fluorescence intensity vs. time to show photobleaching steps of procapsids reconstituted with the dimer. All figures were adapted with permission from the European Molecular Biology Organization.
It is intriguing that the pRNA hexameric ring is configurationally and functionally reminiscent of another family of ATPases, the large AAA+ family (ATPases associated with a variety of cellular activities), to which the phi29 DNA packaging protein gp16 belongs (Fig. 1C).79 The proteins in this family also form a hexameric ring in interactions with DNA or RNA, indicating that this class of nanomachines might possess a similar mechanism in nucleotide contact and DNA/RNA translocation. DNA polymerase, P4 RNA packaging motors, RNA polymerase, helicase, rho factor, DNA polymerase processivity factor, BPV E1 replication initiator, and tens of other nucleotide-binding or translocating proteins also exist as hexamers.55
Nonstructural Components: Packaging Enzyme gp16
The gp16 packaging enzyme contains both Walker A- and B-type consensus ATP-binding sequences; therefore, it likely functions as an ATPase during DNA packaging.51 The A-type sequence of gp16 contains “basic-hydrophobic region-G-X2-G-X-G-K–S-X7-hydrophobic” amino acids. The gp16 enzyme binds and hydrolyzes ATP,49,50,65,80,114 but the role of gp16 in the phi29 DNA packaging motor is still unclear. The hydrophobicity, low solubility, and self-aggregation of phi29 gp16 have long hindered further refinement of the current understanding of its packaging mechanism. For this reason, much contradictory data has been produced regarding ATPase activity, binding location, and the stoichiometry of gp16.42,49,69
It has been observed that gp16 binds DNA without sequence specificity, but specifically to pRNA,79 which implies that the binding properties of gp16 are important determining factors for the biological activity of gp16. It has been long known that gp16 functions during DNA packaging by binding and hydrolyzing ATP to gain energy from the high-energy phosphate bond.49,80,115
Packaging Substrate: Genomic DNA-gp3
Understanding the genomic DNA structure is critical for DNA packaging and will help us to understand the mechanisms of motors, including DNA supercoiling, gyrase activity, or the contact between the wall of the motor channel and the DNA axle. Studies conducted in vitro have revealed that both gaps and single-stranded (ss)DNA will stall or block the DNA packaging, while nicks (breaks) can be tolerated in phi29.91
The question of DNA specificity remains, i.e., how the viral motor packages its own genomic DNA but not nonspecific DNA. Few investigations have reported on the conformational requirements of the DNA substrates that can be packaged. Natural DNA in solution is generally present in the supercoiled or relaxed form, which suggests that the helical nature of the B-form DNA is the preferred conformation. Phi29 DNA-gp3 is treated as a single complex, since the DNA and terminal protein gp3 are covalently linked together. Therefore, it does not require either terminase activity to cut concatemeric DNA into a single copy, as in λ, P2, P4, T3, and T7, or a “headful” mechanism, as observed in T4, P22, and P1.2,43,100,105,136
CONSTRUCTION OF THE PHI29 DNA PACKAGING MOTOR
Construction, Assembly, and Purification of Procapsids
The phi29 procapsid is composed of a scaffolding protein (gp7), a capsid protein (gp8), and a portal protein (gp10) that form the major motor dodecamer channel. The structural genes of the phi29 procapsid were cloned and expressed in E. coli in combinations., In vivo and in vitro studies showed that the scaffolding protein interacts with both the portal vertex and capsid proteins. These results suggest that the scaffolding protein serves as the linkage between the portal vertex and the capsid proteins, and that the portal vertex plays a crucial role in regulating the size and shape of the procapsid. The packaging motor-containing procapsids can be expressed, assembled and purified from E. coli, and are fully functional in DNA packaging in vitro.52
Overproduction and Purification of gp16
In E. coli, gp16 was overproduced from a cloned gene and purified to near homogeneity.49 When over-expressed in E. coli, gp16 aggregates into lumps and loses biological activity. To resolve the aggregated gp16 to homogeneity in a soluble and active form, the gp16 in lumps (inclusion bodies) requires denaturation followed by renaturation.49 To obtain a soluble enzyme, the lumps (inclusion body) were collected by differential centrifugation, denatured with 6 M guanidinium chloride at pH 5.2, and then passed through a Sephadex G150-packed column. Then, gp16 was eluted as a discrete peak with more than 90% purity. The purified gp16 in this peak was inactive in driving the motor. The gp16 required renaturation by dialysis against 10 mM KCl with 50 mM Tris–HCl at pH 7 for 40 min. It was shown that both cysteine residues of gp16 are very important for biological activity, and the use of reducing agents, for example dithiothreitol (DTT), was necessary when guanidinium chloride or urea was used for denaturation. Although such renatured gp16 was biologically active, it reaggregated within 20 min after the renaturation. Co-expression of gp16 with groE was reported to produce soluble and active gp16. But while the coexpression of gp16 with groE solved the problem of aggregation within the cell, it could not solve the problem of self-aggregation after purification.69 Another approach has been developed to obtain affinity-purified, soluble, and highly active gp16. The gene coding for gp16 was fused to a gene coding thioredoxin to be connected to the N-terminus of gp16,65,66 resulting in a chimeric gp16 containing a thioredoxin. Introduction of the redox protein thioredoxin enhanced the solubility and biological activity of the purified gp16. PEG and acetone were found to enhance gp16 solubility and activity.65,66 Recently, a method for the preparation of more soluble gp16 has also been developed by over-expression in Bacillus subtilis.73 As observed by EM, gp16 forms a variety of ring structures depending on the concentration of gp16. The finding is very similar to the ring structure observed for the DNA packaging enzyme gp16 of T4.82
Construction, Synthesis, and Purification of pRNA
Biologically active pRNA can be purified from four sources: (1) separation from procapsids purified from phi29-infected Bacillus subtilis; (2) purification from Bacillus subtilis containing plasmids carrying the pRNA gene and its native promoter; (3) isolation from an in vitro transcription of pRNA by T7 RNA polymerase; and (4) solid state chemical synthesis. Of these, the isolation from an in vitro transcription with T7 RNA polymerase is the simplest and most convenient. A DNA template of the wild-type or mutant pRNA for the in vitro transcription can be generated by polymerase chain reaction (PCR) using a primer containing the T7 promoter.
Defined In Vitro DNA Packaging System Using the Motor Synthesized In Vitro
The defined in vitro phi29 DNA packaging system first developed in 198649 forms the basic platform for further development, modification, and refinement. The entire DNA packaging motor includes procapsid, DNA-gp3, gp16, pRNA, and ATP. Phi29 offers many advantages for the study of viral DNA packaging in general, because up to 90% of the population of available phage DNA can be packaged in this defined in vitro system. The efficiency of DNA packaging can be assayed either by sucrose gradient sedimentation to detect the DNA-filled procapsids,49 or by DNase I protection assay detected by an agarose gel electrophoresis.40,52 The DNA packaging can also be observed directly under a microscope.17,119,123
MECHANISM OF THE PHI29 DNA PACKAGING MOTOR
Single-molecule Studies of DNA Packaging Motors
Recent advances in single-molecule microscopy have provided a new way to understand motor mechanisms through the direct counting of motor components and observing motion events (Fig. 3). This technique allows the analysis of individual motor components, as opposed to averaging the measurements from a massive population of homogenous molecules that are either motionless or in synchronous motion.58,108 As mentioned earlier, single-molecule microscopy has been successfully applied to count the number of pRNAs in the active DNA packaging motor, from which the stoichiometry of pRNA was revealed as a hexamer (Fig. 2).119,150 Unveiling the stoichiometry of motor components that are actively involved in the motor function will make it possible to elucidate the properties of bionanomotors and other biological machines, and it will also help to design novel nanodevices, as well as to imitate natural organs.35
Photobleaching of Single Fluorescene Molecules
Due to the limitation of spatial resolution of optical microscopy, direct detection of phi29 motor or counting of the motor components is challenging, especially for RNA, which is conformationally versatile and structurally flexible. A customized single-molecule dual-viewing total internal reflection fluorescence imaging system (SMDV-TIRF) was constructed to directly count the copy number of RNA within each motor.150 The RNA molecules were labeled with a single fluorophore by in vitro transcription in the presence of a fluorescent AMP. Precise calculation of identical or mixed pRNA building blocks of one, two, three, or six copies within the phi29 DNA packaging motor has been demonstrated by applying a photobleaching assay and evaluated by binomial distribution. The dual-viewing system for excitation and recording at different wavelengths makes it possible to differentiate molecules with different labels.
To image the motor, DNA-packaging intermediates with partially packaged DNA are isolated and utilized to determine the stoichiometry of pRNA. The real time DNA translocation process was directly observed. Single fluorophore imaging clearly identified the quantized photobleaching steps from pRNA labeled with a single fluorophore and concluded its stoichiometry within the motor. Almost all of the motors contained six copies of pRNA before and during DNA translocation, identified by dual-color detection of the stalled intermediates of motors containing Cy3-pRNA and Cy5-DNA.119
Single Molecule Approach to Study the Mechanism of pRNA Hexameric Ring Formation
Many nucleic acid-binding proteins and the AAA+ family form hexameric rings, but the mechanism of hexamer assembly is unclear. Single molecule studies with the SMDV-TIRF elucidated a mechanism suggesting the specificity and affinity in protein/RNA interaction relies on pRNA static ring formation.145 pRNA did not form a ring prior to motor binding. A combined pRNA ring-forming group was very specific for motor binding, but the isolated individual members of the ring-forming group bind to the motor nonspecifically. Only those RNAs that formed a static ring, via the interlocking loops, stayed on the motor. Single interlocking loop interruption resulted in pRNA detachment. Extension or reduction of the ring circumference failed in motor binding. This new mechanism was tested by redesigning two artificial RNAs that formed a hexamer and packaged DNA. The results confirmed the stoichiometry of pRNA on the motor was hexameric with a common multiple of two and three.
Direct Observation of Motor Motion
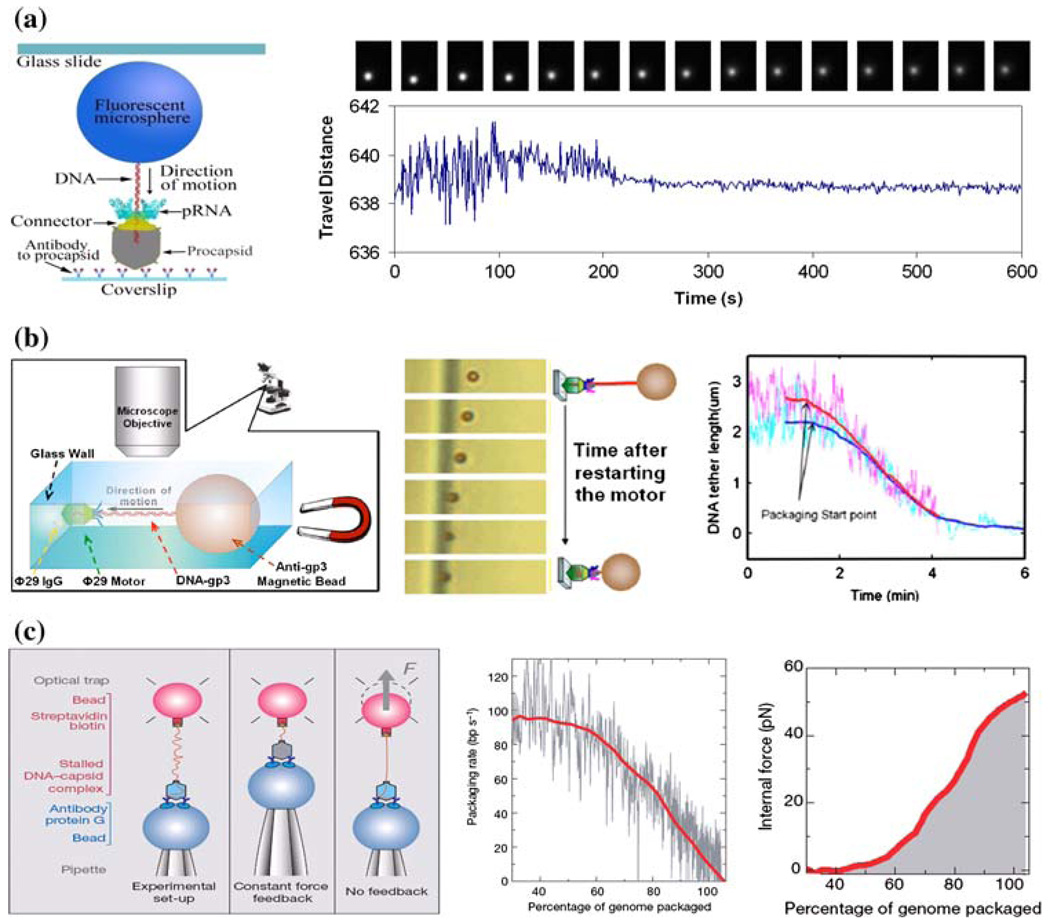
Single-molecule approaches allow the direct observation of physical behaviors to answer many questions, including (1) how chemical energy is converted into physical motion,146 (2) how force is generated,129 (3) how the molecular structure is involved in chemical reactions,152 (4) how the motion starts and continues without interruptions,59 (5) how each motor component responds to the applied force,123 and (6) how the conformational change of each motor component is correlated to the generation of force.19 Covalent attachment of a fluorescent bead to DNA has been used to track the motion of DNA during translocation.119 This attached bead is then able to be visualized through the amplification of the signal using a fluorescence microscope (Fig. 4a). The motion of translocation can then be profiled three-dimensionally (x, y, and z axes), providing information on the orientation of the DNA and the motor. An ATP derivative, γ-S-ATP, proves to be non-hydrolyzable, and thus the DNA packaging intermediates can be stalled using this derivative, restarted by addition of ATP, and observed in real-time by fluorescence microscopy. The migration of DNA into the procapsid during packaging caused the attached microsphere to show a gradual reduction in swing range (Fig. 4a).119 After the DNA was completely packaged, the motion stopped, corresponding to a “zero-distance change” from the reference origin under the CCD camera.
FIGURE 4.
Direct observations of DNA translocation during phi29 DNA packaging by single-molecule studies using (a) fluorescent microsphere-attached phi29 DNA,119 (b) magnetic bead-attached phi29 DNA,17 and (c) optical tweezers for packaging rate and force measurement.123 All figures were adapted with permission from the European Molecular Biology Organization, the American Institute of Physics, and the Nature Publishing Group to the respective citation.
The phi29 DNA packaging behavior has also been observed in bright field by using a relatively simple magnetomechanical system (Fig. 4b). In a microfluidic chamber, the partially packaged phi29 intermediate was immobilized on a glass wall via a phi29-specific antibody. On the opposing end of the DNA, a micronsized magnetic bead was attached to serve as a displacement-indicator, and the DNA was subsequently stretched by the magnetic force. By monitoring the distance between the bead and the glass wall with a conventional optical microscope, it was demonstrated that the motor can transport cargo that is 10,000-fold larger than itself.17
The Force Measurement of the DNA Packaging Motor Using Optical Tweezers
Optical tweezers, or laser traps, have been utilized to catch or trap a small particle by the force generated from laser radiation pressure. When the force from the phi29 DNA packaging motor was investigated, it was found that the motor can package DNA against an internal force of 57 pN, making the phi29 motor the most powerful motor ever constructed to date (Fig. 4c).123 This force measurement also implies that as DNA is tightly packaged, the viral capsid is able to withstand high internal pressure, and it has even been suggested that the tensile strength is comparable to an aluminum alloy.123 It has been shown that the generation of force for translocation occurs as the high energy phosphate bond is broken on ATP to generate the release of phosphate and ADP, rather than ATP binding to the motor. These results, in correlation with previous experiments,115,123 indicate that the sequential action of multiple ATPase subunits power the packaging reaction and each ATPase subunit might complete its ATP hydrolysis cycle before the next cycle starts.
ATP Consumption by the DNA Packaging Motor
Molecular motors need energy to function. Most molecular motors use chemical energy from the hydrolysis of ATP. The active sites on motor proteins bind ATP molecules and catalyze the decomposition to ADP and inorganic phosphate (Pi), thereby releasing a significant quantity of energy, which in turn leads to a conformational change in the motor protein, ultimately resulting in motor movement. This catalytic process repeats with another ATP molecule so that the motor protein can continue the movement. DNA packaging is entropically unfavorable, since the DNA arrangement within the capsid is extremely compact, and the packaged DNA undergoes an approximately 30- to 100-fold decrease in volume compared to that before packaging.53 ATP hydrolysis provides the driving force for viral DNA-packaging motors. The ATP consumption in the phi29 DNA packaging was quantified to be two base pairs of DNA translocation per single ATP hydrolysis event with purified components.51 The individual ATPase activities were found to be stimulated by other motor components, including pRNA and DNA-gp3.42,80,114 These results suggest that the consumption of ATP in the phi29 DNA packaging system is very complicated. ATP is believed to be consumed for the initiation of DNA packaging and translocation, with all components of the packaging system, including pRNA, procapsid, gp16, and DNA-gp3, involved in the generation of maximal ATPase activity.
Energy Conversion in Motor Force Generation
Sequence comparison of the DNA packaging proteins revealed that most of these proteins contain a conserved Walker-A motif, (G/A)XXXXGK(S/T), which is a consensus ATP-binding motif.51 Mutation of a single amino acid at this region could completely abolish ATP binding and hydrolysis, thereby completely inhibiting the DNA packaging. Phi29 pRNA itself does not show ATPase activity, and gp16 itself shows low-affinity ATPase activity.80 However, the pRNA/gp16 complex shows DNA-dependent ATP-ase activity.80 The ATPase activity of gp16 can be stimulated by either pRNA or DNA. Similarly, procapsids with pRNA stimulate the ATPase activity of gp16 by tenfold, as compared to the lack of stimulation by procapsids alone. The ATPase activity was maximally stimulated by all packaging motor components, including pRNA, procapsid, gp16, and DNA-gp3.51,114 The motor translocates DNA containing an extra load of 2.2 µm polystyrene beads at an initial rate of 115–170 base pairs per second as revealed by single-molecule studies.36,123 In another recent report with high resolution optical tweezers, highly coordinated ATP hydrolysis by gp16 was shown to take place in sequential manner to translocate 2.5 base pair of DNA per each ATP hydrolysis.90 These observations indicate that the binding of ATP is essential to induce a conformational change in the motor in order to convert chemical energy (ATP) into physical motion (packaging). The conformational change of the motor component might be correlated with a release of Pi to generate a power stroke on the DNA, rather than the ATP-binding step.123
Models for the Mechanism of the Phi29 DNA Packaging Motor
A number of models have been proposed to describe the mechanism of DNA translocation.
Classic Packaging Models
Many classic models have been proposed to explain the mechanism of the DNA packaging motor. These models were largely based on preliminary understandings of viral structure before the detailed motor structure was revealed. The gyrase-driven packaging model proposed that supercoiled DNA is translocated by DNA gyrase activity of the motor using ATP as an energy source. Recent fluorescence correlation spectroscopy (FCS)-based studies in T499,109 still support this model through observations proving that nicked DNA fails to be packaged. However, recent studies showed that phi29 still packages nicked DNA in vitro,91 implying that the model is only applicable to limited examples. The osmotic ratchet model proposed that the expansion of the procapsid lowers the osmotic pressure inside the capsid, and the connector acts as a ratchet to grab the packaged DNA.113 This model fails to explain how the decreased osmotic pressure can still drive DNA translocation when the internal pressure of the procapsid has increased as it is filled with DNA. In addition, phi29 procapsid expansion is not necessary. The connector rotating thread model, based on the symmetry mismatch between the fivefold portal vertex of the procapsid and the sixfold connector, proposed that the connector rotation produces a driving force to translocate the DNA through the connector channel. Recent reports of single-molecule analysis indicate that preventing the connector from rotating does not inhibit DNA packaging,9,67 raising doubt about the validity of the rotation theory. The supercoiled DNA wrapping model proposed that supercoiled DNA wraps around the portal vertex, and the rotation of the connector allows DNA to pass into the procapsid. This model was based on the finding that the phi29 connector preferentially induces supercoiled DNA rather than the linear form.41 This unique model is not favored, since recent structural data from analysis of partially packaged procapsids by cryo-EM120 has shown that DNA is likely positioned in the central channel of the connector.
Sequential Action Model
The sequential action of phi29 motor components was demonstrated empirically by Chen and Guo in 1997.20 This model was recently confirmed by single molecule studies90 (also see Introduction of the Ref. 90). Chen and Guo20 proposed that the relative motion of two rings could provide a driving force for DNA translocation. Analogous to a car engine, the sequential action of cylinders is a possible way to turn the motor. The finding that a hexameric pRNA complex binds to the connector and that six pRNAs work sequentially provide strong evidence to support the sequential action model. They also predicted that the pRNA contains two domains: one for the connector binding, and the other for the DNA translocation. pRNA binds to the connector leaving the 5′/3′ domain free to interact with gp16, a prediction that was confirmed nine years later.79 It was also proposed that pRNA is part of an ATPase and possesses at least two conformations—a relaxed form and a contracted form. Alternating between contraction and relaxation, each member of the hexameric RNA complex driven by ATP hydrolysis helps to drive the DNA translocation machine20 (Fig. 5a). This process generates torque, driving the DNA translocation machine and consuming one ATP for every 2 bp of DNA translocated.51 Recent single-molecule fluorescence studies further confirmed that indeed pRNA in the actively working packaging motor is of hexameric conformation.119,150 As demonstrated pictorially in Fig. 1C, the connector/pRNA complex resembles hexameric PCNA (proliferating-cell nuclear antigen), and gp16 is an AAA+ type protein that interacts with DNA. The fact that ATP induces a conformational change in pRNA, and that pRNA loop/loop interactions are essential to pass the signal from one pRNA to the other yields support for this model.114 Additionally, recent reports have shown that gp16 binds directly to pRNA and that the binding enhances the ATPase activity of gp16.42,69,79,80 Finally, recent work using optical tweezers for single molecule analysis has also indicated sequential ATPase activity of gp16.90
FIGURE 5.
Proposed phi29 DNA packaging mechanism models. (a) Sequential action model.20 (b) “Inverse Chinese finger puzzle” model proposed by George Oster88 related to connector contraction model. All figures were adapted with permission from American Society for Microbiology and Wiley-VCH Verlag GmbH for the respective citation.
Because many dsDNA bacteriophage viruses share structural similarities, the sequential action model may be applicable for their packaging mechanism. Since most employ terminases to perform DNA packaging, a large subunit of the terminase complexes has been proposed as a counterpart of pRNA.55 A sequential action approach was recently proposed as a packaging model for T4, using the large subunit (gp17) to drive DNA translocation.127 Sequential mechanical motion has also been proposed with ssRNA by the packaging enzyme P4 in the virus phi12.85
Electrodipole Central Channel Model
A 3D crystal structure of the phi29 connector revealed an electronegative interior surface of the central channel.43,120 It was proposed that two lysine residues, separated by 20 Å, formed two lysine rings inside the channel.43 In this scenario, it is likely that, during DNA translocation, contact is established between two phosphates on the adjacent major grooves of DNA contact and each of the lysine residues. The consecutive rotation of the connector by every 6° displaces the lysine-phosphate pairs one base pair apart at a time, consistent with the fivefold/sixfold symmetry mismatch hypothesis.20,60 However, a drawback of this model is its failure to explain the ATP-driving mechanism, since no ATP binding motif has been found in connector or DNA. It is hard to reconcile how DNA translocation is driven by the ATP-generated force acting on the connector or on the DNA to promote its consecutive movement, since the ATP binding sites were found in both gp1651 and pRNA.114
Connector Contraction Model
This model was proposed based on the observation of the 3D structure of the phi29 connector.120 Lengthwise expansion of the connector, led by a 12° rotation of the narrow end of the connector, will change the angle of the long helices, and the wide end of the connector follows the narrow end. It allows structural relaxation and contraction during DNA translocation power stroked by pRNA, which is the stator that uses the procapsid as its standing (footing) base. The symmetry mismatch between pRNA (fivefold) and connector (sixfold) yields discrete rotation of the connector. It was estimated that one complete turn of the connector transferred 60 bp of DNA into the procapsid. In this model, during the consecutive contraction and relaxation cycles, the connector works as an “inverse Chinese finger puzzle,” as proposed by George Oster (Fig. 5b),67,88 to alternatively grip DNA in a coordinated cycle with ATPase activity. However, this model contradicts the finding that pRNA does not use the capsid protein gp8 as its foothold, instead, but binds to the N-terminus of the connector serving as pRNA foothold.7,143 In addition, recent single-molecule studies have confirmed that the pRNA ring is a hexamer,119 which does not lend support to the fivefold portal vertex theory. Moreover, the recent single-molecule analysis of the rotation-prevented connector also undercuts this rotation model.9,67
APPLICATIONS OF PHI29 DNA PACKAGING MOTOR
In nanotechnology, a nanomachine is a mechanical or electromechanical device with nanometer scale dimensions.24,25 Living systems utilize diverse nanoscale machines consisting of protein, DNA, and RNA, with high precision, including motors, arrays, pumps, membrane cores, and valves. Although the structural and conformational complexity of the biomolecules retard practical experimental approaches at the bio-nano interface,18,45,71,92,112,134 remarkable efforts have been exerted to develop nanobiomachines for their potential applications in various related fields. For example, synthetic nanobiomachines, constructed to mimic native structure and function, can be incorporated into conventional nanotechnological applications.39,61 Bottom-up assembly in nanotechnology from building blocks of DNA,33,86,107,147 RNA,1,117,118 and protein46,110,133,139 has definitive advantages over chemically synthesized materials, since biomolecules allow for further modification, such as site-directed modification or specific conjugation with defined stoichiometry. Self-assembly of biomolecules without additional catalysis is another simple, versatile approach for high-volume production of biological arrays.1,33,107,117,118,147 In addition, high precision in their replication and manipulation offers strong advantages in applications of nanotechnology, including tissue engineering, cell scaffolding, drug delivery, sensors, imaging, and nanomedicine.
Model System for the Study of RNA Interactions to Form Dimers and Trimers
The novel dimerization or trimerization process of phi29 pRNA can serve as a simple and stable model system to resolve the structure of pRNA complexes and to investigate the mechanism of universal RNA/ RNA interactions. For example, RNA–RNA interactions are essential to the reaction of tRNA cleavage by RNase P,8,44,98 dimerization of retroviral genomic RNA for translation of viral proteins, reverse transcription of RNA to DNA, encapsidation of viral genomes, the assembly of infectious virions,102,121,128 and replications of the plasmid ColE1 regulated by plasmid-specified small RNAs (RNA I and RNA II) that form complexes by complementary RNA stem– loop interactions.30 This model system can also be used to study RNA–RNA intermolecular interactions followed by the formation of a specific ribonucleoprotein (RNP) particle, called bicoid mRNA 3′ UTR-STAUFEN, that determines the formation of the anterior pattern of the Drosophila embryo during development.34
Model System for Studies of Macromolecular Transportation Across a Wall Barrier of Cell Membrane
Translocation of proteins or nucleic acids or cell membranes is a common process. Similarly, most infected or transfected viral or plasmid DNA must pass nuclear membranes to serve as templates for gene expression.27 The Rev protein of HIV is known to be involved in the translocation of viral mRNA from the nucleus to the cytoplasm through nuclear pores.74,104 Since the phi29 DNA packaging motor contains DNA transportation and encapsidation, the motor can serve as a model system for mechanism studies to provide hints for macromolecular transportation and encapsidation.
Model System for Studies on Nucleic Acid Sliding and Tracking Processes
Most nucleic-acid-binding proteins involve a riding, tracking, or sliding process to function in DNA replication, translocation, recombination, and RNA transcription.31,54,62 Many of them form a hexameric conformation with a ring-shaped morphology to interact with nucleic acids. Helicase,96,140 E. coli transcription termination factor rho,13,38 yeast DNA polymerase processivity factor,11 BPV E1,111 and E. coli DNA polymerase81,124 all belong to this category. Although their ultimate functions are very different from viral DNA packaging, the common fact that their interactions involve nucleic acid and protein, and that their conformation is similar to hexameric pRNA attached to a sixfold symmetrical connector during DNA translocation, indicate that phi29 can serve as a model system to determine a universal mechanism for these motor systems.
Model System for Design of New Antiviral Strategies
By targeting phi29 pRNA in the packaging motor, several methods have been reported for the inhibition of viral replication, including in vitro inhibition of phi29 assembly either by antisense DNA targeting pRNA148 or by mutant pRNA.130–132 High inhibition efficiencies make this system attractive for applications in gene therapy, intracellular immunization, antiviral drug design, or construction of transgenic plants resistant to viral infection. These applications can possibly be achieved through targeting certain viral structural proteins, enzymes, and other RNAs produced during the viral replication cycle.
Applications of the Phi29 DNA Packaging Motor to Nanotechnology
DNA packaging is one of the most fascinating areas in recent bionanotechnology development, due to its relatively simplistic structural studies and ease of in vitro assembly from individually prepared components with high efficiency. It has been suggested that the motor itself and the individual motor components can be used in a number of applications, such as a gene delivery vector,56,57,64,72,83 bottom-up RNA assembly and array,117,118 protein engineering, and single molecule studies.119,150 Possible applications of the motor components include the construction of pRNA or protein arrays that will serve as templates for the erection of patterned supramolecular structures, a possible link between nanotubes and nanoparticles, as well as the attachment of biological moieties and chemical groups to the motor or to the constructed arrays.
pRNA
Through utilization of the novel properties of pRNA, it has been shown that pRNA can be engineered to create a controlled dimer, trimer, or hexamer formation, or even an array formation, as observed by TEM with nanogold-labeled pRNA (Fig. 1D)144 and cryo-AFM (Fig. 6a–6d).117,118 This indicates that pRNA can be used in further construction of suprastructures in applications to nanotechnology. It was also found that pRNA can be used to control the activity of the DNA packaging motor by the addition or discharge of a peptide at the N-terminus of the connector that interacts with the pRNA.126 In addition, pRNA with fluorescence-labeling technology has been successfully adapted to the single-molecule fluorescence technique for stoichiometry studies, as described above, to resolve the long-term debate over five or six copies of pRNA.119,150
FIGURE 6.
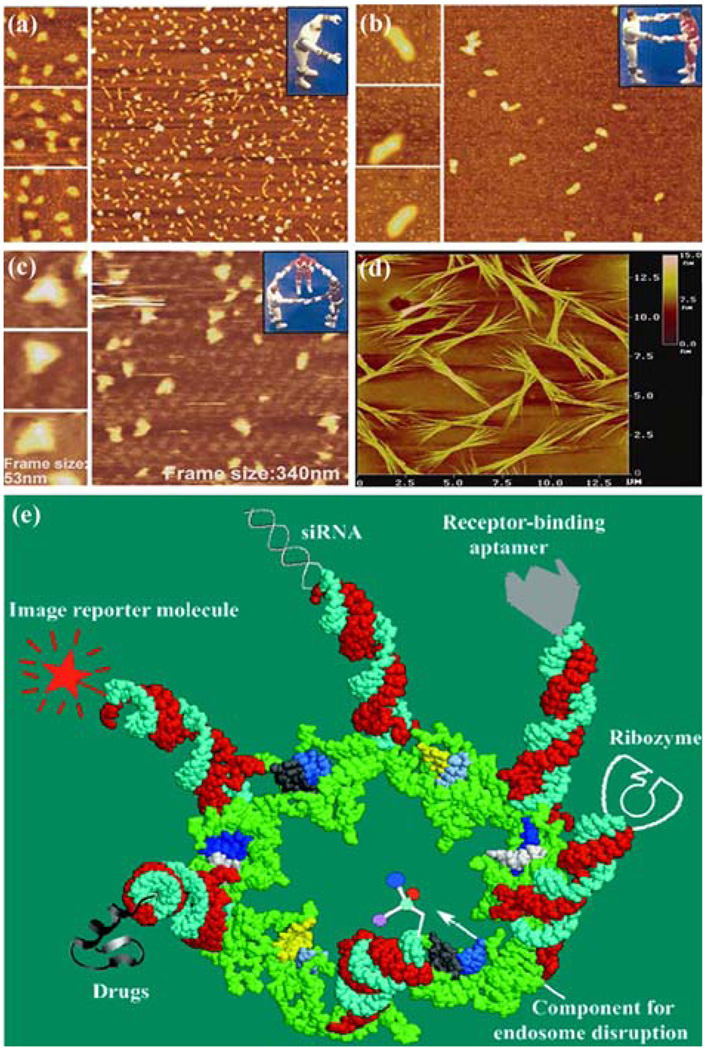
Applications of pRNA for nanotechnology and therapeutic gene and drug delivery. Atomic force microscopy (AFM) images showing (a) monomer, (b) dimer, (c) trimer, and (d) array of engineered pRNA.118 (e) Potential use of pRNA hexamers as polyvalent gene delivery vectors to carry foreign moieties for targeting, therapy, and detection.72 Figures were adapted with permission from the American Chemical Society and Mary Ann Liebert, Inc. for the respective citation.
Connector
Controlled modulation of procapsid assembly, pRNA binding, and procapsid DNA packaging activity were achieved through various artificially constructed phi29 connectors with N- or C-terminal truncations, extensions, or insertions of extra amino acids.14,126 Also, the addition of a cleavable His- or Strep-tag to either the N- or C-terminus of the connector facilitated protein purifications to near homogeneity. Truncations/extensions up to 14 residues at the N-terminus and up to 25 residues at the C-terminus did not affect the connector or procapsid assembly.14 However, procapsids with N-terminal deletion on the connector could not bind pRNA, since the deleted Arg-Lys-Arg residues are essential for pRNA binding.7 A 25-residue deletion and/or 14-residue extension at the C-terminus of gp10 did not affect procapsid assembly. A 42-amino acid extension at the N-terminus did not interfere with the procapsid assembly but significantly decreased the DNA packaging efficiency.14 It has been shown that the wild-type dodecamer connector protein can be used to form arrays with a mixture of single-, double-, or multiple-layered structures.46 Single-layered connectors can be generated by exposing the former crystals over a period of a few weeks to a salt-containing solution of gradually increasing ionic strength.46 Single layer two-dimensional arrays of C- or N-terminal-modified portal vertices can be easily produced by using a lipid monolayer matrix at the air–water interface for the adsorption and growth of 2D protein crystals.144 For nanopatterning, Langmuir deposition or spreading protein solution at the liquid–air interface has been used to produce 2D crystals of proteins.16,92,122 Using the engineered connector array will enable efficient and reproducible transfer of such 2D crystalline protein films onto solid substrates for the design of nanotechnological devices.
Applications of the Phi29 DNA Packaging Motor to Clinical Therapies
The discovery of RNA catalytic activities in 198275 has brought about a revolution in the concept of RNA function, in that RNA can act like an enzyme—thus the term “ribozyme.” Specific ribozymes include RNase P, self-splicing introns, hepatitis delta ribozyme, and hammerhead and hairpin ribozymes. In addition to ribozymes, small interfering RNA (siRNA) has great potential as a therapeutic agent for the treatment of cancer, viral infections, and other genetic diseases. Although Science spotlighted siRNA as the Molecule of the Year in 2002, its therapeutic applications have been hindered by the lack of an efficient, safe, and nonimmunogenic in vivo delivery system to target specific cells.
In contrast, phi29 pRNA provides a model for a ribozyme that acts on DNA. It has been known that the replacement of, or insertion into, the 5′/3′ helical domain does not interfere with dimer formation.21 Thus, end conjugation on pRNA with a chemical moiety or fusion with a receptor-binding RNA aptamer, siRNA, or ribozyme was shown to retain dimer formation without interfering in the function of the inserted moieties.56,57,72,83 Since these are maneuverable and controllable, such properties make pRNA an ideal building block for use as a vector in the delivery of multiple therapeutic RNAs, nanomachine fabrication, pathogen detection, and gene delivery. Through RNA nanotechnology, therapeutic siRNA and receptor-binding RNA aptamers have been engineered into individual motor pRNAs of phi29. The RNA building block harboring the therapeutic molecule was subsequently fabricated into a trimer by utilizing engineered right and left interlocking RNA loops. Incubation of such nanoscale devices containing receptor-binding aptamers or other ligands resulted in binding and co-entry of the trivalent therapeutic particles into cells, subsequently modulating the apoptosis of cancer cells and leukemia model lymphocytes. Animal trials confirmed the specific suppression of tumorigenicity of cancer cells. We have demonstrated that 20- to 40-nm pRNA dimers, trimers, or hexamers can be used as polyvalent vehicles for the delivery of up to six therapeutic molecules to specific cells.56,57,72,83 The use of such antigenicity-free nanoparticles, with sizes of 20–40 nm, will ensure repeated long-term administration and will avoid the problems of the short half-life of small molecules encountered in vivo, due to short retention times and the fact that molecules larger than 100 nm are unable to be delivered into cells.
The polyvalent pRNA complex can deliver up to six kinds of therapeutics to specific cells, as demonstrated in breast cancer, leukemia, lung cancer, and prostate cancer cell lines (Fig. 6e),72 as well as hepatitis B virus infected cells.64 It can simultaneously deliver both a ribozyme and an siRNA or multiple siRNAs against multiple targets or different regions of one target. Thus, this particular system provides unprecedented versatility in constructing polyvalent delivery vehicles by separately constructing individual pRNA subunits with various cargos and mixing them together in any desired combination. Incubation of the chimeric pRNA complex containing receptor-binding aptamers or folate resulted in cell binding and transport of the chimeric pRNA/siRNA, pRNA/ribozyme, or drugs into cells, consequently modulating programmed cell death.56,72,83 The chimeric pRNA complex was found to be processed into functional double-stranded siRNA by Dicer56,72 Target delivery and specificity were brought about by engineering a subunit in the complex to include cell receptor binding ligands for receptor-mediated endocytosis. Included in this process is the conjugation of pRNA with folic acid molecules, which could help target certain kinds of cancer cells that have folate receptors highly expressed on the cell surface.57 Another subunit can carry components to facilitate endosome disruption for the release of therapeutic molecules; and, finally, the other subunits can carry therapeutic siRNA, ribozymes, or pharmacologic compounds to be delivered directly to the cells of interest. The efficiency of this procedure was confirmed in animal trials.72 Using such protein-free, controllable nanoscale RNA particles as therapeutic reagents, which avoid antibiotic response, would allow for long-term administration, thus providing an opportunity for repeated administration and treatment of chronic diseases.
PERSPECTIVES
The phi29 DNA packaging motor serves as an effective model for the study of dsDNA viral packaging. The development of a defined in vitro packaging system utilizing purified phi29 components will aid in the advancement of research into DNA packaging mechanisms, not only for the phi29 system, but also for other genome packaging systems. Specific motor parts and overall motor functions have great potential for use in nanotechnology applications. This will provide a number of technological advances, among which are the targeted delivery of therapeutic agents to cells, precise single-molecule sequencing techniques, and direct attachment of various motor parts to nanodevices. The strong tendency of phi29 pRNA to form dimers, trimers, and hexamers can potentially be harnessed and utilized to construct polyvalent gene/drug delivery vectors or self-assembling arrays that have farranging technological possibilities. It also inspires the development of ultra-high-density memory storage systems and the isolation and separation of multiple pathogens in medical diagnoses. Both the N- and C-terminus of the connector gp10 protein can be easily engineered by deletion, extension, insertion, or tagging to grant additional properties to the DNA packaging motor. Such an engineered connector can also be used as a bottom-up building block to form a single- or multiple-layered array. The gp16 protein can also be engineered to modify its chemical or biological properties, which can then be utilized as an energizer in any type of ATP-consuming works.
These advantages will inspire further assembly of useful nanodevices, such as machineries for in vivo drug delivery, molecular sieves or chips for the diagnosis of diseases, single molecule sensing, or as ultra-high-density data storage systems. In addition, the phi29 nanomotor with engineered components can be a part of a DNA-sequencing apparatus, since the DNA packaging process involves the movement of DNA through a 3.6 nm pore surrounded by six pRNAs, which possesses high potential to be engineered to emit chemical signals. Small packaging RNA can be used to construct nanoparticles as polyvalent vehicles for the delivery of multiple therapeutics, such as siRNA or drug molecules, with specific cell targeting for the treatment of cancers, viral infections, and genetic diseases. By isolating a chemical or metal ion-specific pRNA aptamer mutant using the SELEX (systematic evolution of ligands by exponential enrichment) technique, we can use the pRNA aptamer to detect environmental pathogens or to sense biohazardous chemicals, together with amplified detection signals from fluorescence or radioactive probes labeled to another pRNA moiety. The RNA arrays can be useful as potential parts in nanotechnology for tissue engineering, environmental pathogen detection, and chemical sensing. The same principle can be applied to complement other approaches to using biomolecular self-assembly for precisely arranging particles on a nanometer scale. Furthermore, since engineered motors retain biological DNA packaging activity, a new approach may be applicable to hybrid biological/ nonbiological devices in which nanodevices are powered by the biomimetic motor.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Anne Vonderheide, Linda Keller, Dr. Hui Zhang, Feng Xiao, Dr. Farzin Haque, and Mollie Johnson for their assistance in preparing this review. The work done in the author’s laboratory was supported by NIH Grants GM59944, EB03730, and the NIH Nanomedicine Development Center (NDC) of “Phi29 DNA Packaging Motor for Nanomedicine” (PN2 EY018230) through the NIH Roadmap for Medical Research. Peixuan Guo is the cofounder of Kylin Therapeutics, Inc.
Footnotes
ELECTRONIC SUPPLEMENTARY MATERIAL
The online version of this article (doi:10.1007/ s10439-009-9723-0) contains supplementary material, which is available to authorized users.
REFERENCES
- 1.Afonin KA, Cieply DJ, Leontis NB. Specific RNA self-assembly with minimal paranemic motifs. J. Am. Chem. Soc. 2008;130:93–102. doi: 10.1021/ja071516m. doi:10.1021/ja071516m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agirrezabala X, Martin-Benito J, Valle M, Gonzalez JM, Valencia A, Valpuesta JM, Carrascosa JL. Structure of the connector of bacteriophage T7 at 8A resolution: structural homologies of a basic component of a DNA translocating machinery. J. Mol. Biol. 2005;347:895–902. doi: 10.1016/j.jmb.2005.02.005. doi:10.1016/j.jmb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Aldaye FA, Palmer AL, Sleiman HF. Assembling materials with DNA as the guide. Science. 2008;321:1795–1799. doi: 10.1126/science.1154533. doi:10.1126/science.1154533. [DOI] [PubMed] [Google Scholar]
- 4.Alivisatos AP, Johnsson KP, Peng X, Wilson TE, Loweth CJ, Bruchez MP, Jr, Schultz PG. Organization of nanocrystal molecules using DNA. Nature. 1996;382:609–611. doi: 10.1038/382609a0. doi:10.1038/382609a0. [DOI] [PubMed] [Google Scholar]
- 5.Anderson DL, Reilly B. Morphogenesis of bacteriophage φ29. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and Other Gram-Positive Bacteria: Biochemistry, Physiology, and Molecular Genetics. Washington, DC: American Society for Microbiology; 1993. pp. 859–867. [Google Scholar]
- 6.Andres RP, et al. Self-assembly of a 2-dimensional superlattice of molecularly linked metal clusters. Science. 1996;273:1690–1693. doi:10.1126/science.273.5282.1690. [Google Scholar]
- 7.Atz R, Ma S, Gao J, Anderson DL, Grimes S. Alanine scanning and Fe-BABE probing of the bacteriophage phi29 prohead RNA-connector interaction. J. Mol. Biol. 2007;369:239–248. doi: 10.1016/j.jmb.2007.03.033. doi:10.1016/j.jmb.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baer MF, Reilly RM, McCorkle GM, Hai TY, Altman S, RajBhandary UL. The recognition by RNase P of precursor tRNAs. J. Biol. Chem. 1988;263:2344–2351. [PubMed] [Google Scholar]
- 9.Baumann RG, Mullaney J, Black LW. Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol. Microbiol. 2006;61:16–32. doi: 10.1111/j.1365-2958.2006.05203.x. doi:10.1111/j.1365-2958.2006.05203.x. [DOI] [PubMed] [Google Scholar]
- 10.Black LW. DNA packaging in dsDNA bacteriophages. Ann. Rev. Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. doi:10.1146/ annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 11.Bowers J, Tran PT, Joshi A, Liskay RM, Alani E. MSH-MLH complexes formed at a DNA mismatch are disrupted by the PCNA sliding clamp. J. Mol. Biol. 2001;306(5):957–968. doi: 10.1006/jmbi.2001.4467. doi:10.1006/jmbi.2001.4467. [DOI] [PubMed] [Google Scholar]
- 12.Brust M, Bethall D, Schiffrin DJ, Kiely CJ. Novel gold dithiol nanonetworks with non-metallic electronic properties. Adv. Mater. 1995;7:795–797. doi: 10.1002/adma.19950070907. [Google Scholar]
- 13.Burgess BR, Richardson JP. RNA passes through the hole of the protein hexamer in the complex with the Escherichia coli Rho factor. J. Biol. Chem. 2001;276(6):4182–4189. doi: 10.1074/jbc.M007066200. doi:10.1074/jbc.M007066200. [DOI] [PubMed] [Google Scholar]
- 14.Cai Y, Xiao F, Guo P. N- or C-terminal alterations of motor protein gp10 of bacterial virus phi29 on procapsid assembly, pRNA binding and DNA packaging. Nanomedicine. 2008;4:8–18. doi: 10.1016/j.nano.2007.10.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carrascosa JL, Camacho A, Moreno F, Jimenez F, Mellado RP, Vinuela E, Salas M. Bacillus subtilis phage phi29. Characterization of gene products and functions. Eur. J. Biochem. 1976;66:229–241. doi: 10.1111/j.1432-1033.1976.tb10512.x. doi:10.1111/ j.1432-1033.1976.tb10512.x. [DOI] [PubMed] [Google Scholar]
- 16.Chan YNC, Schrock RR, Cohen RE. Synthesis of single silver nanoclusters within spherical microdomains in block copolymer films. J. Am. Chem. Soc. 1992;114:7295–7296. doi:10.1021/ja00044a051. [Google Scholar]
- 17.Chang CL, Zhang H, Shu D, Guo P, Savran CA. Bright-field analysis of phi29 DNA packaging motor using a magnetomechanical system. Appl. Phys. Lett. 2008;93:153902-1–153902-3. doi: 10.1063/1.3000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Channon K, Bromley EHC, Woolfson DN. Synthetic biology through biomolecular design and engineering. Curr. Opin. Struct. Biol. 2008;18:491–498. doi: 10.1016/j.sbi.2008.06.006. doi:10.1016/j.sbi.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–692. doi: 10.1016/j.cell.2005.06.024. doi:10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 20.Chen C, Guo P. Sequential action of six virus-encoded DNA-packaging RNAs during phage phi29 genomic DNA translocation. J. Virol. 1997;71(5):3864–3871. doi: 10.1128/jvi.71.5.3864-3871.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C, Zhang C, Guo P. Sequence requirement for hand-in-hand interaction in formation of pRNA dimers and hexamers to gear phi29 DNA translocation motor. RNA. 1999;5:805–818. doi: 10.1017/s1355838299990350. doi:10.1017/S1355838299990350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen DN, Erickson SE, Xiang Y, Rossmann MG, Anderson DL. Multifunctional roles of a bacteriophage phi 29 morphogenetic factor in assembly and infection. J. Mol. Biol. 2008;378(4):804–817. doi: 10.1016/j.jmb.2008.02.068. doi: 10.1016/j.jmb.2008.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colvin VL, Goldstein AN, Alivisatos AP. Semiconductor nanocrystals covalently bound to metal surfaces with self-assembled monolayers. J. Am. Chem. Soc. 1992;114:5221–5230. doi:10.1021/ja00039a038. [Google Scholar]
- 24.Craighead HG. Nanoelectromechanical systems. Science. 2000;290:1532–1536. doi: 10.1126/science.290.5496.1532. doi:10.1126/science.290.5496.1532. [DOI] [PubMed] [Google Scholar]
- 25.Cui Y, Lieber CM. Functional nanoscale electronic devices assembled using silicon nanowire building blocks. Science. 2001;291(5505):851–853. doi: 10.1126/science.291.5505.851. doi:10.1126/ science.291.5505.851. [DOI] [PubMed] [Google Scholar]
- 26.Dabbousi BO, Murray CB, Rubner MF, Bawendi MG. Langmuir-Blodgett manipulation of size selected CdSe nanocrystals. Chem. Mater. 1994;6:216–219. doi:10.1021/cm00038a020. [Google Scholar]
- 27.Davis LI. The nuclear pore complex. Ann. Rev. Biochem. 1995;64:865–896. doi: 10.1146/annurev.bi.64.070195.004245. doi:10.1146/annurev.bi.64.070195.004 245. [DOI] [PubMed] [Google Scholar]
- 28.Dujardin E, Peet C, Stubbs G, Culver JN, Mann S. Organization of metallic nanoparticles using Tobacco Mosaic Virus templates. Nano Lett. 2003;3(3):413–417. doi:10.1021/nl034004o. [Google Scholar]
- 29.Earnshaw WC, Casjens SR. DNA packaging by the double-stranded DNA bacteriophages. Cell. 1980;21:319–331. doi: 10.1016/0092-8674(80)90468-7. doi:10.1016/0092-8674(80)90468-7. [DOI] [PubMed] [Google Scholar]
- 30.Eguchi Y, Tomizawa J. Complex formed by complementary RNA stem-loops and its stabilization by a protein: function of CoIE1 Rom protein. Cell. 1990;60:199–209. doi: 10.1016/0092-8674(90)90736-x. doi:10.1016/0092-8674(90)90736-X. [DOI] [PubMed] [Google Scholar]
- 31.Ellison V, Stillman B. Opening of the clamp: an intimate view of an ATP-driven biological machine. Cell. 2001;106(6):655–660. doi: 10.1016/s0092-8674(01)00498-6. doi:10.1016/S0092-8674(01)00498-6. [DOI] [PubMed] [Google Scholar]
- 32.Fang Y, Shu D, Xiao F, Guo P, Qin PZ. Modular assembly of chimeric phi29 packaging RNAs that support DNA packaging. Biochem. Biophys. Res. Commun. 2008;372:589–594. doi: 10.1016/j.bbrc.2008.05.094. doi:10.1016/j.bbrc.2008. 05.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feldkamp U, Niemeyer CM. Rational design of DNA nanoarchitectures. Angew. Chem. Int. Edit. 2006;45:1856–1876. doi: 10.1002/anie.200502358. doi:10.1002/anie.200502358. [DOI] [PubMed] [Google Scholar]
- 34.Ferrandon D, Koch I, Westhof E, Nusslein-Volhard C. RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 1997;16:1751–1758. doi: 10.1093/emboj/16.7.1751. doi:10.1093/emboj/16.7.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fletcher SP, Dumur F, Pollard MM, Feringa BL. A reversible, unidirectional molecular rotary motor driven by chemical energy. Science. 2005;310:80–82. doi: 10.1126/science.1117090. doi:10.1126/science.1117090. [DOI] [PubMed] [Google Scholar]
- 36.Fuller DN, Rickgauer JP, Jardine PJ, Grimes S, Anderson DL, Smith DE. Ionic effects on viral DNA packaging and portal motor function in bacteriophage phi 29. Proc. Natl. Acad. Sci. USA. 2007;104:11245–11250. doi: 10.1073/pnas.0701323104. doi:10.1073/pnas.0701323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gates BD, Xu Q, Stewart M, Ryan D, Willson CG, Whitesides GM. New approaches to nanofabrication: molding, printing, and other techniques. Chem. Rev. 2005;105:1171–1196. doi: 10.1021/cr030076o. doi:10.1021/cr030076o. [DOI] [PubMed] [Google Scholar]
- 38.Gogol EP, Seifried SE, von Hippel PH. Cryoelectron microscopic studies. J. Mol. Biol. 1991;221:1127–1138. doi: 10.1016/0022-2836(91)90923-t. [DOI] [PubMed] [Google Scholar]
- 39.Grigoriev DN, Moll W, Hall J, Guo P. Bionanomotor. Encycl. Nanosci. Nanotechnol. 2003;1:361–374. [Google Scholar]
- 40.Grimes S, Anderson D. In vitro packaging of bacteriophage φ29 DNA restriction fragments and the role of the terminal protein gp3. J. Mol. Biol. 1989;209:91–100. doi: 10.1016/0022-2836(89)90172-1. doi:10.1016/0022-2836(89)90172-1. [DOI] [PubMed] [Google Scholar]
- 41.Grimes S, Anderson D. The bacteriophage phi29 packaging proteins supercoil the DNA ends. J. Mol. Biol. 1997;266:901–914. doi: 10.1006/jmbi.1996.0843. doi:10.1006/jmbi.1996.0843. [DOI] [PubMed] [Google Scholar]
- 42.Grimes S, Anderson D. RNA dependence of the bateriophage phi29 DNA packaging ATPase. J. Mol. Biol. 1990;215:559–566. doi: 10.1016/s0022-2836(05)80168-8. doi:10.1016/S0022-2836(05)80168-8. [DOI] [PubMed] [Google Scholar]
- 43.Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi29 connector particle. J. Mol. Biol. 2002;315(4):663–676. doi: 10.1006/jmbi.2001.5278. doi:10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- 44.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. doi:10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 45.Guo P. RNA nanotechnology: engineering, assembly and applications in detection, gene delivery and therapy. J. Nanosci. Nanotechnol. 2005;5(12):1964–1982. doi: 10.1166/jnn.2005.446. doi:10.1166/ jnn.2005.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo Y, Blocker F, Xiao F, Guo P. Construction and 3-D computer modeling of connector arrays with tetragonal to decagonal transition induced by pRNA of phi29 DNA-packaging motor. J. Nanosci. Nanotechnol. 2005;5:856–863. doi: 10.1166/jnn.2005.143. doi:10.1166/jnn.2005.143. [DOI] [PubMed] [Google Scholar]
- 47.Guo P. Bacterial virus phi29 DNA-packaging motor and its potential applications in gene therapy and nanotechnology. Methods Mol. Biol. 2005;300:285–324. doi: 10.1385/1-59259-858-7:285. [DOI] [PubMed] [Google Scholar]
- 48.Guo P, Erickson S, Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage phi29 DNA. Science. 1987;236:690–694. doi: 10.1126/science.3107124. doi:10.1126/ science.3107124. [DOI] [PubMed] [Google Scholar]
- 49.Guo P, Grimes S, Anderson D. A defined system for in vitro packaging of DNA-gp3 of the Bacillus subtilis bacteriophage phi29. Proc. Natl. Acad. Sci. USA. 1986;83:3505–3509. doi: 10.1073/pnas.83.10.3505. doi:10.1073/pnas.83.10.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo P, Peterson C, Anderson D. Initiation events in in vitro packaging of bacteriophage φ29 DNA-gp3. J. Mol. Biol. 1987;197:219–228. doi: 10.1016/0022-2836(87)90120-3. doi:10.1016/0022-2836(87)90120-3. [DOI] [PubMed] [Google Scholar]
- 51.Guo P, Peterson C, Anderson D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage φ29. J. Mol. Biol. 1987;197:229–236. doi: 10.1016/0022-2836(87)90121-5. doi:10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- 52.Guo P, Rajogopal B, Anderson D, Erickson S, Lee C-S. sRNA of bacteriophage phi29 of B. subtilis mediates DNA packaging of phi29 proheads assembled in E. coli. Virology. 1991;185:395–400. doi: 10.1016/0042-6822(91)90787-c. doi:10.1016/0042-6822(91)90787-C. [DOI] [PubMed] [Google Scholar]
- 53.Guo P, Trottier M. Biological and biochemical properties of the small viral RNA (pRNA) essential for the packaging of the double-stranded DNA of phage φ29. Semin. Virol. 1994;5:27–37. doi:10.1006/smvy.1994.1004. [Google Scholar]
- 54.Guo P, Zhang C, Chen C, Trottier M, Garver K. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol. Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. doi:10.1016/S1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 55.Guo PX, Lee TJ. Viral nanomotors for packaging of dsDNA and dsRNA. Mol. Microbiol. 2007;64:886–903. doi: 10.1111/j.1365-2958.2007.05706.x. doi:10.1111/j.1365-2958.2007.05706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo S, Tschammer N, Mohammed S, Guo P. Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA. Hum. Gene Ther. 2005;16:1097–1109. doi: 10.1089/hum.2005.16.1097. doi:10.1089/hum.2005. 16.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo S, Huang F, Guo P. Construction of folate-conjugated pRNA of bacteriophage phi29 DNA packaging motor for delivery of chimeric siRNA to nasopharyngeal carcinoma cells. Gene Ther. 2006;13(10):814–820. doi: 10.1038/sj.gt.3302716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ha T. Single-molecule fluorescence resonance energy transfer. Methods. 2001;25(1):78–86. doi: 10.1006/meth.2001.1217. doi:10.1006/meth. 2001.1217. [DOI] [PubMed] [Google Scholar]
- 59.Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419(6907):638–641. doi: 10.1038/nature01083. doi:10.1038/nature 01083. [DOI] [PubMed] [Google Scholar]
- 60.Hendrix RW. Symmetry mismatch and DNA packaging in large bacteriophages. Proc. Natl. Acad. Sci. USA. 1978;75:4779–4783. doi: 10.1073/pnas.75.10.4779. doi:10.1073/pnas.75.10.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hess H, Vogel V. Molecular shuttles based on motor proteins: active transport in synthetic environments. J. Biotechnol. 2001;82(1):67–85. doi: 10.1016/s1389-0352(01)00029-0. [DOI] [PubMed] [Google Scholar]
- 62.Hingorani MM, O’Donnell M. Sliding clamps: a (tail)ored fit. Curr. Biol. 2000;10:25–29. doi: 10.1016/s0960-9822(99)00252-3. doi:10.1016/ S0960-9822(99)00252-3. [DOI] [PubMed] [Google Scholar]
- 63.Hoeprich S, Guo P. Computer modeling of three-dimensional structure of DNA-packaging RNA(pRNA) monomer, dimer, and hexamer of phi29 DNA packaging motor. J. Biol. Chem. 2002;277(23):20794–20803. doi: 10.1074/jbc.M112061200. doi:10.1074/jbc.M112061200. [DOI] [PubMed] [Google Scholar]
- 64.Hoeprich S, Zhou Q, Guo S, Qi G, Wang Y, Guo P. Bacterial virus phi29 pRNA as a hammerhead ribozyme escort to destroy hepatitis B virus. Gene Ther. 2003;10:1258–1267. doi: 10.1038/sj.gt.3302002. doi:10.1038/sj.gt.3302002. [DOI] [PubMed] [Google Scholar]
- 65.Huang LP, Guo P. Use of acetone to attain highly active and soluble DNA packaging protein gp16 of phi29 for ATPase assay. Virology. 2003;312(2):449–457. doi: 10.1016/s0042-6822(03)00241-1. doi: 10.1016/S0042-6822(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 66.Huang LP, Guo P. Use of PEG to acquire highly soluble DNA-packaging enzyme gp16 of bacterial virus phi29 for stoichiometry quantification. J. Virol. Methods. 2003;109(2):235–244. doi: 10.1016/s0166-0934(03)00077-6. doi:10.1016/S0166-0934 (03)00077-6. [DOI] [PubMed] [Google Scholar]
- 67.Hugel T, Michaelis J, Hetherington CL, Jardine PJ, Grimes S, Walter JM, Faik W, Anderson DL, Bustamante C. Experimental test of connector rotation during DNA packaging into bacteriophage phi 29 capsids. Plos Biol. 2007;5:558–567. doi: 10.1371/journal.pbio.0050059. doi:10.1371/journal.pbio.0050 059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ibarra B, Caston JR, Llorca O, Valle M, Valpuesta JM, Carrascosa JL. Topology of the components of the DNA packaging machinery in the phage phi29 prohead. J. Mol. Biol. 2000;298:807–815. doi: 10.1006/jmbi.2000.3712. doi:10.1006/jmbi.2000.3712. [DOI] [PubMed] [Google Scholar]
- 69.Ibarra B, Valpuesta JM, Carrascosa JL. Purification and functional characterization of p16, the ATPase of the bacteriophage phi29 packaging machinery. Nucleic Acids Res. 2001;29(21):4264–4273. doi: 10.1093/nar/29.21.4264. doi:10.1093/nar/ 29.21.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jung GY, Johnston-Halperin E, Wu W, Yu ZN, Wang SY, Tong WM, Li ZY, Green JE, Sheriff BA, Boukai A, Bunimovich Y, Heath JR, Williams RS. Circuit fabrication at 17 nm half-pitch by nanoimprint lithography. Nano Lett. 2006;6:351–354. doi: 10.1021/nl052110f. doi:10.1021/nl052110f. [DOI] [PubMed] [Google Scholar]
- 71.Katz E, Willner I. Integrated nanoparticle-biomolecule hybrid systems: synthesis, properties, and applications. Angew. Chem. Int. Edit. 2004;43:6042–6108. doi: 10.1002/anie.200400651. doi:10.1002/anie.200400651. [DOI] [PubMed] [Google Scholar]
- 72.Khaled A, Guo S, Li F, Guo P. Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano Lett. 2005;5:1797–1808. doi: 10.1021/nl051264s. doi:10.1021/ nl051264s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koti JS, Morais MC, Rajagopal R, Owen BA, McMurray CT, Anderson D. DNA packaging motor assembly intermediate of bacteriophage phi29. J. Mol. Biol. 2008;381:1114–1132. doi: 10.1016/j.jmb.2008.04.034. doi:10.1016/j.jmb.2008.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krug RM. The regulation of export of mRNA from nucleus to cytoplasm. Curr. Opin. Cell Biol. 1993;5:944–949. doi: 10.1016/0955-0674(93)90074-z. doi:10.1016/0955-0674(93)90074-Z. [DOI] [PubMed] [Google Scholar]
- 75.Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. doi:10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 76.Lee CS, Guo P. In vitro assembly of infectious virions of ds-DNA phage φ29 from cloned gene products and synthetic nucleic acids. J. Virol. 1995;69:5018–5023. doi: 10.1128/jvi.69.8.5018-5023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee CS, Guo P. Sequential interactions of structural proteins in phage phi29 procapsid assembly. J. Virol. 1995;69:5024–5032. doi: 10.1128/jvi.69.8.5024-5032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee SW, Mao C, Flynn CE, Belcher AM. Ordering of quantum dots using genetically engineered viruses. Science. 2002;296(5569):892–895. doi: 10.1126/science.1068054. doi:10.1126/ science.1068054. [DOI] [PubMed] [Google Scholar]
- 79.Lee TJ, Guo P. Interaction of gp16 with pRNA and DNA for genome packaging by the motor of bacterial virus phi29. J. Mol. Biol. 2006;356:589–599. doi: 10.1016/j.jmb.2005.10.045. doi:10.1016/ j.jmb.2005.10.045. [DOI] [PubMed] [Google Scholar]
- 80.Lee TJ, Zhang H, Liang D, Guo P. Strand and nucleotide-dependent ATPase activity of gp16 of bacterial virus phi29 DNA packaging motor. Virology. 2008;380:69–74. doi: 10.1016/j.virol.2008.07.003. doi:10.1016/j.virol.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leu FP, O’Donnell M. Interplay of a clamp loader subunits in opening the {beta} sliding clamp of E. coli DNA polymerase III holoenzyme. J. Biol. Chem. 2001;276(50):47185–47194. doi: 10.1074/jbc.M106780200. [DOI] [PubMed] [Google Scholar]
- 82.Lin H, Simon MN, Black LW. Purification and characterization of the small subunit of phage T4 terminase, gp16, required for DNA packaging. J. Biol. Chem. 1997;272(6):3495–3501. doi: 10.1074/jbc.272.6.3495. doi:10.1074/jbc.272.6.3309. [DOI] [PubMed] [Google Scholar]
- 83.Liu H, Guo S, Roll R, Li J, Diao Z, Shao N, Riley MR, Cole AM, Robinson JP, Snead NM, Shen G, Guo P. Phi29 pRNA vector for efficient escort of hammerhead ribozyme targeting survivin in multiple cancer cells. Cancer Biol. Ther. 2007;6(5):697–704. doi: 10.4161/cbt.6.5.3962. [DOI] [PubMed] [Google Scholar]
- 84.Lubrich D, Bath J, Turberfield AJ. Templated self-assembly of wedge-shaped DNA arrays. Tetrahedron. 2008;64:8530–8534. doi:10.1016/j.tet.2008.05.135. [Google Scholar]
- 85.Mancini EJ, Kainov DE, Grimes JM, Tuma R, Bamford DH, Stuart DI. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell. 2004;118:743–755. doi: 10.1016/j.cell.2004.09.007. doi:10.1016/j.cell.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 86.Mao C, LaBean TH, Relf JH, Seeman NC. Logical computation using algorithmic self-assembly of DNA triple-crossover molecules. Nature. 2000;407:493–496. doi: 10.1038/35035038. doi:10.1038/35035038. [DOI] [PubMed] [Google Scholar]
- 87.Mbindyo JK, Reiss BD, Martin BR, Keating CD, Natan MJ, Mallouk TE. DNA-directed assembly of gold nanowires on complementary surfaces. Adv. Mater. 2001;13:249–254. doi:10.1002/1521-4095 (200102)13:4≤249::AID-ADMA249≥3.0.CO;2-9. [Google Scholar]
- 88.Mickler M, Schleiff E, Hugel T. From biological towards artificial molecular motors. Chemphyschem. 2008;9:1503–1509. doi: 10.1002/cphc.200800216. doi:10.1002/cphc.200800216. [DOI] [PubMed] [Google Scholar]
- 89.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. doi:10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 90.Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457(7228):446–452. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moll D, Guo P. Translocation of nicked but not gapped DNA by the packaging motor of bacteriophage phi29. J. Mol. Biol. 2005;351:100–107. doi: 10.1016/j.jmb.2005.05.038. doi:10.1016/ j.jmb.2005.05.038. [DOI] [PubMed] [Google Scholar]
- 92.Moll D, Huber C, Schlegel B, Pum D, Sleytr UB, Sara M. S-layer-streptavidin fusion proteins as template for nano patterned molecular arrays. Proc. Natl. Acad. Sci. USA. 2002;99(23):14646–14651. doi: 10.1073/pnas.232299399. doi:10.1073/pnas.232299399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Morton VL, Stockley PG, Stonehouse NJ, Ashcroft AE. Insights into virus capsid assembly from non-covalent mass spectrometry. Mass Spectrom. Rev. 2008;27:575–595. doi: 10.1002/mas.20176. doi:10.1002/mas.20176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Motte L, Billoudet F, Lacaze E, Pileni M. Self-organization of size-selected nanoparticles into three dimensional superlattices. Adv. Mater. 1996;8:1018–1020. doi:10.1002/adma.19960081218. [Google Scholar]
- 95.Murray CBC, Kagan CR, Bawendi MG. Self-organization of CdSe nanocrystals into 3-dimensional quantum dot super lattices. Science. 1995;270:1335–1338. doi:10.1126/science.270.5240.1335. [Google Scholar]
- 96.Niedenzu T, Roleke D, Bains G, Scherzinger E, Saenger W. Crystal structure of the hexameric replicative helicase RepA of plasmid RSF1010. J. Mol. Biol. 2001;306(3):479–487. doi: 10.1006/jmbi.2000.4398. doi:10.1006/jmbi.2000.4398. [DOI] [PubMed] [Google Scholar]
- 97.Nykypanchuk D, Maye MM, van der Lelie D, Gang O. DNA-guided crystallization of colloidal nanoparticles. Nature. 2008;451:549–552. doi: 10.1038/nature06560. doi:10.1038/nature 06560. [DOI] [PubMed] [Google Scholar]
- 98.Oh BK, Pace NR. Interaction of the 3′-end of tRNA with ribonuclease P RNA. Nucleic Acids Res. 1994;22(20):4087–4094. doi: 10.1093/nar/22.20.4087. doi:10.1093/nar/22.20.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Oram M, Sabanayagam C, Black LW. Modulation of the packaging reaction of bacteriophage T4 terminase by DNA structure. J. Mol. Biol. 2008;381:61–72. doi: 10.1016/j.jmb.2008.05.074. doi:10.1016/j.jmb.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Orlova EV, Gowen B, Droge A, Stiege A, Weise F, Lurz R, van Heel M, Tavares P. Structure of a viral DNA gatekeeper at 10 A resolution by cryo-electron microscopy. EMBO J. 2003;22:1255–1262. doi: 10.1093/emboj/cdg123. doi:10.1093/ emboj/cdg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Otero R, Ecija D, Fernandez G, Gallego JM, Sanchez L, Martin N, Miranda R. An organic donor/acceptor lateral superlattice at the nanoscale. Nano Lett. 2007;7:2602–2607. doi: 10.1021/nl070897z. doi:10.1021/nl070897z. [DOI] [PubMed] [Google Scholar]
- 102.Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. USA. 1996;93:5572–5577. doi: 10.1073/pnas.93.11.5572. doi:10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Peterson C, Simon M, Hodges J, Mertens P, Higgins L, Egelman E, Anderson D. Composition and mass of the bacteriophage phi29 prohead and virion. J. Struct. Biol. 2001;135:18–25. doi: 10.1006/jsbi.2001.4375. doi:10.1006/jsbi.2001.4375. [DOI] [PubMed] [Google Scholar]
- 104.Pfeifer K, Weiler BE, Ugarkovic D, Bachmann M, Schroder HC, Muller WE. Evidence for a direct interaction of Rev protein with nuclear envelope mRNA-translocation system. Eur. J. Biochem. 1991;199:53–64. doi: 10.1111/j.1432-1033.1991.tb16091.x. doi:10.1111/j.1432-1033.1991.tb16091.x. [DOI] [PubMed] [Google Scholar]
- 105.Rishovd S, Holzenburg A, Johansen BV, Lindqvist BH. Bacteriophage P2 and P4 morphogenesis: structure and function of the connector. Virology. 1998;245:11–17. doi: 10.1006/viro.1998.9153. doi:10.1006/viro.1998.9153. [DOI] [PubMed] [Google Scholar]
- 106.Robinson MA, Wood JP, Capaldi SA, Baron AJ, Gell C, Smith DA, Stonehouse NJ. Affinity of molecular interactions in the bacteriophage phi29 DNA packaging motor. Nucleic Acids Res. 2006;34:2698–2709. doi: 10.1093/nar/gkl318. doi:10.1093/nar/gkl318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. doi:10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 108.Rueda D, Bokinsky G, Rhodes MM, Rust MJ, Zhuang X, Walter NG. Single-molecule enzymology of RNA: essential functional groups impact catalysis from a distance. Proc. Natl. Acad. Sci. USA. 2004;101:10066–10071. doi: 10.1073/pnas.0403575101. doi:10.1073/pnas.0403575101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sabanayagam CR, Oram M, Lakowicz JR, Black LW. Viral DNA packaging studied by fluorescence correlation spectroscopy. Biophys. J. 2007;93(4):L17–L19. doi: 10.1529/biophysj.107.111526. doi:10.1529/biophysj.107.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sara M, Sleytr UB. S-layer proteins. J. Bact. 2000;182:859–868. doi: 10.1128/jb.182.4.859-868.2000. doi:10.1128/JB.182.4.859-868.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sedman J, Stenlund A. The papillomavirus E1 protein forms a DNA-dependent hexameric complex with ATPase and DNA helicase activities. J. Virol. 1998;72:6893–6897. doi: 10.1128/jvi.72.8.6893-6897.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Seeman NC, Belcher AM. Emulating biology: building nanostructures from the bottom up. Proc. Natl. Acad. Sci. USA. 2002;99 Suppl 2:6451–6455. doi: 10.1073/pnas.221458298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Serwer P. Models of bacteriophage DNA packaging motors. J. Struct. Biol. 2003;141(3):179–188. doi: 10.1016/s1047-8477(02)00628-7. doi:10.1016/ S1047-8477(02)00628-7. [DOI] [PubMed] [Google Scholar]
- 114.Shu D, Guo P. A Viral RNA that binds ATP and contains an motif similar to an ATP-binding aptamer from SELEX. J. Biol. Chem. 2003;278(9):7119–7125. doi: 10.1074/jbc.M209895200. doi:10.1074/jbc.M209895200. [DOI] [PubMed] [Google Scholar]
- 115.Shu D, Guo P. Only one pRNA hexamer but multiple copies of the DNA-packaging protein gp16 are needed for the motor to package bacterial virus phi29 genomic DNA. Virology. 2003;309(1):108–113. doi: 10.1016/s0042-6822(03)00011-4. doi: 10.1016/S0042-6822(03)00011-4. [DOI] [PubMed] [Google Scholar]
- 116.Shu D, Huang L, Guo P. A simple mathematical formula for stoichiometry quantitation of viral and nanobiological assemblage using slopes of log/log plot curves. J. Virol. Meth. 2003;115(1):19–30. doi: 10.1016/j.jviromet.2003.08.015. doi:10.1016/ j.jviromet.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 117.Shu D, Huang L, Hoeprich S, Guo P. Construction of phi29 DNA-packaging RNA (pRNA) monomers, dimers and trimers with variable sizes and shapes as potential parts for nano-devices. J. Nanosci. Nanotechnol. 2003;3:295–302. doi: 10.1166/jnn.2003.160. doi:10.1166/jnn.2003.160. [DOI] [PubMed] [Google Scholar]
- 118.Shu D, Moll D, Deng Z, Mao C, Guo P. Bottomup assembly of RNA arrays and superstructures as potential parts in nanotechnology. 2004;4:1717–1724. doi: 10.1021/nl0494497. doi:10.1021/nl0494497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shu D, Zhang H, Jin J, Guo P. Counting of six pRNAs of phi29 DNA-packaging motor with customized single molecule dual-view system. EMBO J. 2007;26:527–537. doi: 10.1038/sj.emboj.7601506. doi:10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Simpson AA, Leiman PG, Tao Y, He Y, Badasso MO, Jardine PJ, Anderson DL, Rossman MG. Structure determination of the head-tail connector of bacteriophage phi29. Acta Cryst. 2001;D57:1260–1269. doi: 10.1107/s0907444901010435. doi:10.1107/S0907444901010435. [DOI] [PubMed] [Google Scholar]
- 121.Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl. Acad. Sci. USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. doi:10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sleytr UB, Sara M. Bacterial and archaeal S-layer proteins: structure-function relationships and their biotechnological applications. Trends Biotechnol. 1997;15(1):20–26. doi: 10.1016/S0167-7799(96)10063-9. doi:10.1016/S0167-7799(96)10063-9. [DOI] [PubMed] [Google Scholar]
- 123.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. doi:10.1038/ 35099581. [DOI] [PubMed] [Google Scholar]
- 124.Song MS, Dallmann HG, McHenry CS. Carboxyl-terminal Domain III of the delta Subunit of the DNA Polymerase III Holoenzyme Binds delta. J. Biol. Chem. 2001;276(44):40668–40679. doi: 10.1074/jbc.M106373200. doi:10.1074/ jbc.M106373200. [DOI] [PubMed] [Google Scholar]
- 125.Song Q, Ding Y, Wang ZL, Zhang ZJ. Formation of orientation-ordered superlattices of magnetite magnetic nanocrystals from shape-segregated self-assemblies. J. Phys. Chem. B. 2006;110:25547–25550. doi: 10.1021/jp0652695. doi: 10.1021/jp0652695. [DOI] [PubMed] [Google Scholar]
- 126.Sun J, Cai Y, Moll WD, Guo P. Controlling bacteriophage phi29 DNA-packaging motor by addition or discharge of a peptide at N-terminus of connector protein that interacts with pRNA. Nucleic Acids Res. 2006;34(19):5482–5490. doi: 10.1093/nar/gkl701. doi:10.1093/nar/gkl701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. doi:10.1016/ j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 128.Sundquist WI, Heaphy S. Evidence for interstand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic RNA. Proc. Natl. Acad. Sci. USA. 1993;90:3393–3397. doi: 10.1073/pnas.90.8.3393. doi:10.1073/pnas. 90.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Svoboda K, Block SM. Force and velocity measured for single kinesin molecules. Cell. 1994;77(5):773–784. doi: 10.1016/0092-8674(94)90060-4. doi:10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 130.Trottier M, Garver K, Zhang C, Guo P. DNA-packaging pRNA as target for complete inhibition of viral assembly in vitro and in vivo. Nucleic Acids Symp. Ser. 1997;36:187–189. [Google Scholar]
- 131.Trottier M, Guo P. Approaches to determine stoichiometry of viral assembly components. J. Virol. 1997;71:487–494. doi: 10.1128/jvi.71.1.487-494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Trottier M, Zhang CL, Guo P. Complete inhibition of virion assembly in vivo with mutant pRNA essential for phage phi29 DNA packaging. J. Virol. 1996;70:55–61. doi: 10.1128/jvi.70.1.55-61.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Uchida M, Klem MT, Allen M, Suci P, Flenniken M, Gillitzer E, Varpness Z, Liepold LO, Young M, Douglas T. Biological containers: protein cages as multifunctional nanoplatforms. Adv. Mater. 2007;19:1025–1042. doi:10.1002/adma.200601168. [Google Scholar]
- 134.Ulijn RV, Smith AM. Designing peptide based nanomaterials. Chem. Soc. Rev. 2008;37:664–675. doi: 10.1039/b609047h. doi:10.1039/b609047h. [DOI] [PubMed] [Google Scholar]
- 135.Valpuesta JM, Fernandez JJ, Carazo JM, Carrascosa JL. The three-dimensional structure of a DNA translocating machine at 10 A resolution. Structure. 1999;7:289–296. doi: 10.1016/s0969-2126(99)80039-2. doi:10.1016/S0969-2126(99)80039-2. [DOI] [PubMed] [Google Scholar]
- 136.Valpuesta JM, Sousa N, Barthelemy I, Fernandez JJ, Fujisawa H, Ibarra B, Carrascosa JL. Structural analysis of the bacteriophage T3 head-to-tail connector. J. Struct. Biol. 2000;131:146–155. doi: 10.1006/jsbi.2000.4281. doi:10.1006/jsbi.2000. 4281. [DOI] [PubMed] [Google Scholar]
- 137.van Blaaderen A, Ruel R, Wiltzius P. Template-directed colloidal crystallization. Nature. 1997;385(6614):321–324. doi:10.1038/385321a0. [Google Scholar]
- 138.Vossmeyer T, et al. A double diamond superlattice built up of Cd17S4(SCH2CH2OH)(26) clusters. Science. 1995;267:1476–1479. doi: 10.1126/science.267.5203.1476. doi:10.1126/science.267.5203.1476. [DOI] [PubMed] [Google Scholar]
- 139.Wargacki SP, Pate B, Vaia RA. Fabrication of 2D ordered films of tobacco mosaic virus (TMV): processing morphology correlations for convective assembly. Langmuir. 2008;24:5439–5444. doi: 10.1021/la7040778. doi:10.1021/la7040778. [DOI] [PubMed] [Google Scholar]
- 140.West SC. DNA helicases: new breeds of translocating motors and molecular pumps. Cell. 1996;86:177–180. doi: 10.1016/s0092-8674(00)80088-4. doi:10.1016/S0092-8674(00)80088-4. [DOI] [PubMed] [Google Scholar]
- 141.Whetten RL, et al. Nanocrystal gold molecules. Adv. Mater. 1996;8:428–433. doi:10.1002/adma.19960080513. [Google Scholar]
- 142.Xiang Y, Morais MC, Cohen DN, Bowman VD, Anderson DL, Rossmann MG. Crystal and cryoEM structural studies of a cell wall degrading enzyme in the bacteriophage phi29 tail. Proc. Natl. Acad. Sci. USA. 2008;105:9552–9557. doi: 10.1073/pnas.0803787105. doi:10.1073/pnas.0803787105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xiao F, Moll D, Guo S, Guo P. Binding of pRNA to the N-terminal 14 amino acids of connector protein of bacterial phage phi29. Nucleic Acids Res. 2005;33:2640–2649. doi: 10.1093/nar/gki554. doi:10.1093/nar/gki554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xiao F, Sun J, Coban O, Schoen P, Wang JC, Cheng RH, Guo P. Fabrication of massive sheets of single layer patterned arrays using reengineered phi29 motor dodecamer. ACS Nano. 2009;3:100–107. doi: 10.1021/nn800409a. doi:10.1021/ nn800409a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xiao F, Zhang H, Guo P. Novel mechanism of hexamer ring assembly in protein/RNA interactions revealed by single molecule imaging. Nucleic Acids Res. 2008;36(20):6620–6632. doi: 10.1093/nar/gkn669. doi:10.1093/nar/gkn669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yasuda R, Noji H, Yoshida M, Kinosita K, Jr, Itoh H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature. 2001;410(6831):898–904. doi: 10.1038/35073513. doi:10.1038/35073513. [DOI] [PubMed] [Google Scholar]
- 147.Zhang C, Su M, He Y, Zhao X, Fang PA, Ribbe AE, Jiang W, Mao CD. Conformational flexibility facilitates self-assembly of complex DNA nanostructures. Proc. Natl. Acad. Sci. USA. 2008;105:10665–10669. doi: 10.1073/pnas.0803841105. doi:10.1073/pnas.0803841105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang CL, Garver K, Guo P. Inhibition of phage phi29 assembly by antisense oligonucleotides targeting viral pRNA essential for DNA packaging. Virology. 1995;211:568–576. doi: 10.1006/viro.1995.1439. doi:10.1006/viro.1995.1439. [DOI] [PubMed] [Google Scholar]
- 149.Zhang F, Lemieux S, Wu X, St.-Arnaud S, McMurray CT, Major F, Anderson D. Function of hexameric RNA in packaging of bacteriophage phi29 DNA in vitro. Mol. Cell. 1998;2:141–147. doi: 10.1016/s1097-2765(00)80123-9. doi:10.1016/ S1097-2765(00)80123-9. [DOI] [PubMed] [Google Scholar]
- 150.Zhang H, Shu D, Huang F, Guo P. Instrumentation and metrology for single RNA counting in biological complexes or nanoparticles by a single molecule dual-view system. RNA. 2007;13:1793–1802. doi: 10.1261/rna.587607. doi:10.1261/rna.587607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zheng NF, Bu XH, Feng PY. Self-assembly of novel dye molecules and [Cd-8(SPh)(12)](4+) cubic clusters into three-dimensional photoluminescent superlattice. J. Am. Chem. Soc. 2002;124:9688–9689. doi: 10.1021/ja020480p. doi:10.1021/ ja020480p. [DOI] [PubMed] [Google Scholar]
- 152.Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S. A single-molecule study of RNA catalysis and folding. Science. 2000;288(5473):2048–2051. doi: 10.1126/science.288.5473.2048. doi:10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.